Abstract
The hematopoietic system produces a large number of highly specialized cell types that are derived through a hierarchical differentiation process from a common stem cell population. miRNAs are critical players in orchestrating this differentiation. Here, we report the development and application of a high-throughput microfluidic real-time quantitative PCR (RT-qPCR) approach for generating global miRNA profiles for 27 phenotypically distinct cell populations isolated from normal adult mouse hematopoietic tissues. A total of 80,000 RT-qPCR assays were used to map the landscape of miRNA expression across the hematopoietic hierarchy, including rare progenitor and stem cell populations. We show that miRNA profiles allow for the direct inference of cell lineage relations and functional similarity. Our analysis reveals a close relatedness of the miRNA expression patterns in multipotent progenitors and stem cells, followed by a major reprogramming upon restriction of differentiation potential to a single lineage. The analysis of miRNA expression in single hematopoietic cells further demonstrates that miRNA expression is very tightly regulated within highly purified populations, underscoring the potential of single-cell miRNA profiling for assessing compartment heterogeneity.
Keywords: RT-qPCR, stem cell, hematopoiesis, microfluidic, single cell
Hematopoiesis is a complex process through which a hierarchy of increasingly specialized cell populations originate from a common pluripotent hematopoietic stem cell. New blood cells (1010 to 1011) are produced daily in adult humans, and the regulated output of this enormous number of cells involves multiple levels and mechanisms of control. The latter includes cell–cell interactions and soluble cytokine-activated signaling events, transcription factor activation, and epigenetic changes leading to altered gene expression programs (1). The relatively recent discovery of microRNAs (miRNAs) and their ability to regulate many differentiation processes has introduced new complexity into understanding the regulation of hematopoiesis (2).
miRNAs are short RNA molecules, 19–25 nucleotides in length, that play key roles in regulating gene expression by inhibiting translation and/or triggering degradation of target messenger RNAs (3). There are hundreds of miRNAs found in a given mammalian genome, and a single miRNA can regulate hundreds of genes (4). miRNAs thus act as master regulators of transcriptional programs, and their expression patterns are thought to reflect biological relationships between hematopoietic lineages. A growing body of work studying miRNA expression in abundant cell populations, including both normal and malignant cells, has made it clear that miRNAs play a critical role in hematopoiesis and blood cancers (4–7).
Generating a comprehensive atlas of miRNA expression patterns, and how they change during the initial stages of hematopoietic stem cell differentiation in particular, has been impeded by the scarcity of primitive cells and the limited sensitivity and high cost of current profiling methodologies. Low RNA yields pose a major constraint to quantifying miRNAs by using established methods such as Northern blotting, cloning, microarrays, and deep sequencing, all of which rely on the availability of microgram RNA quantities. Real-time quantitative PCR (RT-qPCR) provides the highest sensitivity of miRNA quantification, is capable of distinguishing mature and precursor miRNA, and produces fewer false-positives and reduced bias when compared with microarray or sequencing approaches (8, 9). Here, we combine the sensitivity and specificity of multiplexed RT-qPCR with the economy of scale and throughput of microfluidics to allow for global miRNA profiling from limited cell populations and single cells. We apply this method to map the expression patterns of 288 miRNA species in 27 different hematopoietic subsets drawn from all parts of the hematopoietic hierarchy.
Results
miRNA Profiling in Rare and Single Cells Using Microfluidic qPCR.
We sought to apply newly available microfluidic technology to generate a unified and comprehensive dataset of miRNA expression across the hematopoietic tree. To achieve this goal, we developed and validated a protocol for miRNA profiling in rare cell types that is based on multiplexed stem-loop RT-qPCR (10), followed by high-throughput nanoliter volume qPCR on microfluidic arrays (48.48 Dynamic Array; Fluidigm). All protocol development and testing was performed by using freshly derived primitive adult mouse bone marrow cells engineered to express a NUP98-HOXD13 (ND13) fusion gene (11). These cells possess a phenotype that reflects immature (c-kit+) and mature (Mac1+) murine bone marrow cells and, thus, are a suitable representative of the murine hematopoietic system for which miRNA expression has been characterized in depth (12). Our protocol consists of three steps: multiplexed RT, multiplexed cDNA preamplification, and nanoliter volume simplex qPCR using TaqMan probes (Fig. S1 and SI Materials and Methods). To permit the global analysis of limited samples, we selected 96× multiplexing as an optimal tradeoff between sensitivity and throughput. To cover 288 miRNA targets, each sample was split into three reverse transcription (RT) reactions performed by using unique pools of 96 RT primers (Dataset S1). The generated cDNA from each pool was then subjected to 18 cycles of PCR before analysis on microfluidic qPCR arrays. Comparison of the number of miRNAs detected from purified RNA and cell lysates established that cell inputs of 1,024 cells were sufficient to detect the majority (>95%) of miRNA present in a given sample. These cell numbers are well-matched to achievable purification yields of rare primitive cells.
To account for possible variations in assay efficiency and sensitivity, we included external calibration standards consisting of 10× serial dilutions of synthetic miRNAs (mirVana miRNA Reference Panel v9.1; ABI). Analysis of these standards revealed that assay efficiency varied considerably (Dataset S2) and that sensitivity ranged between 10 and 105 molecules per reaction. Variations in assay efficiency were found independent of multiplexing level and were observed even for serial dilutions of miRNA standards in single-plex reactions performed in conventional tubes (Fig. S2).
We further extended our protocol to measure miRNA expression in single cells. Nine highly expressed miRNA species were measured from a 2-fold dilution series of purified RNA template ranging from 16 single cell equivalents (320 pg) to 1/16 of a cell equivalent (1.25 pg) at varying multiplexing levels of 12×, 48×, 96×, and 312× (Fig. S3). These experiments established that 12× multiplexing provided reliable detection of abundant miRNA directly from single cell amounts of RNA with technical variation below one CT value and sensitivity better than 1 pg of whole RNA (Fig. 1).
Fig. 1.
Single-cell sensitivity and variation. (A) Technical variation in qPCR of single-cell lysates. Real-time fluorescent curves show reproducibility of qPCR detection of miR-19b from three single ND13 cells using the 12× multiplexing protocol. Five technical replicates from each cell are shown. (B) Cell lysate dilution series. Dilution series of whole RNA from ND13 cells were assayed for miR-19b by using the 12× multiplexing protocol showing sensitivity down to 0.1 pg (≈1/100 single cell equivalent). Blue and red points indicate duplicate measurements from 2 ND13 samples. (C) Single-cell measurements of six miRNAs from seven different ND13 cells using the 12× multiplexing protocol along with cell lysates at 10× and 100× single-cell equivalents. Error bars represent the SD of five replicates.
Profiling 288 miRNAs in 27 Hematopoietic Subpopulations.
We then applied our protocol to the profiling of miRNA expression in select populations representing the hematopoietic hierarchy. A total of 27 cell populations, chosen to represent a large spectrum of differentiated and primitive cell types, were tested for their expression of 288 different miRNAs (Fig. 2A). A list of cell surface markers used in the purification of the subsets studied and the miRNAs assayed is included as supporting information (Dataset S1 and Table S1). Cell populations were isolated from bone marrow, peripheral blood, spleen, thymus, and peritoneal washes of normal adult C57BL/6J mice. RNA extracts were prepared from at least three sets of independently isolated cells and each extract was analyzed in triplicate with two exceptions: E-SLAM (stem) cells and mature thymocytes, where only a single replicate produced an acceptable signal (see SI Materials and Methods for details). All qPCR measurements were performed in duplicate. In total, we assembled six runs in which 288 assays were tested against seven calibration and 41 biological samples derived from 27 purified hematopoietic cell samples including representatives of all hematopoietic lineages, as well as various stem and progenitor cell populations (Fig. 2A).
Fig. 2.
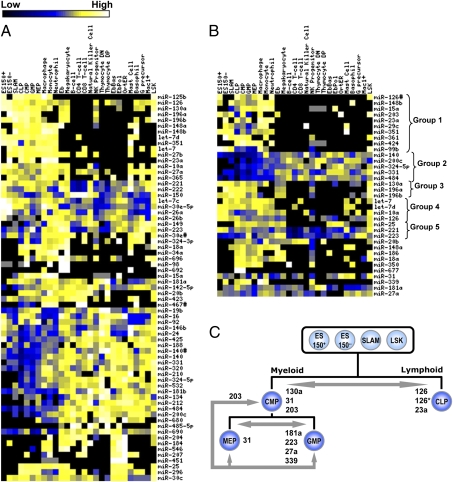
miRNA profiling of the hematopoietic hierarchy. (A) Hierarchical lineage relation of analyzed populations. The relation of 27 purified subpopulations representing all hematopoietic lineages is shown. These cell types include 12 myeloid populations (red), 7 lymphoid populations (green), and 8 progenitor and stem cell populations (blue). (B) Phylogenetic tree reconstructed from miRNA expression data. The grouping of cell populations based on miRNA expression profiling reveals a clear separation of myeloid, lymphoid, and stem cell/progenitor populations with four distinct subgroups present in the myeloid branch. Circles indicate the number of miRNA species detected in each population. Bootstrap values are indicated next to branch nodes.
Altogether, 82,924 independent qPCR reactions were performed. One hundred eighty-seven of all 255 tested miRNAs were detected in multiple biological replicates of at least one of the 27 cell populations examined (Dataset S3). The remaining 68 miRNAs were regarded as not expressed at detectable levels (Datasets S1 and S2). Twenty-six of the 187 detected miRNAs were found in all cell populations tested, 46 were found in <25% of the 27 cell types, and 51 where found in >75% of these. Log2-ratio measurements of technical and biological deviation in our dataset were 1.08 and 2.02, respectively, with a total set deviation of 2.6, indicating that biological variability is the main source of measurement error (Fig. S4).
Comparison of miRNA Expression Profiles in Different Cell Types.
Recent work suggests that miRNAs are modulated during differentiation and act to lock in specific gene expression programs (13, 14). The mean number of miRNAs detected in each population was 96 and varied between 49 and 148 (Fig. 2B). Terminally differentiated thymocytes exhibited the lowest number of detected miRNAs (49 miRNAs in CD4+CD8+ thymocytes and 61 miRNAs in CD4−CD8− thymocytes). This finding was comparable with the results obtained for primitive cells (69 miRNAs in EPCR+150+CD48− and 49 miRNAs in EPCR+150−CD48−) and Mac1+ cells (54 miRNAs). By comparison, a more diverse range of miRNAs was identified in monocytes (148), peripheral macrophages (145), and common myeloid progenitor cells (136).
To further explore the relation of differentiated state and the pattern of miRNA expression, we constructed a hierarchical tree based on the similarity of miRNA expression profiles (Fig. 2B). The 27 purified cell types in this study were found to group into six main branches: stem cells and multipotent progenitors, lymphoid cells, and four distinct branches of myeloid cells. Analysis of the miRNAs that were differentially expressed in the members of these six branches revealed further patterns that appeared to be differentiation-associated (Fig. 3). The 10 miRNAs that showed the greatest differential expression in a pairwise comparison of the six branches of cells are presented in Dataset S4. Several miRNAs were found to be generally up-regulated in stem cell and progenitor populations relative to more differentiated cell types. These miRNAs included miR-125b, miR-196a, miR-196b, miR-130a, let-7d, miR-148b, and miR-351 (Fig. 3A). Most specific to these cells was miR-125b, which was detected in two of three stem cell populations, all progenitors, and only two other cell types including peripheral macrophages and monocytes. Expression of miRNA-125b was also enriched in the branch containing monocytes, macrophages, and neutrophils, a pattern that was shared by several other miRNAs, including miR-126*, miR-148b, miR-15a, miR-203, miR-23a, miR-23b, miR-29c, miR-351, miR-361, miR-424, miR-99b (Fig. 3B, group 1). Examples of down-regulated miRNAs in stem cell and progenitor populations include miR-140, miR-200c, miR-484, miR-331, and miR-324–5p (Fig. 3B, group 2). Notably, miR-29a was found to be up-regulated in highly enriched HSC populations ES 150− and ES 150+ relative to SLAM populations or other progenitors (15). No miRNAs were expressed in all differentiated cells but not stem cells and progenitors. However, several miRNAs observed to be down-regulated in primitive cells, including miR-484, miR-200c, miR-331, miR-320, miR-210, miR-324–5p, miR-212, and miR-690, are expressed widely across differentiated cell types and may be candidates for differentiation-specific markers.
Fig. 3.
miRNA expression in different hematopoietic lineages. (A) Most differentially expressed miRNAs between the six branches of cell types identified in Fig. 2B. (B) Examples of observed miRNA expression patterns across various hematopoietic populations. (C) Differential expression of miRNAs between early progenitor populations. Upregulated miRNAs are listed adjacent to the arrows indicating the pair-wise comparison of cell populations.
We also identified groups of miRNAs whose expression patterns were similar in many different cell populations. For example, we found miR-130a, miR-196a, and miR-196b to be expressed in stem cell and progenitor populations and then down-regulated upon commitment to the lymphoid lineages (Fig. 3B, group 3) (16). All three of these miRNAs were also found in megakaryocyte and erythroblast populations. Other miRNAs that were strongly down-regulated in lymphoid lineages included let-7, let-7d, miR-126, miR-148b, miR-351, and miR-10a (Fig. 3B, group 4).
A comparison of all myeloid cell populations with all lymphoid cell populations revealed three miRNAs that are strongly up-regulated in myeloid cells. These miRNAs include miR-25, miR-221, and miR-223 (2, 17) (Fig. 3B, group 5). Other patterns of miRNA expression were revealed when selected myeloid populations were compared. For example, miR-20b is expressed in all progenitors, nearly all lymphoid cells, and in monocytes and granulocytes, but was not detected in stem cell populations, terminal erythroid cells, granulocyte precursors, or histamine-secreting cells. As another example, miR-148a was found to be highly expressed in all terminally differentiated granulocytes, macrophages, and monocytes, and in all progenitor cells, NK progenitors, and erythroblasts (Fig. 3B).
To identify miRNA species that might be involved in early stages of hematopoietic cell differentiation, we compared profiles obtained for stem cell and different progenitor populations. CMPs and CLPs are thought to represent the stage at which restriction to either the myeloid or lymphoid lineages has just occurred. Interestingly, several miRNAs were found to be up-regulated in these cells by comparison with more primitive cell types. These miRNAs include miR-15a, miR-18a, miR-186, miR-25, miR-350, and miR-424. In addition, miR-677 was detected repeatedly in CLPs only (in 2/4 samples) and in CMPs only (in 3/5 samples) and may be specific for these progenitors. The most differentially expressed miRNAs between the CMPs and CLPs include miR-130a (2.4-fold, P = 0.001), miR-31 (6.2-fold, P = 0.003), and miR-203 (6.4-fold, P = 0.01), which are up-regulated in CMPs and miR-126 (−2.8-fold, P = 0.01), miR-126* (−7.7-fold, P = 0.005), and miR-23a (−7.4-fold, P = 0.002), which are up-regulated in CLPs (Fig. 3C). miR-203 was further found to be down-regulated (7.0-fold, P = 10−5) upon differentiation of CMPs into either of the next level of multipotent progenitors, i.e., GMPs and MEPs. Within these cell populations, miR-181a (6.7-fold, P = 0.005), miR-223 (9.1-fold, P = 0.0001), miR-27a (5.4-fold, P = 0.016), and miR-339 (6.4-fold, P = 0.004) were found to be expressed at a higher level in GMPs relative to MEPs, with the opposite pattern for miR-31 (−5.5 fold, P = 0.019).
Comparison of miRNA signatures from four stages of erythroid cell development provides a snapshot of miRNA regulation through sequential steps of differentiation along a single lineage (Fig. 4). Erythroblasts, the first erythroid-restricted population studied, group most closely with megakaryocytes and exhibit a distinct miRNA signature from the three more mature erythroid populations tested (EbPol, EbBas, OrtER). Through differentiation, the total number of detected miRNAs in erythroid cells gradually decreases from 106 in erythroblasts to 76 in the population of orthochromatic erythroblasts and reticulocytes (OrtER). However, this trend was paralleled by a marked increase in the level of expression of several other miRNAs (miR-151, miR-152, miR-184, miR-187, miR-212, miR-30a-3p, miR-30e-5p, miR-451). In particular, we found miR-451 expression, previously reported to increase during red cell maturation (14), to increase specifically in the latest stage of erythroid cell maturation and was not detected in any other populations tested. These studies also showed that several miRNAs are progressively down-regulated during erythroid cell differentiation and are lost in the terminal OrtER population. These miRNAs include miR-126, miR-29a, and miR-696. Interestingly, these trends of increased and decreased miRNA expression can be extended into the MEP population (miR-126, miR-152, miR-184, miR-187, miR-29a, miR-30a-3p, and miR-451; Fig. 4). Further, these miRNAs were also expressed in megakaryocytes at similar levels to erythroblasts.
Fig. 4.
miRNA expression during erythroid differentiation. Several miRNAs are progressively up- or down-regulated during differentiation from erythroblasts to terminal erythrocytes. Megakaryocytes exhibit a similar expression pattern to erythroblasts.
Single-Cell Analyses.
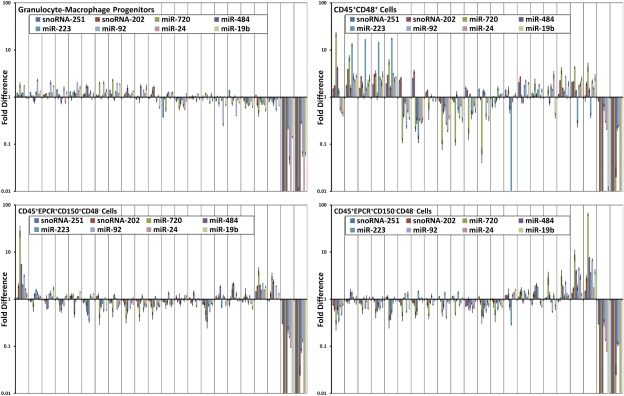
Although most populations in our study were at least 90% pure as determined by repeat phenotype analysis, some are known to comprise functionally distinct subtypes. It was therefore of interest to examine the extent of variability in miRNA expression by single-cell analyses. To investigate this issue, we tested single cells from three populations that represented cells that were expected to show a high degree of functional homogeneity, a high degree of functional diversity, and highly enriched populations of stem cells where some functional diversity has been reported (18, 19). GMPs (Lin−c-Kit+Sca1−CD34+FcγRhi cells) were chosen as the candidate homogeneous cell type (20) to enable a measure of the inherent variability of miRNA expression between functionally similar cells. CD45+CD48+ cells were chosen as the candidate functionally diverse population since they include almost all hematopoietic populations except for stem cells (21) and, hence, provide a positive control for miRNA diversity between individual cells. Two related stem cell populations were then also tested at the single-cell level. This population contains ≈40% of durable self-renewing stem cells (CD45+EPCR+CD150+CD48− or ES 150+), and a population in which ≈30% of the cells are finite self-renewing stem cells (CD45+EPCR+CD150−CD48− or ES 150−) (18, 19). In each case, we measured the expression of six highly expressed miRNAs (miR-720, miR-484, miR-223, miR-92, miR-24, and miR-19) and two snoRNAs (snoRNA-251, snoRNA-202) in 20 single cells from each group.
The results are shown in Fig. 5. As expected, CD45+CD48+ cells exhibited the largest degree of variability with miRNA expression differences of >100-fold between individual cells. Despite the small number of cells there appears to be at least three main subpopulations with highly similar expression profiles. miR-223 exhibits the highest degree of variability in the CD45+CD48+ population, consistent with its role in myeloid lineage differentiation and its expected presence in granulocytes and their precursors (17). snoRNA-251 and snoRNA-202 expression varied by ≈4-fold across the population (Fig. 5). The variability observed within the GMPs was much less, with not more than a 2-fold variation in the level of any miRNA among all 20 cells tested and no evidence of any subpopulations within the GMP sample. The variability in snoRNA-251 and snoRNA-202 expression is also well below that observed in the CD45+/CD48+ sample. The ES150+ and ES150− populations exhibited similar levels of heterogeneity that were intermediate to that seen in the GMP and CD45+CD48+ populations. Both stem cell populations contain obvious outliers that exhibit general up-regulation of miRNA expression. In particular, miR-720, a miRNA that we found to be expressed broadly across the hematopoietic hierarchy, is up-regulated by >50-fold in a minority of these cells.
Fig. 5.
Variation of miRNA expression in single cells. Expression levels of 6 miRNAs and 2 sno-RNAs in 20 single cells are plotted for four sorted populations including GMPs—a population of functionally homogeneous cells; CD45+CD48+—cells that include many cell types; CD45+EPCR+CD150+CD48− cells (ES150+)—a population of stem cells of which a high proportion show durable self-renewal abilities; and CD45+EPCR+CD150−CD48− (ES150−) cells—a population of stem cells of which a high proportion show finite self-renewal abilities. All data are plotted as a log-fold ratio to the average of the population. Two NTC samples are shown on the right side of each graph.
Discussion
Our data reveals several general features of miRNA regulation during the process of hematopoietic differentiation. First, analysis of globally profiled miRNAs allowed for the reconstruction of the accepted hierarchical relationships between different phenotypically defined hematopoietic populations previously arranged according to their proliferative and differentiation functional properties. Second, analysis of their miRNA expression patterns showed that cell populations group by differentiation state with progenitor and stem cell populations closely related and a dramatic reprogramming upon commitment to a single lineage. Finally, our single-cell measurements reflect the known functional heterogeneity of the various purified populations examined and show that miRNA expression levels are very tightly regulated within functionally homogeneous cells.
Although it has been shown that miRNA profiles provide a signature of cellular state (22, 23), this property has not been demonstrated for a large number of primary and hierarchically related cell types. Comparison of 27 cell populations within a single unified data set reveals that miRNA patterns reflect both cell differentiation state and lineage potential. Terminally differentiated cell types generally exhibited miRNA expression patterns with high similarity between populations related in function and lineage. For instance, all lymphoid cell types form a single branch with CD4+ and CD8+ populations clustering closely with B cells, natural killer cells, and natural killer cell progenitors. This grouping of functionally related cells is also apparent within the myeloid population. However, these cells were more diverse in miRNA expression, possibly reflecting a greater functional diversity. The three most differentiated erythroid populations formed a single cluster. Mast cells and basophils, both of which are responsible for the secretion of histamine, group together with complex populations of Mac1+Gr1− and Mac1+Gr1+ cells. Cells associated with phagocytosis and innate immunity, including neutrophils, peripheral macrophages, and monocytes form a third group. Although mast cells and basophils are most closely associated with each other, their relatively low similarity, as measured by actual distance between populations on a phylogenetic tree, supports the long debated distinction between these populations (24). The division of neutrophilic and basophilic granulocytes is unexpected on the basis of their lineage relationship but possibly reflects their distinct roles in immune responses.
The dramatic remodeling of miRNA expression upon commitment to a specific lineage suggests that there is a stem cell signature of miRNA expression, as has been shown for mRNA expression (25–27). Comparison of populations that have varying purity of HSCs (45% in ES 150+, 35% in ES 150−, 30% in SLAM, 2% in LSK, <<2% in CMP/CLP/GMP/MEP) reveals a trend in which fewer miRNA are detected in the most enriched populations (Fig. 2). However, this trend may be in part due to increased purity of these populations or lower cell numbers for the most highly enriched samples. Similar trends are not observed in more differentiated cell types and, thus, we do not find clear evidence of a progressively increasing number of miRNA species expressed through differentiation.
The large divergence of miRNA expression patterns between stem cell/progenitor populations and all terminally differentiated cell types is difficult to reconcile with a single-step differentiation process. miRNAs are thought to have increased stability relative to mRNA, which would imply that their expression profiles may be more stable than transcriptional signatures. Thus, there likely exist many intermediate cellular states that have not been captured in this analysis. This feature reflects the different resolution of sorting strategies and the disproportionate amount of effort invested on the refinement of purification strategies for isolating progenitor and stem cell populations. Evidence of such transitions is found in erythroid differentiation (Fig. 4) and in the observation that several miRNAs found in stem cell and progenitor populations also persist in NK progenitors, at attenuated levels, but are not detected in any differentiated lymphoid populations. Time course studies of miRNA expression from in vitro differentiation assays, ideally with single-cell resolution, may allow for direct measurement of these transitions. New purification strategies would be needed to identify such transient populations in vivo.
Analysis of miRNA across 27 populations did not reveal any miRNA species that are specific to a single cell type. Instead, we find that many miRNAs display bi- and sometimes trimodal expression patterns that reflect their membership in larger categories. For example, several miRNA species, including miR-130a, miR-196b, let-7d, and miR-125b, are generally up-regulated in stem cell and progenitor populations, are not detected in any lymphoid cells, and are present in only a subset of myeloid cells. This result suggests that miRNA orchestrate hematopoiesis by modulating multiple signaling pathways that may be context-specific to that cell type. This multifunctional role has been best described for miR-155 in myeloid and lymphoid cells (28). Understanding the role of miRNA expression in differentiation will therefore require systems-level analysis rather than reductionist approaches to identify single driving miRNA species.
Our comparisons of sorted populations represent ensemble averages of thousands of single cells. The varying degree of heterogeneity of each purified cell population must therefore be considered when interpreting relative miRNA expression levels. Here, we have shown that single-cell measurements of purified cell populations reflect the functional heterogeneity of these populations. Notably, single-cell measurements of six highly expressed miRNA in GMPs, which are thought to be highly homogeneous, reveal remarkably low variability (<2-fold and consistent with the estimated technical variation of the method), suggesting miRNA levels are very tightly regulated within biologically homogeneous cell populations. This behavior may be contrasted to mRNA expression levels, which are subject to transcriptional bursts and large stochastic fluctuations (29). The tighter regulation of miRNA is understandable given their global role in posttranscriptional regulation, increased stability relative to mRNA, and high copy numbers. Single-cell miRNA profiling of sorted populations may therefore prove to be a novel and powerful approach for evaluating the purity of populations and for identifying new subclasses of cells that are not resolved by known surface markers, including cells present in malignant populations or other disease states.
Materials and Methods
Detailed protocols for FACS purification of cell populations, miRNA detection, and single-cell measurements are provided as SI Materials and Methods. Details of bioinformatics analysis and statistical testing are also provided as SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Bob Jones (Fluidigm Corporation) for providing devices and reagents used in this study.
Footnotes
Conflict of interest statement: C.L.H. is a member of the Scientific Advisory Board of Fluidigm Corporation and has a financial interest in this company.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009320107/-/DCSupplemental.
References
- 1.Ceredig R, Rolink AG, Brown G. Models of haematopoiesis: Seeing the wood for the trees. Nat Rev Immunol. 2009;9:293–300. doi: 10.1038/nri2525. [DOI] [PubMed] [Google Scholar]
- 2.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: Key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Odenike O, Rowley JD. Leukaemogenesis: More than mutant genes. Nat Rev Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillis AJ, et al. High-throughput microRNAome analysis in human germ cell tumours. J Pathol. 2007;213:319–328. doi: 10.1002/path.2230. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Gelfond JA, McManus LM, Shireman PK. Reproducibility of quantitative RT-PCR array in miRNA expression profiling and comparison with microarray analysis. BMC Genomics. 2009;10:407. doi: 10.1186/1471-2164-10-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller CW, et al. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27:1013–1023. doi: 10.1038/nbt.1585. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohta H, et al. Near-maximal expansions of hematopoietic stem cells in culture using NUP98-HOX fusions. Exp Hematol. 2007;35:817–830. doi: 10.1016/j.exphem.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuchenbauer F, et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stadler B, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan M, Miller CP, Papayannopoulou T, Stamatoyannopoulos G, Song CZ. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp Hematol. 2007;35:1015–1025. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han YC, et al. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popovic R, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 18.Kent DG, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009;113:6342–6350. doi: 10.1182/blood-2008-12-192054. [DOI] [PubMed] [Google Scholar]
- 19.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 21.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Viswanathan SR, et al. microRNA expression during trophectoderm specification. PLoS ONE. 2009;4:e6143. doi: 10.1371/journal.pone.0006143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113:4586–4594. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arinobu Y, Iwasaki H, Akashi K. Origin of basophils and mast cells. Allergol Int. 2009;58:21–28. doi: 10.2332/allergolint.08-RAI-0067. [DOI] [PubMed] [Google Scholar]
- 25.Kim YC, et al. The transcriptome of human CD34+ hematopoietic stem-progenitor cells. Proc Natl Acad Sci USA. 2009;106:8278–8283. doi: 10.1073/pnas.0903390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehtonen A, et al. Gene expression profiling during differentiation of human monocytes to macrophages or dendritic cells. J Leukoc Biol. 2007;82:710–720. doi: 10.1189/jlb.0307194. [DOI] [PubMed] [Google Scholar]
- 27.Shim MH, Hoover A, Blake N, Drachman JG, Reems JA. Gene expression profile of primary human CD34+CD38lo cells differentiating along the megakaryocyte lineage. Exp Hematol. 2004;32:638–648. doi: 10.1016/j.exphem.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Georgantas RW, 3rd, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci USA. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.