Abstract
The epidermal growth-factor receptor (EGFR) tyrosine kinase inhibitor erlotinib has been proven to be highly effective in the treatment of nonsmall cell lung cancer (NSCLC) harboring oncogenic EGFR mutations. The majority of patients, however, will eventually develop resistance and succumb to the disease. Recent studies have identified secondary mutations in the EGFR (EGFR T790M) and amplification of the N-Methyl-N′-nitro-N-nitroso-guanidine (MNNG) HOS transforming gene (MET) oncogene as two principal mechanisms of acquired resistance. Although they can account for approximately 50% of acquired resistance cases together, in the remaining 50%, the mechanism remains unknown. In NSCLC-derived cell lines and early-stage tumors before erlotinib treatment, we have uncovered the existence of a subpopulation of cells that are intrinsically resistant to erlotinib and display features suggestive of epithelial-to-mesenchymal transition (EMT). We showed that activation of TGF-β–mediated signaling was sufficient to induce these phenotypes. In particular, we determined that an increased TGF-β–dependent IL-6 secretion unleashed previously addicted lung tumor cells from their EGFR dependency. Because IL-6 and TGF-β are prominently produced during inflammatory response, we used a mouse model system to determine whether inflammation might impair erlotinib sensitivity. Indeed, induction of inflammation not only stimulated IL-6 secretion but was sufficient to decrease the tumor response to erlotinib. Our data, thus, argue that both tumor cell-autonomous mechanisms and/or activation of the tumor microenvironment could contribute to primary and acquired erlotinib resistance, and as such, treatments based on EGFR inhibition may not be sufficient for the effective treatment of lung-cancer patients harboring mutant EGFR.
Keywords: epidermal growth-factor receptor, nonsmall cell lung cancer
In recent years, rapid advances in our understanding of the molecular events required for tumor onset and progression have led to the development of cancer agents referred to as molecular-targeted therapies. Because they specifically target the product of selective cancer mutations that is required for cancer-cell survival, they are thought to become invaluable therapeutic tools in the treatment of cancer. Specifically in the case of lung cancer, much excitement has been generated by the finding that patients harboring oncogenic epidermal growth-factor receptor (EGFR) mutations highly benefit from treatment with selective inhibitors (i.e., erlotinib and gefitinib) (1–4). Erlotinib and gefitinib are members of a class of quinazolium-derived agents that inhibit the EGFR pathway by binding in a reversible fashion to the EGFR ATP pocket domain (5). Remarkably, retrospective studies showed a striking correlation between occurrence of certain EGFR oncogenic mutations and erlotinib/gefitinib responses. The presence of deletions in exon 19 of EGFR or EGFR L858R missense substitutions is, in fact, found in more than 80% of nonsmall cell lung cancer (NSCLC) patients that respond to erlotinib or gefitinib treatment (6). However, as in the case of other targeted therapies, the emergence of resistance presents a major hurdle for their successful utilization. Clinical data, in fact, have shown that, in the majority of the cases, responses to drug treatment are transient and within a short period, patients that initially responded progress or relapse with resistant disease. The acquisition of an additional mutation in exon 20 of EGFR resulting in a threonine-to-methionine substitution at position 790 (T790M mutation) and/or amplification of c-MET can account for ∼50% of cases of erlotinib-acquired resistance (7). However, the mechanisms that lead to resistance in the remaining cases are unknown.
Results
Erlotinib Resistant-Derived Cells Display Mesenchymal-Like Features and Increased Metastatic Potential.
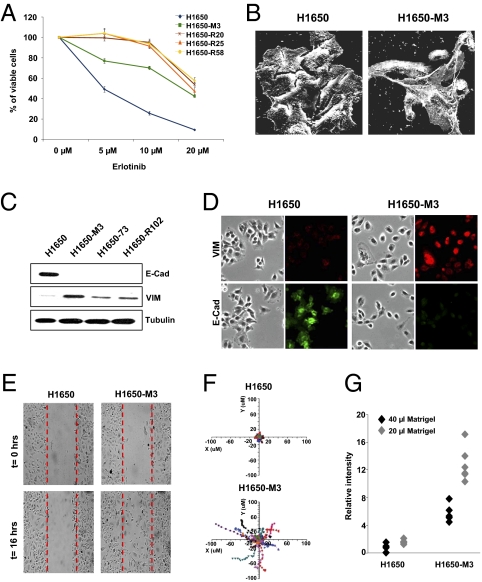
To study molecular mechanisms of gefitinib and erlotinib resistance in NSCLC, we have developed a cell-based system using the broncho-alveolar cancer cell line H1650. This cell line harbors an oncogenic deletion within the EGFR (delE746-A750) and has a one-half maximal inhibitory concentration (IC50) to gefitinib or erlotinib treatment of ∼5 μM. By culturing this cell line in the presence of a constant high concentration of erlotinib, we have been able to isolate cell lines capable of growing in the presence of up to 20 μM of the drug (Fig. 1A). Interestingly, ∼13% of the erlotinib-resistant cells displayed morphological appearances of mesenchymal cells (i.e., 16 of a total of 123 colonies examined). These striking morphological features (Fig. 1B) were associated at the molecular level with an increased expression of the mesenchymal protein Vimentin and a decreased expression of the epithelial marker E-cadherin (Fig. 1 C and D). In addition, resistant cells displayed enhanced motility (Fig. 1 E and F) and Matrigel invasion (Fig. 1G) compared with parental cells.
Fig. 1.
Erlotinib-resistant cells are characterized by mesenchymal-like features and an increased metastatic potential. (A) Parental and resistant cell lines were treated with erlotinib at the indicated concentrations. Viability of cells was measured after 72 h of treatment by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. The percentage of viable cells was calculated relative to untreated control. (B) As shown by scanning electron microscopy, striking morphological differences were readily observed in certain erlotinib-resistant cells (i.e., 13% of erlotinib-derived resistant cells). (C) Western blot analysis of cell extracts with E-cadherin and Vimentin antibodies and (D) immunofluorescence staining of cells labeled with E-cadherin and Vimentin antibodies. (E) Images from a time-lapse sequence of erlotinib-sensitive (H1650) and erlotinib-resistant cells (H1650-M3) migrating to heal a scratch. The images were taken immediately after scratching the cell monolayer (time = 0 h) and after 16 h. (F) Tracking the movement of single cells (n = 12) revealed differences in intrinsic cell motility. Each trace represents the movement of a single cell, whereas individual dots designate a frame of 10 min. (G) A modified Boyden-chamber assay using filters coated with different volumes of Matrigel (i.e., 20 μL and 40 μL) showed an increased invasive potential of the erlotinib-resistant cells relative to parental cells. Each dot represents an individual replicate (n = 5).
Erlotinib-Resistant Cells Have a Distinct Gene-Expression Profile and Are Characterized by an Increased Activation of TGF-β Axis.
In NSCLC, resistance to erlotinib treatment has been associated with the acquisition of EGFR secondary mutations (EGFR T790M mutation) and overexpression and/or activation of c-Met (8). None of these factors were deregulated in any of the selected erlotinib-resistant cells with mesenchymal-like features that we examined (SI Appendix, Fig. S1). To identify the molecular mechanisms responsible for the acquired resistance to erlotinib, we performed gene-expression profile analysis of the H1650 parental cells and a resistance-derived cell line. Compared with the parental cells, H1650-M3 had increased expression of genes previously associated with epithelial-to-mesenchymal transition (e.g., SNAI, zinc finger E-box binding homeobox (ZEB)2, Vimentin, CD44, TGF-β1, and TGF-β2), increased motility/invasion (e.g., TGF-β1/2, matrix metallopeptidase (MMP)2, and thrombospondin (THBS)1), resistance to apoptosis (e.g., XAF1) as well as a lung metastatic signature (e.g., homeobox B (HOXB)2, S100A4, S100A2, and Tenascin C) (SI Appendix, Fig. S2). Interestingly, many of the differentially expressed genes were previously shown to be regulated by TGF-β. ELISA also indicated that the increase in TGF-β1 and TGF-β2 mRNA expression seen in erlotinib-resistant cells correlated with higher levels of protein secretion (SI Appendix, Fig. S3A). Notably, the levels of expression and secretion of both TGF-β 1 and 2 did not change on erlotinib treatment (SI Appendix, Fig. S3B); this was accompanied by increased levels of SMAD2 and SMAD3 phosphorylation (SI Appendix, Fig. S3C) as well as an augmented nuclear translocation of SMAD2 and SMAD4 in the erlotinib-resistant H1650-M3 cells compared with the H1650 cells (SI Appendix, Fig. S3D). Based on these findings, we concluded that the erlotinib-resistant cells are characterized by an up-regulation of TGF-β–mediated signaling.
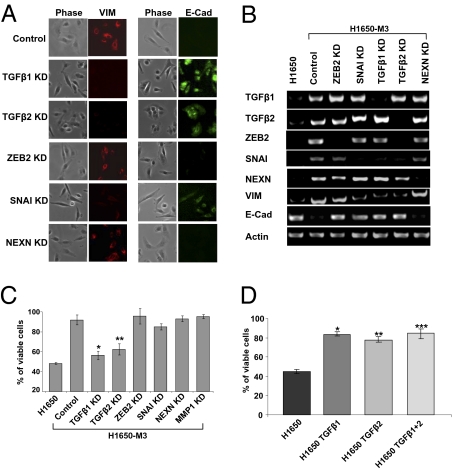
TGF-β1 and TGF-β2 Are Necessary and Sufficient for Erlotinib Resistance, EMT, and Increased Activation of the IL-6 Axis.
To determine the contributions of TGF-β signaling pathways in mediating erlotinib resistance and EMT, we used RNA interference-based technology to inhibit the expression TGF-β1 and TGF-β2 as well as SNAI and ZEB2 (two TGF-β axis components previously described to be master regulators of EMT). An shRNA-targeting nexillin (NEXN), a gene expressed at higher levels in the erlotinib-resistant cells than parental cells but not regulated by TGF-β, was used as a control. Decreasing the expression of TGF-β1 and TGF-β2 in the erlotinib-resistant H1650-M3 cells resulted in morphological changes typical of mesenchymal-to-epithelial transition (Fig. 2A) as well as decreased cell motility and invasion (SI Appendix, Fig. S4 A and B). These changes were accompanied at the molecular level with a decreased expression of Vimentin, SNAI, and ZEB2 as well as an increased expression of E-cadherin in TGF-β1 or 2 knockdown erlotinib-resistant cells (Fig. 2 A and B). Consistent with the role of SNAI in EMT, reducing the expression of SNAI phenocopied the effect of TGF-β knockdown on levels of E-cadherin and Vimentin expression as well as on the impairment of cell motility and invasion (Fig. 2 A and B and SI Appendix, Fig. S4 A and B). Surprisingly, inhibition of ZEB2 expression caused only a significant increase in E-cadherin mRNA level with no change in other measured parameters (Fig. 2 A and B and SI Appendix, Fig. S4 A and B). Interestingly, although neither ZEB2 nor SNAI knockdown modified the cells sensitivity to erlotinib, diminishing the expression of TGF-β1 and TGF-β2 resulted in the restoration of cell erlotinib sensitivity comparable with that of the parental H1650 cell line (Fig. 2C). Not only were TGF-β1 and 2 required for the observed phenotypes, but we also found that paracrine stimulation with either TGF-β1, TGF-β2, or both in combination was sufficient to induce EMT and increase cell motility and Matrigel invasion (SI Appendix, Fig. S4 C–E) as well as erlotinib resistance (Fig. 2D). Notably, all these phenotypes required the constant presence of these factors, because their removal resulted in a reversion from mesenchymal- and erlotinib-resistant to epithelial- and erlotinib-sensitive. Our data, thus, clearly indicate that the activation of the TGF-β axis is required and sufficient for the acquisition of a mesenchymal-like morphology, increased motility and invasion ability, and erlotinib resistance. Because neither ZEB2 nor SNAI knockdown modify the cells sensitivity to erlotinib, we also concluded that distinct TGF-β axis components contribute differentially to the erlotinib resistance and the EMT/ invasion program. Therefore, although a correlation between EMT and erlotinib resistance can be observed, the two programs are clearly distinct and mediated by different signal transduction pathways.
Fig. 2.
The activation of the TGF-β1 and TGF-β2 signaling is required and sufficient for EMT, increased cell motility and invasion, and erlotinib resistance. (A) To determine the contributions of TGF-β signaling pathways in mediating erlotinib resistance and EMT, we used RNA interference-based technology to inhibit the expression of TGF-β1 and TGF-β2 as well as SNAI and ZEB2. A shRNA-targeting nexillin (NEXN) was used as a control. Two independent shRNA constructs were used to generate cell lines in which the expression of each gene of interest was stably knocked down. Although similar results were obtained with both shRNAs, the figure depicts the result obtained with one shRNA only. RT-PCR analysis showed a dramatic decrease in the expression of the targeted genes. (B) Changes in the expression of Vimentin and E-cadherin observed by RT-PCR were confirmed by immunostaining analysis. (C) Erlotinib-resistant cells in which TGF-β1 and TGF-β2 expression was silenced displayed sensitivity to erlotinib comparable with that of the parental cells (H1650). The data shown represent the mean value of the percentage of viable cells ± SD (n = 6; P < 0.0001). (D) Erlotinib-sensitive (H1650 cells) cells treated for 8 d with TGF-β1, TGF-β2, or TGF-β1 and TGF-β2 in combination displayed an increased resistance to erlotinib. Cell viability was measured by MTT assay. The percentage of viable cells was calculated relative to untreated cells. The bars shown are the means ± SD (n = 4), and the differences between the treated and control cells are significant (P < 0.0001, two-tailed Student t test).
IL-6 Is Increasingly Expressed in Erlotinib-Resistant Cells and Required for Their Survival.
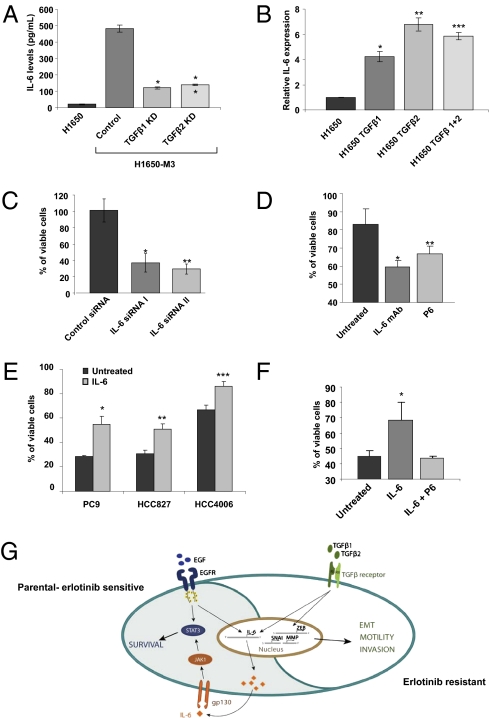
Among the genes that we found to be increasingly expressed in the erlotinib-resistant cells, we noticed a dramatic up-regulation of IL-6 and genes reported to be regulated by IL-6 (SI Appendix, Fig. S2). The increased expression of IL-6 resulted in an augmented IL-6 secretion (Fig. 3A). IL-6 expression depended on TGF-β–mediated signaling, because TGF-β1 or TGF-β2 knockdown cells have a reduced expression of IL-6 and treating cells with either TGF-β1 and/or 2 drastically increased the expression of IL-6 (Fig. 3 A and B). We were particularly intrigued by this observation, because Gao et al. (9) recently showed that NSCLC cells expressing mutant EGFR are dependent on the IL-6 axis for their long-term proliferation/survival. We, therefore, hypothesized that an increased autocrine stimulation of the IL-6/gp130/STAT3 pathway through TGF-β could unleash the cells from their dependency on EGFR. To this end, we measured the viability of erlotinib-derived resistant cells on inhibition of IL-6–mediated signaling. Reduction of the level of IL-6 by siRNA as well as inhibition of the IL-6 axis by means of an IL-6 neutralizing antibody or a JAK1/2 inhibitor not only significantly decreased IL-6–mediated signaling (SI Appendix, Fig. S5A) but also the cell viability (Fig. 3 C and D). Together, these findings suggest that although the erlotinib-resistant cells have become unleashed from their EGFR activity dependence, they still rely on IL-6–mediated signaling for their survival.
Fig. 3.
IL-6 is responsible for the EGFR-independent STAT3 phosphorylation observed in the erlotinib-resistant cells. (A) TGF-β1 and 2 are required for the expression of IL-6. The chart represents IL-6 levels in the media secreted by H1650, H1650-M3 (control) and TGF-β1, or TGF-β2 knockdown cells determined by an ELISA. The data are the means ± SD (n = 4), and the Student t test showed significant difference (t < 0.05). (B) TGF-β treatment is sufficient to induce the expression of IL-6. The H1650 cells were treated for 2 d with TGF-β1, TGF-β2, or both in combination, and the RNA was isolated and subjected to a quantitative RT-PCR test. The data shown represent the relative expression level (means ± SD; n = 6). (C) Blockage of IL-6 signaling decreased cell viability. Cells were transfected with the indicated siRNA. After 2 d, the cells viability was determined by MTT assay and expressed relative to untreated cells (n = 4). (D) The chart shows the percentage of viable cells relative to control in the presence of 10 μg/mL IL-6 neutralizing antibody and 2 μM pan-Jak inhibitor (P6). (Data shown are the means ± SD; P < 0.02). (E) IL-6 is sufficient to modify sensitivity of cells to erlotinib treatment. Viability of PC9, HCC827, and HCC4006 cells was determined in the presence of 0.01 mM erlotinib. Of note, PC9, HCC827, and HCC4006 cells have different basal-cell sensitivities to erlotinib. Cell viability by IL-6 treatment is shown with the means of the percentage of viable cells ± SD (n = 6; P ≤ 0.0002). (F) Treatment with the JAK1/2 inhibitor P6 restored the cell sensitivity to erlotinib. Cells were pretreated with IL-6 (10 ng/mL) or IL-6 and P6 (2 μM) and then, treated with erlotinib (5 μM). After 72 h, cell viability was assessed. Each bar represents the average of four replicates ± SD (P < 0.01). (G) Schematic representation of our model system. Paracrine or autocrine stimulation of the TGF-β axis is sufficient to lead to acquisition of mesenchymal-like morphology, increased motility and invasion ability, and increased erlotinib resistance. However, in erlotinib-sensitive cells, mutant EGFR regulates the expression of IL-6; in erlotinib-resistant cells, TGF-β drives the expression of IL-6 independently of the activation of EGFR and unleashes the cells from their EGFR activity dependency.
IL-6 Is Sufficient to Modify Sensitivity of Cells to Erlotinib Treatment.
Because both the parental, sensitive cells as well as the resistant cells express similar levels of IL-6 receptor, we then speculated that stimulation of cells with IL-6 might decrease sensitivity to erlotinib treatment in cells expressing mutant EGFR. To explore this, we measured the effect of IL-6 on the viability of multiple NSCLC cell lines (HCC827, PC9, and HCC4006) that express mutant EGFR. Likely as a consequence of the accumulation of different genetic and epigenetic abnormalities, these cell lines, despite harboring somatic-activating EGFR mutations, display different erlotinib sensitivities with IC50s ranging from 5 μM in the case of H1650 to 0.001 μM in the case of HCC827 (10). Despite these differences in all cases as shown in Fig. 3E and SI Appendix, Fig. S5B, the presence of IL-6 in the medium was sufficient to diminish the sensitivity of various cell lines to erlotinib. Notably, in these experimental settings, IL-6 treatment did not substantially affect the cell-proliferation rates but rather, decreased their apoptotic response. To test whether the effect of IL-6 was indeed caused by the activation of the gp130-STAT3 axis but not a decreased bioavailability of erlotinib, we measured the effect of IL-6 treatment on the activation of key components of the EGFR signaling pathway in cells treated with erlotinib. Although IL-6 treatment did not have any effect on erlotinib-mediated inhibition of v-akt murine thymoma viral oncogene homolog (AKT) activation, it did impair inhibition of STAT3 phosphorylation (SI Appendix, Fig. S5C). Furthermore, treatment with the P6 JAK1/2 inhibitor restored the cell sensitivity to erlotinib, even in the presence of IL-6 (Fig. 3F). In summary, our data indicate that, independently of the complexity of the genomes of various NSCLC cell lines, IL-6 activation of the gp130/JAK pathway was sufficient to decrease sensitivity to erlotinib. Therefore, we propose a model in which paracrine or autocrine stimulation of the TGF-β axis is sufficient for acquisition of mesenchymal-like morphology, increased motility and invasion ability, and increased erlotinib resistance. (Fig. 3G).
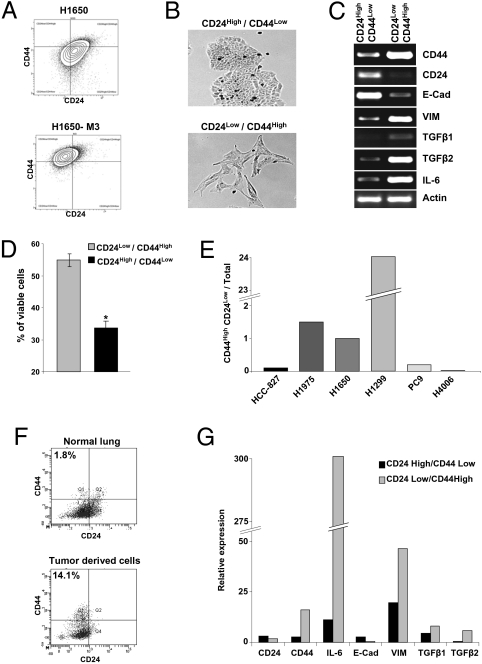
Erlotinib-Resistant Mesenchymal-Like Cells Are Present in Cell Lines and Tumors Before Erlotinib Treatment.
Gene-expression profile analysis revealed that CD44 expression was higher and CD24 expression was lower in the erlotinib-resistant cells compared with the parental cells (SI Appendix, Fig. S2). Because a subpopulation of cells that are CD44high/CD24low mesenchymal highly motile and invasive has been previously described to exist within both primary breast carcinomas as well as certain breast cancer-derived cell lines, we reasoned that erlotinib-resistant mesenchymal-like cells could exist as a subpopulation within certain lung carcinomas and as such, could have been present in the parental sensitive H1650 cell line before erlotinib treatment. By FACS analysis, we were able to confirm the differential expression of CD44 and CD24 in the erlotinib-resistant compared with parental cells (Fig. 4A) as well as assess the feasibility of using FACS to separate our putative target cells from the parental population. Using bivariant plotting of CD44 and CD24 staining, we were able to identify a subpopulation of cells with features similar to those of the erlotinib-resistant derived cells within the parental H1650 population before drug treatment. Specifically, we found that in the erlotinib-naïve H1650 cells, a CD44high/CD24low-enriched cell fraction was characterized by a mesenchymal appearance (Fig. 4B), increased expression of Vimentin, TGF-β1, TGF-β2, and IL-6, and decreased levels of E-cadherin (Fig. 4C). Importantly, these cells were also more resistant to erlotinib treatment compared with the CD44low/CD24high fraction (Fig. 4D). The presence of CD44high/CD24low cells was not limited to the H1650 cell line. Populations with similar properties were also present in several other cell lines. (Fig. 4E). To exclude the possibility that the occurrence of erlotinib-resistant mesenchymal-like cells was caused by artificial in vitro growth conditions and/or events that occurred during establishment of the cell lines, we extended our analysis to cells obtained from patients with NSCLC. As shown in Fig. 4F and SI Appendix, Fig. S6 A–C, we were able to identify CD44high/CD24low cells in preparations from early-stage erlotinib-naïve human NSCLC tumors as well as the bone marrow of patients with NSCLC. The CD44high/CD24low cells, like H1650-M3 erlotinib-resistant cells, had decreased expression of E-cadherin and higher levels of expression of IL-6, Vimentin, TGF-β1, and TGF-β2 (Fig. 3G). Importantly, to make sure that these cells were bona fide tumor cells, we also sorted them based on their positivity for the epithelial marker EpCAM (11) and their lack of expression of the cell-surface markers CD45 and CD31 (SI Appendix, Fig. S6D).
Fig. 4.
Erlotinib-resistant mesenchymal-like cells are present in cell lines and tumors before erlotinib treatment. (A) H1650 cells and H1650-M3 cells were analyzed by flow cytometry for CD44 and CD24 expression. (B) The bright field image of the sorted cells showed that the CD44high/CD24low population of the H1650 cells has distinct morphology and like H1650-M3 cells, resembles mesenchymal cells. (C) RT-PCR analysis of sorted cells indicated that cells in the CD44high/CD24low population of the H1650 cells have increased expression of Vimentin, TGF-β1, TGF- β2, and IL-6 and diminished expression of E-cadherin relative to most H1650 cells. (D) Similar to the erlotinib-resistant H1650-M3 cells, the CD44high/CD24low fraction of H1650 cells is more resistant to erlotinib treatment than the unsorted H1650 cells. Sorted cells were treated with 5 μM of erlotinib for 72 h. Cell viability was assessed by MTT assay and expressed relative to untreated cells (n = 6; P < 0.0001). (E) Flow-cytometry analysis for CD44 and CD24 of cells harboring mutant EGFR (i.e., HCC827, H1975, PC9, and HCC4006 cells) and the H1299 cells that are wild-type for the EGFR gene revealed the presence of subpopulations of cells that are CD44high/CD24low. The chart represents the percentage of CD44high/CD24low cells relative to total number of viable cells. (F) NSCLC harbors CD44high/CD24low subpopulations. Dissociated tumor cells were sorted for CD45, CD31, and EpCAM and analyzed for CD44 and CD24 expression. (G) Real-time PCR analysis was used to compare CD45−CD31−EpCAM+CD44highCD24low cells with CD45−CD31−EpCAM+CD44lowCD24high cells.
Inflammation-Induced IL-6 Expression Decreases the Tumor Sensitivity to Erlotinib.
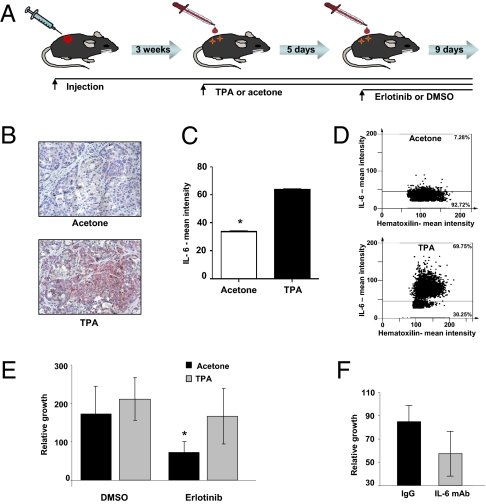
Whereas these data implicate a cell-autonomous mechanism behind the ontogeny of erlotinib-resistant mesenchymal-like cells, the finding that TGF-β stimulation was sufficient for induction of EMT and decreased sensitivity to drug led us to hypothesize that the tumor microenvironment may also play a role in emergent resistance through paracrine signaling. Because inflammation has already been shown to augment expression levels of IL-6 and TGF-β (12), this concept could provide intriguing insights into the degree of heterogeneity seen in the response of NSCLC tumors harboring mutant EGFR to erlotinib treatment and the occurrence of acquired erlotinib resistance. To test the hypothesis, we used a mouse-model system previously described by Vasunia et al. (13) in which cutaneous inflammation and secretion of TGF-β and IL-6 are induced by topical treatment of the mouse epidermis with low dosage of 12-O-tetradecanoyl-phorbol-13-acetate (TPA) (13). To provide further evidence that inflammation decreases erlotinib response, we also induced inflammation by treatment with LPS. Nu/Nu mice were injected in the flank with NSCLC cells expressing mutant EGFR. When the tumors reached a volume of ∼100 mm3, we topically treated the epidermis with TPA or in the case of LPS, subcutaneous injection for 5 d. Mice were then treated with erlotinib for 9 d at a dosage equivalent to the one used in patients (Fig. 5A). Immunostaining of tumor sections using an anti–IL-6 antibody confirmed that TPA augmented IL-6 expression in the tumors (Fig. 5 B–D). By comparing tumor burden after different treatment conditions, we found that TPA and LPS treatments dramatically reduced the response to erlotinib treatment (Fig. 5E and SI Appendix, Fig. S7A). In accordance with our previous in vitro observations, immunostaining with Ki67, a commonly used marker of cell proliferation, and TUNEL assay (to assess apoptosis) showed that inflammation was able to attenuate the effects of erlotinib in vivo, primarily through the inhibition of apoptosis rather than by increase in cell proliferation (SI Appendix, Fig. S7 B–D). To establish a causal link between TPA treatment, induction of IL-6, and diminished erlotinib response, we decreased IL-6 bioavailability by injecting mice with neutralizing antibody specific to this cytokine. Cotreatment of mice decreased the effect of TPA and increased tumor sensitivity to targeted therapy (Fig. 5F).
Fig. 5.
Inflammation induces IL-6 expression, which decreases the tumor sensitivity to erlotinib. (A) Schematic figure of the mouse model used in our studies. Mice were injected with H1650 cells s.c. After ∼3 wk, when the tumor reached a volume of ∼100 mm3, mice were treated topically with TPA or vehicle (acetone) every other day. At day 5, the mice were treated with erlotinib or DMSO by oral gavage for 9 d. At this time, the tumor burden was evaluated. Each time point represents the average of at least six tumors. (B) TPA treatment induces expression of IL-6. Formalin-fixed, paraffin-embedded sections (4 μm) were stained for IL-6 and counterstained with hematoxilin. (C) Protein expression was measured by defining regions of interest (ROI) using automated cell acquisition and quantification software for immunohistochemistry (Histoquest). (D) The IL-6 mean intensities in tumors from a representative animal treated with TPA and a vehicle-treated animal are shown. (E) TPA treatment decreases the tumors response to erlotinib treatment. The chart represents growth percentage of tumors. Each column represents the mean volume of eight different tumors. Of note, similar effects were observed on treatment with LPS (SI Appendix, Fig. S8). (F) The effects of TPA are, in part, caused by an increased secretion of IL-6. Before erlotinib treatment, mice were injected intraperitoneally with IL-6 neutralizing antibody (0.08 mg/g) or as a control, with IgG (0.08 mg/g). After 8 d, tumor burden was evaluated, and the averages of four separate tumors were charted.
Discussion
In summary, by selecting cells in high concentration of erlotinib, we identified a subpopulation of erlotinib-resistant cells with mesenchymal morphology characterized by an EGFR-independent augmented secretion of TGF-β1 and TGF-β2. We determined that the increased autocrine secretion of TGF-β was sufficient to activate a complex program that lead to acquisition of mesenchymal-like morphology, increased motility and invasion ability, and increased erlotinib resistance. In the latter case, we provided evidence that an up-regulation of IL-6 secretion by TGF-β was sufficient to unleash cells harboring mutant EGFR from their EGFR dependency, which was manifested by their decreased sensitivity to erlotinib treatment (Fig. 3A). By using a surface-marker signature derived from the erlotinib-resistant cells, we, in addition, showed that cells that are mesenchymal- and erlotinib-resistant were already present in NSCLC-derived cell lines as well as in early-stage treatment-naive tumors. These data, thus, indicate that cell-autonomous mechanisms could generate subpopulations of cells intrinsically resistant to erlotinib treatment. However, because both IL-6 and TGF-β are secreted factors prominently produced during the inflammatory response, the activation of the tumor microenvironment could also contribute to erlotinib resistance. By use of a mouse-model system in which inflammation was induced either by topical treatment with low concentration of TPA or with LPS, we were indeed able to show that the induction of inflammation was successful in stimulating IL-6 secretion and decreasing the tumor response to erlotinib treatment. Thus, our data provide compelling evidence indicating that acquired resistance to molecular-targeted therapies could arise not only as a consequence of genetic and/or epigenetic heterogeneity within cancer cells but also through the activation of the tumor microenvironment. Because we identified cells that are intrinsically resistant to erlotinib in NSCLC before treatment, it is also tempting to speculate that this same mechanism of erlotinib-acquired resistance could explain the heterogeneity of primary erlotinib responses observed in patients. In fact, although the majority of patients harboring similar EGFR oncogenic mutations do respond to erlotinib treatment, overall responses can vary from 5% to 90%, and remission could span 3 mo to more than 5 y. Notably, several studies have already reported increased levels of IL-6 in ∼30% of NSCLC (14).
Why IL-6 is required for the survival of the cancer cells is yet not clear. Our data seems to indicate a role of IL-6 in protecting cells from apoptosis (SI Appendix, Fig. S5B). This seems to be consistent with current literature. Catlett-Falcone et al., (15) in fact, showed that IL-6–induced STAT3 protects myeloma cells from tumor necrosis factor receptor superfamily, member 6 (FAS)-induced apoptosis by up-regulating the expression of B-cell CLL/lymphoma 2-like protein X isoform L (BCL-X)L. Similarly, Haga et al. (16) show that constitutively active STAT3 provided protection against FAS-mediated liver injury, likely through an augmented expression of the antiapoptotic proteins FLICE-like inhibitory protein (FLIP), BCL-2, and BCL-XL. In agreement with a role of IL-6/STAT3-mediated survival, we observed an up-regulation of Surivin, BCL-X1, and BCL-2 in the erlotinib-resistant cells.
Interestingly, the erlotinib-resistant cells characterized in this study also have an increased metastatic potential. They display mesenchymal-like features, are highly motile and invasive, and can be found with an increased representation in the bone marrow of patients compared with primary tumors. Notably, many of the genes that we see increasingly expressed in the resistant cells compared with the parental line have already been associated with a metastatic signature and/or with poor prognosis. These genes include HOXB2, S100A4, S100A2, Tenascin C, sushi domain containing (SUSD), melanoma cell adhesion molecule (MCAM), and transcription factor 4 (TCF-4).
Notably, although EMT has been previously reported to be associated with erlotinib resistance (17–19), our data indicate that the programs that lead to mesenchymalization and drug resistance are distinct. Although we found TGF-β to be required and sufficient for EMT, invasion and motility as well as induction of an increased expression of IL-6 and reduction of the expression of the EMT master regulator SNAI only impaired EMT/motility/invasion but did not change the cells sensitivity to erlotinib. Furthermore, treatment of cells with IL-6 increased their resistance to erlotinib but did not induce EMT (SI Appendix, Fig. S8). Our data are consistent with the observation that certain cell lines harboring mutant EGFR (i.e., PC9), despite being extremely sensitive to erlotinib, display mesenchymal-like features.
In conclusion, our data provide compelling evidence indicating that resistance to molecular-targeted therapies could arise not only as a consequence of genetic and/or epigenetic changes within cancer cells but also through the activation of the tumor microenvironment. Thus, the contribution of selective and adaptive mechanisms adds a layer to the complexity of cancer-drug resistance and poses challenges for the clinical use of molecular-targeted therapies. In particular, it clearly indicates that, in the case of lung tumors driven by mutant EGFR, treatment based only on the inhibition of EGFR will not be effective and suggests the intriguing possibility that adjunctive therapies designed to either control inflammation and/or decrease the bioavailability of IL-6 may provide effective means to improve response to EGFR TKI treatment. Interestingly, clinical trials combining Cox2 inhibitors (e.g., rofecoxib and celecoxib) and gefitinib/erlotinib have already shown encouraging results.
Materials and Methods
Cell Culture.
H1650 (H1650), HCC827, H1975 (NCI-H1975), H1299 (NCI-H1299), and HCC4006 were obtained from the American Type Culture Collection repository. The PC9 cell line was a gift from Jeff Engelman (MGH, Charlestown, MA). All of the cell lines were cultured in Roswell Park Memorial Institute medium supplemented with 5% FBS, glutamine, penicillin, and streptomycin. The lentiviral packaging cell line HEK293T was cultured in DMEM containing 10% FBS, penicillin, streptomycin, and sodium pyruvate.
Antibodies and Reagents.
The following antibodies were used in this study: mouse anti–E-cadherin antibody (BD Transduction Laboratories), monoclonal anti-Vimentin antibody (RV202; Santa Cruz Biotechnology), mouse anti–β-Tubulin antibody (2-28-33; Santa Cruz Biotechnology), mouse anti-STAT3 antibody (124H6; Cell Signaling Technology), and rabbit anti–phospho-STAT3 antibody (D3A7; Cell Signaling Technology). The Smad molecules were detected with the Phospho-Smad antibody Sampler Kit from Cell Signaling Technology. Erlotinib hydrochloride was purchased from LGM Pharmaceuticals, Inc. Recombinant IL-6 (rhIL6), anti–IL-6 monoclonal antibody (MAB206), and anti–IL-6R antibody (AB-227-NA) were obtained from R&D Systems. Recombinant TGFβ1/2 was purchased from Sigma-Aldrich. Human recombinant EGF was purchased from Millipore. The JAK 1/2 inhibitor tetracyclic pyridone 2-tert-butyl-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinoline-7-one, pyridone 6 (P6) was purchased from Calbiochem.
SI Appendix has further descriptions of the experimental procedures described in this paper. Human tissues were obtained from CT Surgery Department, Weill Cornell Medical College, and patient consent was obtained according to approved IRB protocols from the institution.
Supplementary Material
Acknowledgments
We thank Drs. Linda Van Aelst and Greg Hannon for their precious insights and Elizabeth Nakasone, Maria Pineda, and Anna Saborowski for their helpful suggestions. The Flow Cytometry and Microscopy shared resources at Cold Spring Harbor Laboratory for their assistance. This work was possible thanks to the generous support of Damon-Runyon Cancer Research Foundation, Robertson Found, Swim for America, and Diane Emdin Sachs Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009472107/-/DCSupplemental.
References
- 1.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Maemondo M, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Wakeling AE, et al. ZD1839 (Iressa): An orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–5754. [PubMed] [Google Scholar]
- 6.Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–595. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 7.Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 8.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 9.Gao SP, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandhi J, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS ONE. 2009;4:e4576. doi: 10.1371/journal.pone.0004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eramo A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Novoa JM, Nieto MA. Inflammation and EMT: An alliance towards organ fibrosis and cancer progression. EMBO Mol Med. 2009;1:303–314. doi: 10.1002/emmm.200900043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasunia KB, Miller ML, Andringa S, Baxter CS. Induction of interleukin-6 in the epidermis of mice in response to tumor-promoting agents. Carcinogenesis. 1994;15:1723–1727. doi: 10.1093/carcin/15.8.1723. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi T, et al. Involvement of interleukin-6 in the elevation of plasma fibrinogen levels in lung cancer patients. Jpn J Clin Oncol. 1998;28:740–744. doi: 10.1093/jjco/28.12.740. [DOI] [PubMed] [Google Scholar]
- 15.Catlett-Falcone R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 16.Haga S, et al. Stat3 protects against Fas-induced liver injury by redox-dependent and -independent mechanisms. J Clin Invest. 2003;112:989–998. doi: 10.1172/JCI17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson S, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 18.Yauch RL, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11:8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 19.Barr S, et al. Bypassing cellular EGF receptor dependence through epithelial-to-mesenchymal-like transitions. Clin Exp Metastasis. 2008;25:685–693. doi: 10.1007/s10585-007-9121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.