Abstract
Lateral segregation of cell membranes is accepted as a primary mechanism for cells to regulate a diversity of cellular functions. In this context, lipid rafts have been conceptualized as organizing principle of biological membranes where underlying cholesterol-mediated selective connectivity must exist even at the resting state. However, such a level of nanoscale compositional connectivity has been challenging to prove. Here we used single-molecule near-field scanning optical microscopy to visualize the nanolandscape of raft ganglioside GM1 after tightening by its ligand cholera toxin (CTxB) on intact cell membranes. We show that CTxB tightening of GM1 is sufficient to initiate a minimal raft coalescence unit, resulting in the formation of cholesterol-dependent GM1 nanodomains < 120 nm in size. This particular arrangement appeared independent of cell type and GM1 expression level on the membrane. Simultaneous dual color high-resolution images revealed that GPI anchored and certain transmembrane proteins were recruited to regions proximal (< 150 nm) to CTxB-GM1 nanodomains without physical intermixing. Together with in silico experiments, our high-resolution data conclusively demonstrate the existence of raft-based interconnectivity at the nanoscale. Such a linked state on resting cell membranes constitutes thus an obligatory step toward the hierarchical evolution of large-scale raft coalescence upon cell activation.
Keywords: cholera toxin, membrane heterogeneity, near-field scanning optical microscopy, raft ganglioside GM1, single-molecule detection
Advances in microscopy techniques applied to biology have led to the conviction that cell membranes are not merely a sea of lipids and proteins (1, 2). Instead, the membrane is a more complex system where individual components preorganize into spatially distinct compartments (3–5) and give rise to important strategic advantages for protein function and signaling (6). The organization of proteins and lipids into compartments, or the shielding of these assemblies from other proteins, predetermines specific interactions in the steady state. Important players in this compartmentalization are hypothesized to be lipid rafts, defined as transient nano-sized entities enriched in cholesterol, sphingolipids (such as the ganglioside GM1) and glycosylphosphatidylinositol anchored proteins (GPI-AP) (6–8). Activation and/or clustering of raft constituents will amplify and strengthen preexisting interactions and mediate signal transduction across the membrane (6). However, there is no consensus yet on the spatio-temporal regimes regulating lipid rafts in the resting state. This is mainly because their evaluation requires tools compatible with live cell imaging and capable of resolving heterogeneities at the nanoscale, a spatial regime not accessible to standard fluorescence microscopy. Nevertheless, recent advances in biophysical techniques have led to the identification of cholesterol-dependent nanoassemblies, supporting their existence in living cells (9–12).
A vast amount of research has convincingly demonstrated that lipid-lipid interactions induce two-phase coexistence in model lipid membranes and giant unilamellar vesicles prepared from cell membrane lipid extracts (13, 14). However, such phase coexistence has remained so far largely unresolved on biological membranes. Interestingly, recent studies show that the cell membrane in all its complexity is fully capable to phase segregate into a micrometer-sized two-phase fluid-fluid system, upon a temperature decrease (15), or through GM1 tightening by its ligand cholera toxin-β (CTxB) at physiological temperatures (16), provided that the membrane is separated from the influence of the cortical cytoskeleton. Based on these results, it has been proposed that an underlying selective connectivity mediated by cholesterol must exist among membrane rafts even at the resting state. This connectivity will thus be responsible for the large-scale phase segregation induced far beyond the valency of initial GM1 tightening through CTxB (16, 17). Yet, most of the experimental proof for such raft connectivity is based on the visualization of the end stage of an activated condition and in the absence of the cytoskeleton and/or membrane traffic, where the transient rafts are amplified to coalesce into larger, stable micrometer-sized raft domains. It is only at this stage that standard fluorescence microscopy is able to observe this segregation.
To visualize nanoscale cell membrane organization we can nowadays benefit from techniques that allow subdiffraction optical resolution (18, 19). Among these, near-field scanning optical microscopy (NSOM) takes advantage of the evanescent field exiting a subwavelength excitation source, being therefore particularly suited for nanoscale optical imaging on intact biological membranes (12, 19). The lateral resolution of the technique depends essentially on the subwavelength aperture size and is typically better than 80 nm. Recently, we have exploited NSOM to demonstrate the existence of preformed cholesterol-dependent nanoscale platforms composed by GPI-AP nanodomains and transmembrane proteins in the resting state (12). Here we sought to investigate whether GM1 tightening by CTxB is sufficient to initiate a minimal raft coalescence unit and to recruit raftophilic proteins to specific sites on fully intact cell membranes. High-resolution NSOM imaging revealed the formation of cholesterol-dependent CTxB-GM1 nanodomains on both monocytes and dendritic cells. Importantly, raftophilic proteins did not physically intermixed at the nanoscale with CTxB-GM1 nanodomains but converged within a characteristic distance. In silico experiments showed that this spatial scale corresponded to multicomponent connective regions on the cell membrane where raftophilic proteins probed the length scale of these raft-like regions. Collectively, our high-resolution data and in silico experiments demonstrate the existence of raft-based interconnectivity at the nanoscale and suggest that this specific organization constitutes the basis for large-scale raft coalescence upon cell activation.
Results
Association of Raftophilic Proteins and Raft Components by Secondary Antibody Crosslinking.
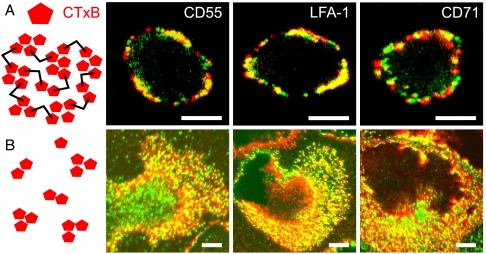
To investigate the partitioning of proteins in lipid rafts by means of standard confocal microscopy, membrane components are commonly crosslinked with antibodies (Ab). Colocalization between two membrane components after crosslinking is then taken as an indication for their a priori association; i.e., before crosslinking (20). Representative confocal images of fixed monocytes after secondary Ab crosslinking of GM1 bound CTxB are displayed in Fig. 1A for three different proteins investigated. As expected, the images show micrometer-sized fluorescent patches clearly resolvable by standard confocal microscopy. CD55 (a GPI-AP) and LFA-1 (a transmembrane protein from the integrin family) show significant overlap with the highly crosslinked GM1 lipids (Fig. 1A), whereas less overlap is observed for CD71 [a nonraft protein (10, 12)]. Indeed, the colocalization coefficient C renders values of: 0.65 ± 0.06 and 0.64 ± 0.07 for CD55 and LFA-1 respectively and 0.26 ± 0.08 for CD71, where C = 1 corresponds to full colocalization and C = 0 to uncorrelated distribution.
Fig. 1.
Secondary crosslinking enforces raft colocalization. (A) Representative confocal images of monocytes labeled with CD55, LFA-1, and CD71 (green) copatched with CTxB (red) crosslinked with secondary antibodies. (B) Representative confocal images of monocytes in the absence of secondary anti-CTxB crosslinking. Scale bars: 5 μm.
To reveal the initial stages of raft coalescence at the nanoscale without secondary Ab crosslinking, we next applied single-molecule-sensitive confocal microscopy where GM1 lipids were only locally tightened by the pentameric unit CTxB (Fig. 1B). The distinct micrometer-sized patch-like structures seen in presence of the secondary Ab crosslinking are now absent and instead, a high-packing density of all the components is observed. This, together with the limited lateral resolution, results in a large overlap of CTxB-GM1 fluorescence with any of the three proteins investigated, irrespectively of their putative (or not) association to rafts. Accordingly, the C values obtained from multiple images are 0.40 ± 0.01 and 0.47 ± 0.06 for CD55 and LFA-1 respectively, and 0.46 ± 0.15 for CD71. These results indicate that CTxB on its own is not sufficient to induce large-scale changes on the cell membrane as opposed to secondary Ab crosslinking. These images furthermore highlight the utter need for higher spatial resolution.
CTxB-Tightened GM1 Forms Nanodomains on Intact Cell Membranes.
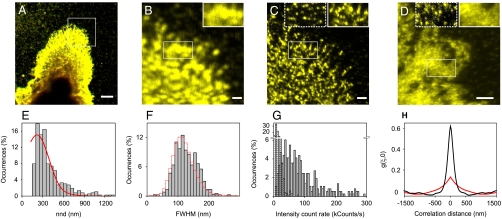
To map the nanolandscape of GM1 after CTxB tightening we performed extensive nanoscale imaging of CTxB-GM1 using a combined single-molecule confocal/NSOM setup working in aqueous conditions (12). Representative confocal and NSOM images of CTxB-GM1 on the cell membrane of fixed monocytes stretched on poly-L-lysine (PLL) coated coverslips are shown in Fig. 2 A–C. The diffraction-limited resolution (∼300 nm) and sensitivity of our confocal setup already reveals heterogeneities of CTxB bound to GM1 on the cell membrane (Fig. 2B) that become strikingly distinguishable when imaging the same area with NSOM (Fig. 2C). The marked increase in resolution of NSOM (∼100 nm in this case) reveals numerous well-defined CTxB-GM1 spots decorating the cell surface.
Fig. 2.
GM1 forms nanodomains upon CTxB tightening. (A) Representative confocal image of CTxB-GM1 at the cell surface of a fixed monocyte. Scale bar: 5 μm. (B) Confocal zoom-in of the area highlighted in A. (C) Counterpart NSOM image. (D) Representative NSOM image of CTxB-GM1 after MβCD treatment. Scale bars: 1 μm. Insets on the most right sides in B and C highlight the increase in resolution afforded by NSOM. Insets on the most left sides in C and D highlight single CTxB molecules on the glass substrates demonstrating equal sensitivity and NSOM resolution on untreated and MβCD treated samples (see also Fig. S3). (E) Histogram of the nnd distribution of CTxB-GM1 spots (bars) and simulations of random organization (red). (F) Size distribution of individual CTxB-GM1 spots (bars) and of individual CTxB on the glass (red) obtained with the same NSOM probe. (G) Intensity count rate distribution of CTxB-GM1 over multiple cells (gray) and of single CTxBs nonspecifically attached to the glass (dash). (H) Autocorrelation functions of a representative MβCD treated sample (red) compared with that of an untreated sample (black).
To investigate the spatial distribution pattern exhibited by CTxB-GM1 spots we performed nearest neighbor distribution (nnd) analysis. The resulting nnd was further compared with simulations of random organization (Fig. 2E) using experimental spot densities (see Materials and Methods). The large overlap between the experimental and simulated random distribution demonstrates that the CTxB-GM1 spots are randomly scattered on the cell membrane with a characteristic interdomain separation of ∼300 nm. This random distribution is markedly different to the nanoscale-biased distribution of typical raft-associated proteins such as GPI-APs that exhibit a hierarchical organization in resting conditions (12, 21). In addition, the average density of CTxB-GM1 spots from multiple NSOM images on different cells rendered a value of (4.8 ± 0.7) spots/μm2.
We further determined the size distribution of all individual CTxB-GM1 spots by measuring their full-width at half maximum (FWHM). The distribution peaks at 123 (σ = 37) nm (Fig. 2F). This value is close to the NSOM probe’s aperture indicating that the real size of the CTxB-GM1 spots lies below this value. Because the size of membrane rafts resides in the range of 10–200 nm (8), our results suggest that local CTxB tightening of GM1 lipids induces a minimal raft coalescence unit.
To quantify the extent of CTxB tightening, we measured the fluorescence intensity of individual CTxB-GM1 spots on the cell membrane and compared it to that of scarcely scattered single CTxB molecules on the glass substrate (Fig. 2G and Fig. S1A). As intensity is proportional to the number of molecules, the large number of occurrences at high intensity values on the cell surface compared to single CTxB molecules on glass indicates that multiple molecules are present in each spot evidencing the organization of CTxB-GM1 in nanodomains. To provide further proof for nanodomain formation we compared the experimental data with simulations to estimate the random probability of “apparent” clustering taking into account the NSOM probe size, the experimental CTxB density and the labeling variability of each CTxB (See Fig. S1 B and C and SI Text). At the given experimental conditions, simulated and experimental distributions deviate from each other significantly, confirming that the CTxB-GM1 nanodomains observed are indeed real. Although we could not assess the exact position of each single CTxB inside its corresponding fluorescent spot, we can estimate its molecular proximity within the nanodomains by calculating the average intermolecular distance r using the relation r ∼ 1/(2ρ1/2)) under the assumption of random distribution. In here, ρ is the molecular density of each spot given by its physical size (Fig. 2F) and the number of CTxB-GM1 per spot (Fig. 2G). Under our experimental conditions r < 25 nm (because the spot size < 120 nm), a value considerably smaller than the 300 nm measured for the nnd between CTxB-GM1 nanodomains (Fig. 2E). Collectively, these data confirm the local enrichment of GM1 lipids in nanodomains of characteristic size below 120 nm and demonstrate that “phase segregation” is induced only slightly beyond the valency of initial GM1 tightening through CTxB.
CTxB-GM1 Nanodomain Formation Is Independent on Cell Type and GM1 Expression Levels.
To assess whether this CTxB-GM1 type of arrangement is common to cells other than monocytes, we also quantified brightness and size of CTxB-GM1 spots on antigen-presenting dendritic cells, which exhibit ∼30-fold lower GM1 expression (Fig. S2 A and B). Remarkably, our results confirm the persistence of small CTxB-GM1 nanodomains containing in this case ∼2 CTxB molecules per spot and with larger nnd values resulting from the lower GM1 expression on these cells (Fig. S2 C–F). This confirms that CTxB induces GM1 segregation into local nanoenvironments independent of cell type or GM1 expression level. These data are also in line with a recent model that favors the view of membrane rafts having an upper size limit, with the number of rafts increasing upon GM1 content (22).
CTxB-Tightened GM1 Nanodomains Require Cholesterol for Their Integrity.
Cholesterol has been largely recognized as a key player in raft formation and interconnectivity (6, 16). To enquire whether the formed CTxB-GM1 nanodomains are cholesterol dependent, we performed CTxB induced GM1 tightening after cholesterol depletion by means of methyl-β-cyclodextrin (MβCD). NSOM images on MβCD treated samples showed more irregular fluorescence as compared to the clear punctuated pattern observed on untreated samples (Fig. 2D). However, as also reported by others, our NSOM images did not show full cluster disruption (23). In fact, some larger patches and local heterogeneities were still present on the membrane above a continuous fluorescence level (Fig. S3). We note that NSOM imaging on MβCD treated samples was performed on smaller areas and with lower scanning speeds to avoid sample perturbation while guaranteeing that image resolution and sensitivity are not compromised (Fig. S3 and SI Text). Furthermore, single dye tracking experiments on fixed cells after cholesterol extraction confirmed complete fixation of CTxB-GM1 molecules (Fig. S4).
To quantitatively assess the effect of cholesterol depletion we applied image correlation analysis (24). The normalized two-dimensional correlation was calculated and fitted with a two-dimensional Gaussian function of the form: g = g(0,0) exp(r2/w2) where r is the shift from the image center (0,0) and w is the characteristic width of the Gaussian function at 1/e of its maximum. In here, g(0,0) is directly proportional to the degree of clustering and w is a measure for the size of the clusters. The resulting values of g(0,0) decrease from 0.65 ± 0.11 for untreated cells to 0.15 ± 0.03 upon MβCD treatment (Fig. 2H). Moreover, the width of the autocorrelation functions becomes broader on the MβCD treated samples (w = 700 ± 300 nm vs. w = 210 ± 30 nm on untreated samples), demonstrating that cholesterol depletion significantly disturbs the local nanometer-scale ordering of CTxB-GM1 on the cell membrane. Taken together, the punctuated local order at the nanoscale and its cholesterol dependence demonstrate that CTxB tightening of GM1 lipids creates raft-based nanoplatforms on intact cell membranes.
CTxB-GM1 Nanodomains Recruit Raftophilic Proteins Without Physical Intermixing.
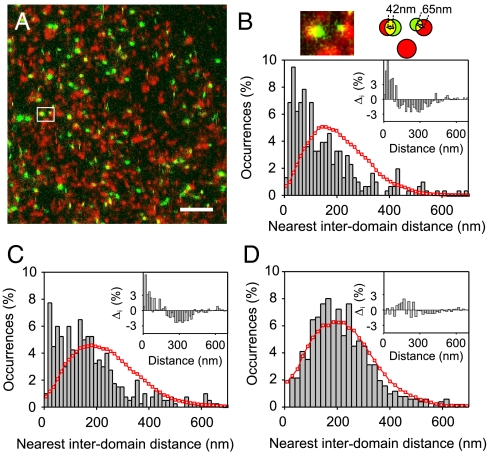
The individual CTxB-GM1 nanodomains do not seem to preassemble into higher order structures as evidenced by their random distribution on the cell surface (Fig. 2E). This raises the question of how other raft-like components are spatially distributed with respect to CTxB-GM1 nanodomains. A representative dual color NSOM image of CD55 and CTxB-GM1 is shown in Fig. 3A. Surprisingly, and in contrast with the artificially induced copatching data shown in Fig. 1A, no spatial overlap between these two raft constituents was observed at the nanoscale (C = 0.17 ± 0.03). Similarly, we did not observe colocalization of LFA-1 and CTxB-GM1 nanodomains (Fig. S5A, with C = 0.20 ± 0.08) entirely consistent with our previous results where no colocalization between LFA-1 and GPI-APs was observed in the resting state (12). These results indicate that CTxB tightening of GM1 is not sufficient to drive the coalescence of LFA-1 and GM1 into supramolecular assemblies. Similar lack of colocalization was observed between the nonraft marker CD71 and CTxB-GM1 nanodomains (Fig. S5B, with C = 0.11 ± 0.07). As a positive control for true colocalization at the nanoscale, we imaged doubly labeled CD71 using primary and secondary antibodies (Fig. S6). As expected, a clear colocalization value (C = 0.75 ± 0.07) is obtained. Furthermore, to exclude the possibility that this lack of nanoscale colocalization might result from protein or CTxB-GM1 nanodomain diffusion during and after cell fixation, we performed fluorescence recovery after photobleaching and single particle tracking experiments (see SI Text). Importantly, these two controls confirmed complete fixation of CTxB-GM1 at the nanoscale (Fig. S4).
Fig. 3.
Raftophilic proteins reside proximal to CTxB-GM1 nanodomains. (A) Dual color excitation/detection NSOM image of CD55 (green) and CTxB-GM1 nanodomains (red). Scale bar: 1 μm. (B–D) i-nnd distributions of (B) CD55, (C) LFA-1, (D) CD71 to its closest CTxB-GM1 nanodomain (bars) together with simulations of random spatial distribution of proteins and CTxB-GM1 nanodomains (red). The insets correspond to the difference between experimental data and simulations. Experimental and simulated random distributions are significantly different for B and C with P = 0.019 and P = 0.015 respectively, and similar for D with P = 0.41.
To uncover any potential spatial correlation between the proteins under study and the CTxB-GM1 nanodomains, we performed interdomain nearest neighbor distribution analysis (i-nnd) on the dual color NSOM images with localization accuracies on the determination of the (x, y) positions ∼2–5 nm (see Materials and Methods). Remarkably, the i-nnd for both CD55 and LFA-1 with respect to CTxB-GM1 nanodomains showed a clear deviation toward shorter distances compared to random organization of these receptors with respect to CTxB-GM1 nanodomains (Fig. 3 B and C). Indeed, the insets in Fig. 3 B and C, showed an increasing number of positive residuals at distances < 150 nm. A 35–50% of both LFA-1 and CD55 receptors respectively were found within a < 50–100 nm neighborhood of CTxB-GM1 nanodomains. These results thus reveal a preferential spatial proximity of raftophilic proteins to the CTxB-GM1 nanodomains, but without physical intermixing. On the other hand, the experimental i-nnd of CD71 (Fig. 3D) overlapped well with that of random organization of the receptor with respect to GM1, entirely consistent with its classification as nonraft marker (10, 12).
In Silico Experiments Support the Existence of Raft Interconnectivity at the Nanoscale.
The biological functions of both LFA-1 and CD55 are dependent on intact membrane rafts because cholesterol depletion severely compromises the function of both receptors (12, 25). This raises the possibility that the observed nanoscale proximity between raftophilic proteins and the CTxB-tightened GM1 nanodomains is likely caused by an underlying raft-connectivity. In this case, the proximal distances extracted from the NSOM images should then correspond to the spatial length scale of interdomain connectivity. Because recent reports support the idea that cholesterol plays a key organizational role of such raft-based connectivity (16, 26), in our case extraction of cholesterol will likely result in the disappearance of the observed proximity. Unfortunately, cholesterol depletion also perturbs the local tightening effect of CTxB on GM1 (Fig. 2H). In these conditions, the point pattern analysis required to derive the i-nnd distributions cannot be performed and the interdomain distances cannot be evaluated. Therefore, to test our hypothesis and to gain deeper insight on the spatial scale of raft connectivity on fully intact cells we turned to in silico experiments.
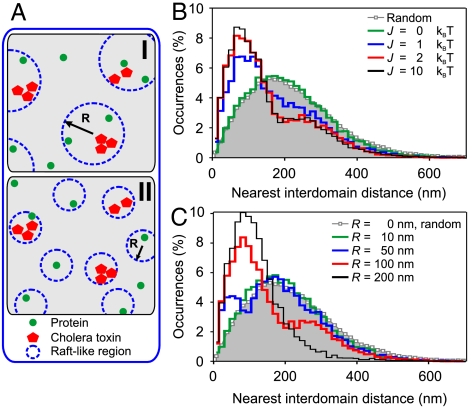
The simulations relied on the representation of the cell membrane as a two-dimensional square lattice system (see SI Text). The model assumed (i) the existence of raft-like regions with size R randomly distributed on the lattice; (ii) 25% of the total area is covered by raft-like lipids, according to reported estimations from Prior et al. (3); and (iii) CTxB-GM1 nanodomains always reside inside raft-like regions. In addition, the square lattice was seeded with a number of proteins that diffuse until equilibration following a standard Monte Carlo (MC) algorithm (see SI Text). Protein affinity for a raft-like region was quantified by the interaction parameter J (in kBT units). Fig. 4A illustrates two examples for different values of R. Running the MC simulations rendered an image comparable to the experimental ones (see SI Text and Fig. S7A). Subsequently, the i-nnd distances were extracted from the images in similar fashion as for the experimental data. It is important to clarify that our model is simply based on the assumption that there are raft-like regions of size R on the cell membrane enriched with CTxB-GM1 and that proteins have an affinity, given by J, to partition (or not) into these regions. Thus, the model implicitly assumes the existence of connective regions on the membrane kept by cholesterol.
Fig. 4.
Coarse-grained MC simulations. (A) Schematic representation of the end-state of MC simulations for two raft-like regions of different size R. (B) Resulting i-nnd distributions for R = 100 nm and different interaction values J. (C) Resulting i-nnd distributions at J = 2 and for different sizes of the raft-like regions.
We first tested the model by considering raft-like regions of R = 100 nm and assuming no specific affinity for the protein to partition in these regions; i.e., J = 0. As expected, the i-nnd followed a Poissonian distribution (Fig. 4B, J = 0 and Fig. S7D), in close resemblance to the CD71:GM1 experimental data (Fig. 3D). By increasing J, we approached the case of raftophilic proteins. This caused the i-nnd to develop into a bimodal-like distribution; i.e., the appearance of a subpopulation shifted at shorter distances corresponding to protein-lipid pairs inside the same raft regions, and a concomitant decrease in the distances at larger values (Fig. 4B and Fig. S7 B and C). These distributions are qualitatively similar to those experimentally obtained for CD55:GM1 and LFA-1:GM1 (Fig. 3 B and C). Importantly, increasing J above a certain threshold value did not lead to an increase on the subpopulation of proximal occurrences, neither shifting the characteristic distances involved, as similar histograms were obtained for J = 2 and 10 (Fig. 4B). These results indicate that once protein affinity for a raft-like region is established by a given interaction value J, partitioning of the proteins to these regions evolves without energy cost to the system.
We further investigated how the i-nnd depends on R. R = 0 corresponds to the situation where there is no cholesterol-maintained connectivity on the membrane. In this case the resulting i-nnd distribution showed random patterns between CTxB-GM1 nanodomains and the protein of interest (Fig. 4C). For R ≤ 10 nm the regions are exceedingly small so that the probability of both proteins and CTxB-GM1 nanodomains to reside in a given region is very low and the i-nnd resembled again the random case (Fig. 4C). On the other extreme, for R > 200 nm, the regions contained multiple CTxB-GM1 nanodomains and proteins. In this case, the distribution also resembled the random case but fully shifted to shorter distances. Remarkably, only a certain range of sizes R = 100–120 nm resulted in bimodal distributions qualitatively similar to the experimental histograms (Fig. 4C and Fig. S7 B and D) underscoring the importance of a critical spatial length scale for the raft-like regions.
The nanoscale proximity between raftophilic proteins and the stabilized CTxB-GM1 nanodomains observed experimentally can thus be understood in terms of an interdomain connectivity within the raft-like regions where the protein probes the length scale of this connective region. Indeed, fitting the short-distance distribution of the experimental data for CD55:CTxB-GM1 to a Gaussian renders the characteristic size of this region, with a peak at 53 (σ = 42) nm. This value is remarkably similar to the size range of the raft-like regions explored in our simulations (Fig. S7 B and C), indicating that the experimental interdomain distances directly map the extent of the nanoscale compositional connectivity on the cell membrane. In the case of LFA-1:CTxB-GM1 we did not attempt to fit the shorter i-nnd values because the distribution is rather broad most probably reflecting the lower affinity of LFA-1 for rafts as compared to the GPI-AP CD55 (12).
Discussion
Recently Lingwood et al. (16) elegantly demonstrated that cell membranes at physiological temperatures contain all the ingredients to segregate into two distinct micrometer-sized phases provided that the membrane is separated from the influence of cortical actin and in the absence of membrane traffic. The workers thus validated a concept in which global domain coalescence on biological membranes is primarily based on the organization principle that these clustered components possess a predefined level of cholesterol-based connectivity in the resting state. Because biological membranes are also crowded with proteins, including transmembrane ones, it has been further hypothesized that proteins will, via chemical and/or physical specificity, increase this lipid-based connectivity (26).
Nevertheless, one might ask whether such cholesterol-based interdomain connectivity exists in biological membranes in the presence of the cytoskeleton. The challenge is to observe this phase segregation in intact cells because cytoskeletal constraints will impose an upper limit in the nanometer range. By exploiting superresolution NSOM, we resolved individual CTxB-tightened GM1 nanodomains on fully intact cell membranes. These nanodomains represented the minimum coalescent unit and depended on cholesterol for their integrity. Furthermore, they recruited both certain transmembrane and lipid anchored proteins without physical intermixing, but not the transferring receptor CD71, a classical nonraft marker, thus demonstrating a raft-based compositional selectivity in the presence of an intact cytoskeleton, where the capacity to phase separate is more tightly controlled (Fig. 5). Importantly, in silico experiments substantiated our experimental observations and underscored the relevance of a critical spatial scale for nanoscale compositional connectivity on the cell membrane.
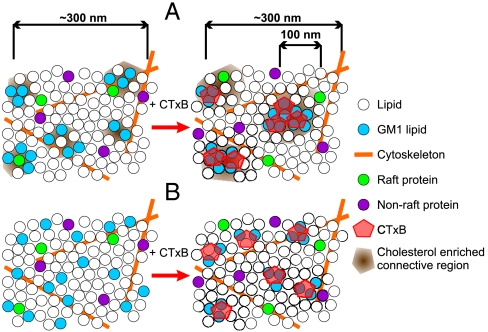
Fig. 5.
Schematic representation of underlying nanoscale connectivity on fully intact cell membranes. (A) Distribution of raft-like and nonraft-like lipids and proteins in the unperturbed state (Left), where raft-like entities have already a cholesterol-dependent affinity for each other (12). Upon GM1 tightening by CTxB, a connective region of ∼100 nm is formed, with raft-like entities being selectively recruited (Right). (B) In the absence of interconnectivity; i.e., through cholesterol depletion, there is no selective nm-scale proximity in the resting state and raft coalescence does not take place.
Interestingly, whereas for some integrins posttranslational modification mediating association with rafts has been determined (27), no clear acylation motives are recognizable in the cytoplasmic or transmembrane regions of LFA-1 (28). These results appear to indicate that the inclusion of LFA-1 as transmembrane protein in the lipid-cholesterol connective regions is more dominated by physical interactions rather than by chemical affinities. Incidentally, LFA-1 interacts with cortical actin in its extended form (29) and actin requirement has also been implicated in cholesterol-based nanoheterogeneity (21) suggesting that the propensity of LFA-1 to interact with the cytoskeleton and its affinity for membrane rafts are interrelated.
Although CTxB labeling was performed at 4 °C, an absolutely necessary step to prevent endocytosis, our results indicate that temperature is not the determining factor inducing the formation of CTxB-GM1 nanodomains, playing, if at all, a minor role on biasing membrane ordering. First, CTxB-GM1 nanodomain formation is dependent on cholesterol. Indeed, cholesterol depletion under identical sample preparation conditions significantly affected the local ordering of CTxB-GM1 on the membrane, demonstrating that CTxB-GM1 nanoclustering depends on cholesterol integrity of the cell membrane and it is not the result of sample preparation. Second, the raft-like regions as observed in this work are structurally equivalent to those naturally occurring on intact cell membranes labeled after fixing at 37 °C (12). In fact, we recently showed that GPI-APs labeled at 37 °C exhibited a similar spatial proximity to other raftophilic proteins (12), indicating that the proximity effect is a connective property of cell membranes and not induced by temperature. Third, the immobilization of CTxB-GM1 at 4 °C and after fixation at 4 °C excludes the possibility that ordered CTxB-GM1 nanoenvironments will diffuse to associate with other lipid clusters leading to the formation of artificial and/or larger clusters.
The ability of cholesterol to promote a more ordered lipid phase is generally accepted as the thermodynamic driving force for raft organization. Recent observations suggest that dynamic, small-scale raft domains may correspond to composition heterogeneities when the lipid mixture is in the one-phase (mixed) region but close to the transition boundary (30). Within this scenario, small perturbations of the cell state due to out-of-equilibrium inputs involving little energy cost may drive the membrane mixture to strong changes in its phase stability, even promoting the appearance of micrometer-sized raft-like domains. As a first, yet fundamental, step to understand more complex patterning events/functions, our Monte Carlo approach focuses on a simplified equilibrium description of the membrane. More complicated modeling schemes that include nonequilibrium processes, and realistic kinetics can eventually account for the aforementioned dynamical behavior in full. However, according to our results, such level of detail seems unnecessary to grasp the essence underlying the experimental observations reported here.
In summary, we have demonstrated cholesterol-dependent local enrichment of GM1 lipids at the nanoscale after CTxB binding. These nanoscale assemblies recruited GPI-anchored and certain transmembrane proteins without intermixing of the individual raft components. As such, our data is entirely consistent with the existence of raft-based interconnectivity at the nanoscale. Such a connective condition on resting cell membranes appears thus as a pre-requisite for the global coalescence of rafts upon cell activation.
Materials and Methods
Cell Culture.
The monocytic THP-1 cell line was cultured in RPMI medium 1640 Dutch modification medium supplemented with 10% FCSand antibioticantimycotic from Gibco. Immature dendritic cells (DC) were obtained from monocytes isolated from buffy coats of healthy individuals. Prior to antibody staining, cells were allowed to adhere to PLL coated coverslips to allow extensive cell stretching.
Antibody Staining.
Cells were labeled with ice-cold 10 μg/mL CTxB for 30 min on ice to prevent CTxB internalization. LFA-1 was labeled with 10 μg/mL mAb TS2/4 (neutral monoclonal Ab). CD55 was labeled with the anti-CD55 mAb and CD71 with the anti-CD71 mAb (10 μg/mL). Details on cell labeling procedures and fixation conditions are further described in SI Text.
Cholesterol Depletion.
To deplete the cell from cholesterol the cells were treated using MβCD (10 mM at 37 °C). Methodology is described in more detail in SI Text.
Near-Field Scanning Optical Microscopy.
Experiments were performed on a custom-built confocal/NSOM microscope optimized for dual color single-molecule fluorescence in aqueous conditions (12). Homemade NSOM probes had apertures of 80–100 nm in diameter. Fixed cells were first imaged in confocal mode. A selected area of the cell surface was further inspected by confocal and then the excitation light was focused onto the back-end of the NSOM probe. Only well-stretched areas of the cell membrane were inspected by NSOM to exclude potential height artifacts on the fluorescence image. Scanning speeds and imaged areas were adjusted to minimize tip interaction with the samples under study (2 μm/s–6 μm/s and areas ranging from (2.5 × 2.5) to (8 × 8) μm2. Typically 3–10 images from different cells were taken at each experimental condition.
Image Analysis.
Unprocessed images were analyzed using custom-made software that determines spot size, brightness and (x, y) position. Spot sizes were determined using the FWHM of a 2D Gaussian fit to the intensity profile. Brightness of each spot (kcounts/s) was defined as the background-corrected average of the intensity over all pixels within the FWHM. The (x, y) coordinates of each individual spot were determined using the center of mass of the FWHM with a localization accuracy σ ∼ FWHM/(N)1/2 where N is the number of counts in each spot after background subtraction (31). For the samples under study σ ∼ 2.4 nm (for CTxB-GM1: FWHM = 123 nm, N = 2588); σ ∼ 2.1 nm (for CD71: FWHM = 82 nm, N = 1563); σ ∼ 3.6 nm (for LFA-1: FWHM = 85 nm, N = 562) and σ ∼ 4.7 nm (for CD55: FWHM = 80 nm, N = 286). The i-nnd distributions between the protein of interest and the CTxB-GM1 spots were determined from the (x, y) coordinates of each protein spot to the (x, y) coordinates of the nearest CTxB-GM1 spot. Simulations of random i-nnd distributions were performed using experimental spot densities as derived from multiple images and placing both protein and CTxB-GM1 spots at random (x, y) positions with respect to each other. Experimental spot densities for each experimental and simulated condition are given in SI Text.
Supplementary Material
Acknowledgments.
We thank B. Joosten for technical assistance, and C. G. Figdor and F. Sagués for useful discussions. This work was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) Veni Grant 916.66.028 and the Human Frontier Science Program (A.C.), EC-Research Training Network IMMUNANOMAP and EC-New and Emerging Science and Technology-BIO-LIGHT-TOUCH (M.F.G.-P., T.S.v.Z., C.M.), Spanish Ministry of Science and Technology Grants MAT2007-66629-C02-01 (M.F.G.-P.), FIS2009-11104 (J.B.), and FIS2006-03525 (R.R., J.G.), and Generalitat de Catalunya Grants 2009 SGR 597 (M.F.G.-P.) and 2009 SGR 01055 (to R.R., J.G., J.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003876107/-/DCSupplemental.
References
- 1.Vereb G, et al. Dynamic, yet structured: The cell membrane three decades after the Singer–Nicolson model. Proc Natl Acad Sci USA. 2003;100:8053–8058. doi: 10.1073/pnas.1332550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 3.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma P, et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 5.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein–protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 7.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 8.Pike LJ. Rafts redefined: a report on the Keystone symposium on lipid rafts and cell function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 10.Lasserre R, et al. Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat Chem Biol. 2008;4:538–547. doi: 10.1038/nchembio.103. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki KGN, et al. GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: Single-molecule tracking study 1. J Cell Biol. 2007;177:717–730. doi: 10.1083/jcb.200609174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Zanten TS, et al. Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc Natl Acad Sci USA. 2009;106:18557–18562. doi: 10.1073/pnas.0905217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown RE. Sphingolipid organization in biomembranes: What physical studies of model membranes reveal. J Cell Sci. 1998;111:1–9. doi: 10.1242/jcs.111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich C, et al. Lipid rafts reconstituted in model membranes. Biophys J. 2001;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumgart T, et al. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lingwood D, Ries J, Schwille P, Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc Natl Acad Sci USA. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond AT, et al. Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc Natl Acad Sci USA. 2005;102:6320–6325. doi: 10.1073/pnas.0405654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lippincott-Schwartz J, Manley S. Putting super-resolution fluorescence microscopy to work. Nat Methods. 2009;6:21–23. doi: 10.1038/nmeth.f.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Zanten TS, Cambi A, Garcia-Parajo MF. A nanometer scale optical view on the compartmentalization of cell membranes. BBA-Biomembranes. 2010;1798:777–787. doi: 10.1016/j.bbamem.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goswami D, et al. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 2008;135:1085–1097. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao M, Mukherjee S, Maxfield FR. Cholesterol depletion induces large scale domain segregation in living cell membranes. Proc Natl Acad Sci USA. 2001;98:13072–13077. doi: 10.1073/pnas.231377398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen NO, Hoddelius PL, Wiseman PW, Seger O, Magnusson KE. Quantitation of membrane-receptor distributions by image correlation spectroscopy—Concept and application. Biophys J. 1993;65:1135–1146. doi: 10.1016/S0006-3495(93)81173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart AD, Eustace HE, McKee TA, Brown TDK. A novel cell entry pathway for a DAF-using human enterovirus is dependent on lipid rafts. J Virol. 2002;76:9307–9322. doi: 10.1128/JVI.76.18.9307-9322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 27.Gagnoux-Palacios L, et al. Compartmentalization of integrin alpha6 beta4 signaling in lipid rafts. J Cell Biol. 2003;162:1189–1196. doi: 10.1083/jcb.200305006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calderwood DA. Integrin activation. J Cell Sci. 2004;117:657–666. doi: 10.1242/jcs.01014. [DOI] [PubMed] [Google Scholar]
- 29.Dustin ML, Bivona TG, Philips MR. Membranes as messengers in T cell adhesion signaling. Nat Immunol. 2004;5:363–372. doi: 10.1038/ni1057. [DOI] [PubMed] [Google Scholar]
- 30.Veatch SL, Soubias O, Keller SL, Gawrisch K. Critical fluctuations in domain-forming lipid mixtures. Proc Natl Acad Sci USA. 2007;104:17650–17655. doi: 10.1073/pnas.0703513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson RE, et al. Precise nanometer localization analysis for individual fluorescence probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.