Abstract
The epithelial-to-mesenchymal transition (EMT) produces cancer cells that are invasive, migratory, and exhibit stem cell characteristics, hallmarks of cells that have the potential to generate metastases. Inducers of the EMT include several transcription factors (TFs), such as Goosecoid, Snail, and Twist, as well as the secreted TGF-β1. Each of these factors is capable, on its own, of inducing an EMT in the human mammary epithelial (HMLE) cell line. However, the interactions between these regulators are poorly understood. Overexpression of each of the above EMT inducers up-regulates a subset of other EMT-inducing TFs, with Twist, Zeb1, Zeb2, TGF-β1, and FOXC2 being commonly induced. Up-regulation of Slug and FOXC2 by either Snail or Twist does not depend on TGF-β1 signaling. Gene expression signatures (GESs) derived by overexpressing EMT-inducing TFs reveal that the Twist GES and Snail GES are the most similar, although the Goosecoid GES is the least similar to the others. An EMT core signature was derived from the changes in gene expression shared by up-regulation of Gsc, Snail, Twist, and TGF-β1 and by down-regulation of E-cadherin, loss of which can also trigger an EMT in certain cell types. The EMT core signature associates closely with the claudin-low and metaplastic breast cancer subtypes and correlates negatively with pathological complete response. Additionally, the expression level of FOXC1, another EMT inducer, correlates strongly with poor survival of breast cancer patients.
Keywords: cancer stem cells, Twist, Snail, FOXC1
The epithelial-to-mesenchymal transition (EMT) is a process in which adherent epithelial cells shed their epithelial characteristics and acquire, in their stead, mesenchymal properties, including fibroblastoid morphology, characteristic gene-expression changes, increased potential for motility, and in the case of cancer cells, increased invasion, metastasis, and resistance to chemotherapy (1, 2). Recent studies have linked EMTs with both metastatic progression of cancer (3–5) and acquisition of stem-cell characteristics (6, 7), leading to the hypothesis that cancer cells that undergo an EMT are capable of metastasizing through their acquired invasiveness and, following dissemination, through their acquired self-renewal potential, which enables them to spawn the large cell populations that constitute macroscopic metastases.
EMTs can be induced in vitro by exposing certain normal and neoplastic epithelial cells to various growth factors, including TGF-β1, hepatocyte growth factor, and PDGF (1, 8). Downstream of each of these growth factors and their cognate receptors lies an array of transcription factors (TFs), each of which is capable, on its own, of inducing an EMT. These TFs include the homeobox protein Goosecoid (Gsc) (9), the zinc-finger proteins Snai1 (Snail) and Snai2 (Slug) (10–12), the basic helix-loop-helix protein Twist1 (Twist) (3), the forkhead box proteins FOXC1 (8, 13) and FOXC2 (14), and the zinc-finger, E-box-binding proteins Zeb1 and Sip1 (Zeb2) (8, 15).
In addition to TFs, members of the miR-200 family of microRNAs are down-regulated during an EMT (16). This down-regulation results, in turn, in the up-regulated expression of several critical target genes, notably Zeb1 and Zeb2. Expression of any one of these TFs or down-regulation of the miR-200 family in an appropriate epithelial cell suffices to induce an EMT (17, 18). Moreover, many of these TFs are expressed concomitantly in the mesenchymal cells that have passed through an EMT. The overlapping and unique contributions of each inducer to the EMT program have not been adequately explored.
Recent microarray analyses have allowed stratification of clinical breast cancers into a large number of distinct subtypes, such as luminal, basal-like, and HER2+ (19–21). Yet, other subtypes of tumors have recently been delineated by us and others (22, 23). These distinctions have proven to be useful in predicting responses to therapy, time to metastasis, and survival.
In the present study, we assayed gene expression signatures (GESs) in human mammary epithelial cells (HMLE) induced to undergo an EMT by expressing Gsc, Snail, Twist, or TGF-β1 or by knocking down expression of E-cadherin (24), and found that EMTs induced by these methods induce an overlapping set of changes in gene expression, which we term the “EMT core signature.” We compared the EMT core signature with signatures that define breast cancer subtypes and found a close association with the claudin-low and metaplastic breast cancer subtypes.
Results
Interaction of EMT-Related TFs as a Regulatory Network.
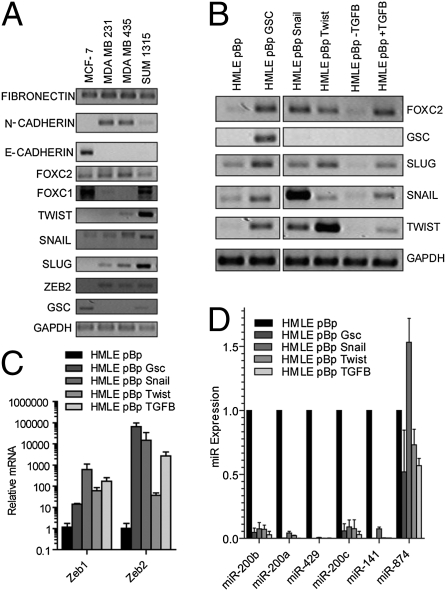
To elucidate the network of interactions between EMT regulatory factors, we first assessed the expression of known EMT inducers and the genes known to be regulated during EMT in various breast cancer cell lines. To do so, we assayed four breast cancer cell lines for expression of the following TFs known to promote EMTs: Gsc (9), FOXC1 (8, 13), FOXC2 (14), Zeb1, Zeb2 (25, 26), Slug (10, 27), Snail (11, 28), and Twist (3), as well as other genes associated with either the mesenchymal state [N-cadherin (29) and fibronectin (30)] or the epithelial state [E-cadherin (31)]. Relative to the luminal epithelial cell line, MCF-7, the MDA-MB 231, MDA-MB 435, and SUM 1315 basal B cell lines consistently expressed higher levels of Snail, Slug, and Zeb2, but not Gsc or FOXC1 (Fig. 1A).
Fig. 1.
Expression of EMT marker genes in breast cancer cell lines and nontransformed EMT-induced human mammary epithelial cells. (A) Expression of EMT marker genes was measured by semiquantitative RT-PCR performed on RNA extracted from MCF-7, MDA-MB 231, MDA-MB 435, and SUM1315 cells. (B–D) HMLE cells were transduced with a retrovirus overexpressing the indicated gene and expression of the indicated gene was measured by semiquantitative RT-PCR (B) or quantitative PCR (C and D). GAPDH (C) or U6 snoRNA (D) was amplified for normalization. Error bars represent the SD from three independent experiments.
To further understand the interactions among EMT-inducing TFs, we tested how overexpression of a single EMT-inducing TF affects the expression of other TFs in this network, thereby establishing an “EMT interactome.” To do so, we used immortalized HMLE cells that express high levels of E-cadherin and low levels of vimentin, similar to the MCF-7 breast cancer cell line and contrasting with the more mesenchymal MDA-MB 231 and SUM1315 breast cancer cell lines (Fig. S1). HMLE cells were induced to undergo EMTs through overexpression of Gsc, Snail, Twist, or TGF-β1. We then used RT-PCR to confirm the expression levels of these genes, as well as various genes known to be associated with EMT programs (Fig. 1B and Fig. S2).
Among the EMT-inducing TFs, expression of FOXC2, Slug, Zeb1, and Zeb2 was consistently elevated in response to overexpression of Gsc, Snail, Twist, or TGF-β1, suggesting that these four genes operate downstream of Gsc, Snail, Twist, and TGF-β1 in the EMT interactome (Fig. 1 B and C). In contrast, Gsc was not up-regulated by overexpression of the other EMT inducers, suggesting an independent mode of transcriptional regulation (Fig. 1B).
Recent findings have demonstrated that EMT programs can also be induced by inhibiting members of the miR-200 family of microRNAs (17, 18, 32). We wished to determine the location of these miRNAs in the EMT interactome. The miR-200 family and Zeb transcription factors form a mutually inhibitory, double negative-feedback loop (16–18, 33). Consistent with a role for Zeb1 and Zeb2 in miR-200 repression, all miR-200 family members were repressed by the forced expression of either Gsc, Snail, Twist, or TGF-β1 (Fig. 1D). These results reinforce the findings that in addition to interactions of the EMT-inducing TFs, repression of the miR-200 family of microRNAs accompanies and participates in execution of the EMT program.
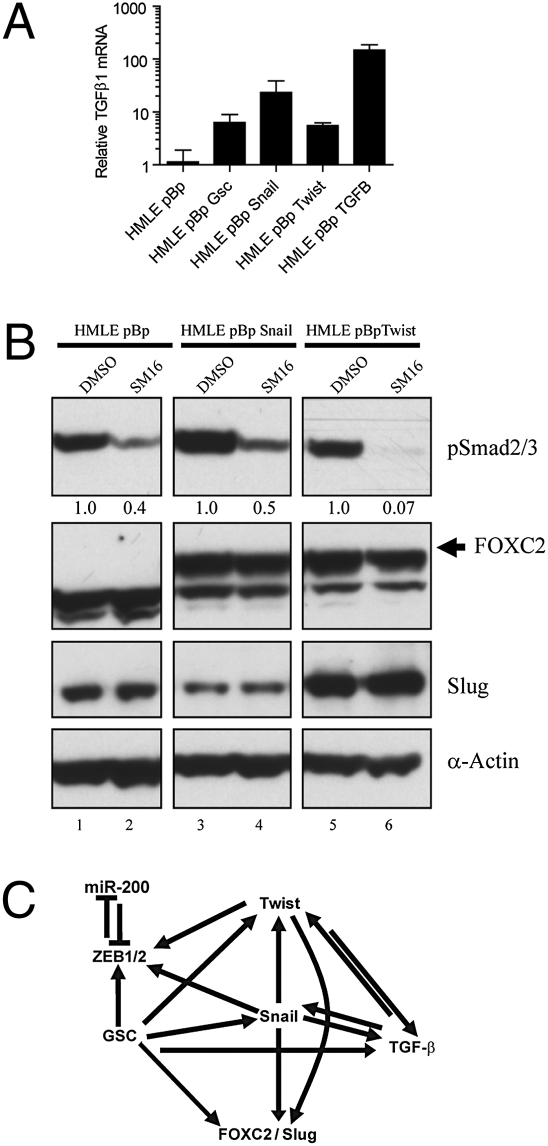
A widely accepted model of the EMT during tumorigenesis proposes that TGF-β1, produced by the tumor microenvironment, promotes tumor progression by inducing expression of the EMT-inducing TFs (34). Indeed, treatment of HMLE cells with TGF-β1 for a period of 12 d induced an EMT (14) and the expression of a number of EMT-inducing TFs (Fig. 1B). We undertook to determine whether another type of feedback loop operated here, specifically whether induction of an EMT achieved by overexpressing EMT-inducing TFs yields, in turn, the expression of TGF-β1. For this experiment, we measured the expression of TGF-β1 mRNA by quantitative RT-PCR (qRT-PCR) in HMLE cells expressing either an empty vector control, Gsc, Snail, Twist, or TGF-β1. In all cases, expression of TGF-β1 mRNA increased by more than 5-fold compared with control cells (Fig. 2A).
Fig. 2.
TGF-β1 is up-regulated by EMT inducers, but is not required for up-regulation of FOXC2 or Slug. (A) HMLE cells were transduced with a retrovirus overexpressing the indicated genes and expression of TGF-β1 mRNA was measured by quantitative RT-PCR. GAPDH was amplified for normalization. (B) HMLE cells transduced with a retrovirus overexpressing the indicated gene were treated with DMSO or SM16, and a TGF-β signaling inhibitor and gene expression was assayed by Western blot for the indicated proteins. Relative levels of pSmad2/3 were calculated by densitometry and listed beneath the bands. α-Actin was used as a loading control.
Because TGF-β1 sufficed to induce an EMT, we investigated whether the TGF-β1 expressed in response to an EMT operates in an autocrine manner to induce and maintain expression of downstream EMT effectors, specifically FOXC2 and Slug. Because Snail and Twist are known to induce a complete EMT, including expression of FOXC2, Slug, and TGF-β1, we blocked TGF-β1 signaling in these cells using SM16, a compound developed to inhibit the TGF-β1 pathway by interfering with the type I receptor signaling (35). As anticipated, treating HMLE cells expressing either an empty control vector, Snail, or Twist with SM16 significantly reduced the levels of phosphorylated SMAD2/3 by 50 to 90%, compared with DMSO (Fig. 2B). Under these conditions, neither the Snail- nor Twist-expressing cells altered expression of FOXC2 or Slug protein (Fig. 2B). This finding indicated that induction of an EMT by Snail or Twist does not depend on ongoing TGF-β1 autocrine signaling and suggests, instead, that Snail and Twist can induce FOXC2 and Slug through alternative mechanisms, quite possibly involving only intracellular signaling.
Hierarchical Clustering of GESs from EMTs Induced by Gsc, Snail, Twist, TGF-β1, and by Down-Regulating E-Cadherin.
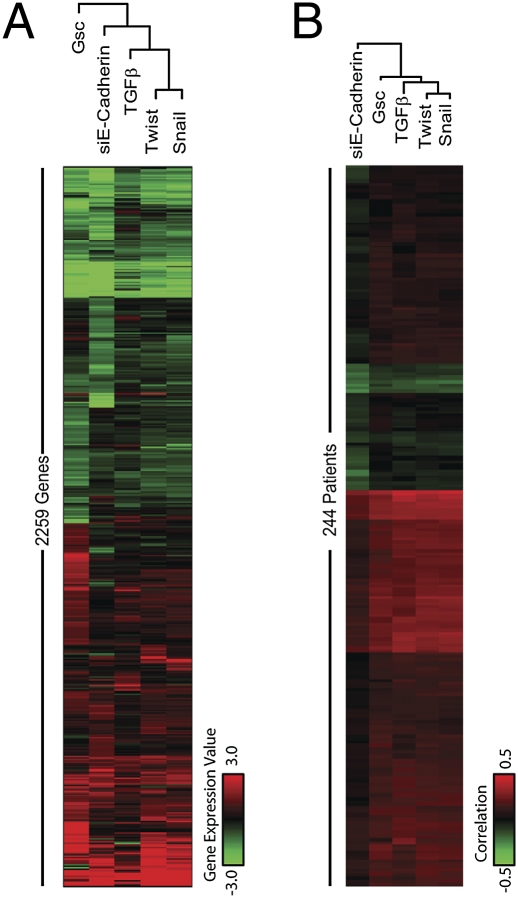
Because various inducers of EMT seemed to be capable of transactivating a common set of downstream effectors, we extended these comparisons by determining the larger effects of these TFs on overall gene expression within cells. We began by deriving individual GESs of the cells induced to undergo EMT by forced expression of Gsc, Snail, Twist, or TGF-β1, or by knocking down E-cadherin, and cataloguing genes exhibiting at least a 2-fold up- or down-regulation under any of the conditions of EMT induction. Cells in which E-cadherin was experimentally down-regulated were also included in these analyses, as we previously demonstrated that this alteration could also serve to induce an EMT in these cells (24). We then performed hierarchical clustering of these GESs to measure their degree of similarity (Fig. 3A).
Fig. 3.
Clustering of the individual EMT-inducer gene expression profiles based on their similarity to each other and to a large cohort of breast cancer patient gene-expression samples. (A) Heatmap of gene expression profiles in each sample. Values represent the log2 ratio over control. The diagram above each heatmap shows the similarities of EMT-inducer profiles to each other. (B) Heat map of correlations of each EMT-inducer profile with the gene expression profiles of patient tumors in a large cohort of breast cancer patients (49). Pearson correlation coefficients were calculated for all of the EMT-inducer–patient pairs and plotted as a heat map, in which red indicates a highly significant positive correlation, green indicates a highly significant negative correlation and black indicates a weak or absent correlation.
Of the five methods used to induce EMTs in this analysis, Snail and Twist generated the most similar GESs, consistent with the fact that Twist is a direct target gene of the Snail transcription factor (36), and Gsc showed the most distinct GES (Fig. 3A), consistent with the observation that Gsc was not induced by expression of either Snail, Twist, or TGF-β1 (Fig. 1B). Not surprisingly, expression of TGF-β1 or knockdown of E-cadherin produced GESs that diverged slightly from both Snail and Twist, likely because of the fact that, even though either method induces an EMT, neither involves ectopic expression of a TF and, therefore, may affect its downstream targets less directly.
EMT Core Signature.
Localized paracrine signals arising in the tumor microenvironment appear to be important in inducing an EMT in nearby cancer cells; accordingly, only a minority of cells within a tumor may display characteristics of having entered into or passed through an EMT. These observations complicate attempts to identify groups of cells within a tumor that have undergone an EMT. For this reason, we sought to identify a GES common to several known EMT-inducing signals. Establishment of such a core signature should prove useful in the future for identifying the small subset of cells that have undergone an EMT within a tumor, even if such an EMT is induced by a currently unknown inducer of this program.
To identify such a signature, we reanalyzed the microarray-based gene-expression changes from HMLE cells expressing Gsc, Snail, Twist, or TGF-β1 or an siRNA targeting E-cadherin. We identified an EMT core signature consisting of 159 genes that were down-regulated and 87 genes that were up-regulated at least 2-fold by all of these EMT-inducing signals (Table S1). Several of these gene-expression changes were validated by qRT-PCR (Fig. S3). As expected, the epithelial adhesion molecule E-cadherin (CDH1) was down-regulated in all samples and the mesenchymal markers N-cadherin (CDH2), vimentin, and the invasion-associated protease, matrix metalloproteinase (MMP2), were commonly up-regulated (Table S1). In addition, Zeb1, one of the TFs capable of orchestrating an EMT, was commonly up-regulated.
Overexpression of Snail has been shown to down-regulate the expression of the cell-cycle protein cyclin D2 (CCND2) (37), and indeed we found that CCND2 was down-regulated in the EMT core signature. Cells undergoing an EMT are known to be resistant to apoptosis (38–41). Consistent with this finding, the proapoptosis gene BIK was down-regulated in all samples. In addition, genes with a Zeb1 binding site present in their promoters were enriched in the set of genes down-regulated by EMT, including the gene discoidin domain receptor 1 (DDR1), which encodes an RTK involved in E-cadherin localization and distinguishes basal A from basal B cell lines (42–45), and the gene follistatin (FST), which is a TGF-β antagonist (Tables S2 and S3) (46–48).
Contributions of EMT-Inducing TFs to Breast Cancer.
We next attempted to use existing GESs of various types of breast cancer to uncover possible connections between the various TFs and breast cancer pathogenesis. We sought to understand the relatedness of individual EMT-inducers and their respective GESs to the gene-expression profiles with individual breast carcinomas in a large (244 patient) cohort of these tumors (49). The gene-expression profiles derived from many tumors displayed a high correlation to GESs derived from Gsc, Snail, Twist, and TGF-β1, but correlated less strongly with the signature derived from knocking down E-cadherin (Fig. 3B). As predicted from previous observations, tumors with a high correlation to the Snail GES also displayed a high correlation to the Twist GES. In addition, the gene-expression profiles derived from each tumor in this dataset tend to correlate with the GESs derived from more than one EMT inducer. Moreover, the expression signature of individual tumors did not make it possible to resolve between alternative mechanisms of EMT induction (i.e., Twist-, Snail-, TGF-β1-, or Gsc-induced EMT) in these breast cancers. Nonetheless, the GESs of individual EMT-inducers could be used to assay for the occurrence of an EMT in a breast tumor, even if the EMT was induced by a still-unknown inducer and activation of this transdifferentiation program occurred in only a minority of the cells within each tumor. Importantly, the possibility that stromal elements within the tumor samples rather than the carcinoma cells themselves resulted in the detection of a mesenchymal GES could not be discounted.
Correlation of EMT Core Signature with the Basal B, Claudin-Low, and Metaplastic Signatures.
We also wished to determine how the EMT core signature relates to the GESs of various subtypes of breast cancer. We compared the mean expression values of genes up- or down-regulated by an EMT against the GES of various breast cancer cell line subtypes (50) and found that the EMT core signature most strongly correlated with the signature derived from basal B cell lines (Fig. S4). This subtype is characterized by high vimentin expression and a stem cell-like expression profile (45, 50), similar to cells that have undergone an EMT; in contrast, the signatures derived from the basal A and luminal subtypes (45, 50) (Fig. S4) were not closely related to the EMT core signature.
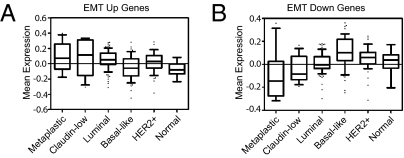
We also measured the association of the EMT core signature to GESs derived from breast cancer subtypes defined by Hennessy et al. (23) and against the GES of a cohort of metaplastic tumors that had been previously analyzed using the same microarray platform (22). The up- and down-regulated genes from the EMT core signature were also significantly up- and down-regulated in both the metaplastic and claudin-low breast cancers, but not in other breast cancer subtypes (Fig. 4 A and B). This finding is consistent with recent reports that have linked low E-cadherin expression, indicative of passage through an EMT, to clinically encountered breast tumors of either the claudin-low or metaplastic subtype (22, 23). Because passage through an EMT has also been linked with acquisition of stem cell characteristics (6), this suggests that the neoplastic cells in these tumors contain relatively high proportions of cancer stem cells.
Fig. 4.
The core EMT signature correlates with metaplastic and claudin-low breast cancers. (A and B) Gene-expression data were plotted as box plots for the mean expression of the EMT-up genes (A) and the EMT-down genes (B) by subtype using the dataset from Hennessey et al. (23) with the addition of 12 metaplastic tumors. Subtypes were called as in Herschkowitz et al. (22). The list was derived using Significance Analysis of Microarrays and cut off at the top ∼1,155 probes, 544 up and 611 down. Next, the genes were extracted in the dataset and averaged in each tumor (up and down separately). The one-way ANOVA significance for each plot was P < 0.0001.
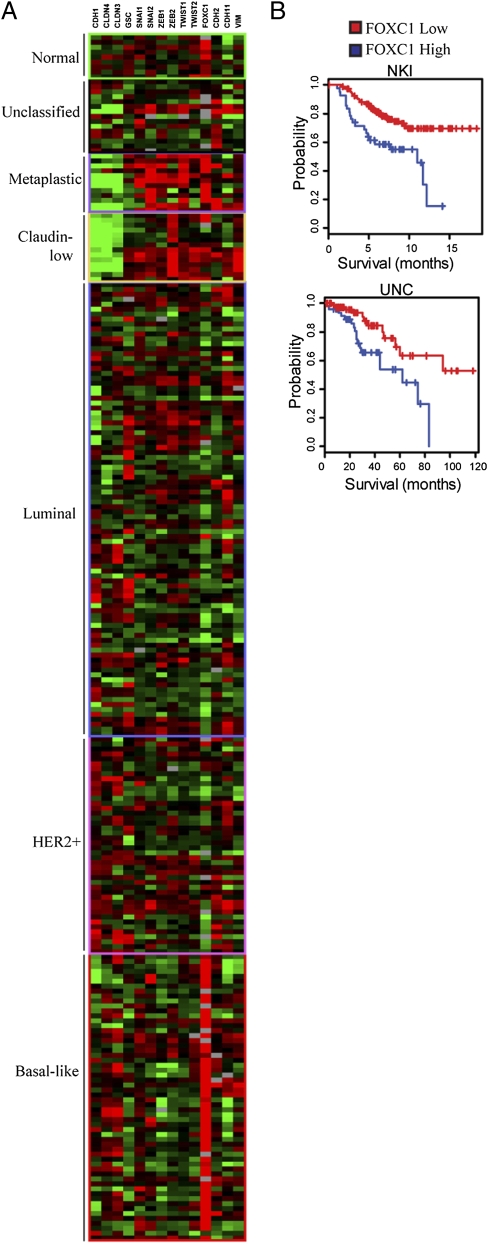
We also assayed the expression of the mRNAs encoding EMT-inducing TFs in breast cancers that had been classified into basal-like, HER2+, luminal, claudin-low, and metaplastic subtypes (22, 23). We found that the Snail, Slug, Zeb1, Twist1, and FOXC1 TFs were up-regulated in metaplastic tumors, and Zeb2 was most commonly up-regulated in claudin-low tumors (Fig. 5A). Both metaplastic and claudin-low tumor subtypes showed significant down-regulation of E-cadherin mRNA expression (CDH1) as well as claudin 4 (Fig. 5A). Although nearly all claudin-low tumors express high levels of Zeb2, only half of these tumors concomitantly expressed high levels of other EMT-inducing genes (Snail, Twist, FOXC1) (Fig. 5A). Strikingly, nearly all basal-like breast tumors, but not luminal or HER2+ tumors, display consistently high expression of the EMT-inducer FOXC1, which is known to be associated with increased cell motility and invasion and decreased expression of E-cadherin (13) (Fig. 5A and Fig. S5).
Fig. 5.
EMT-inducing genes are up-regulated in metaplastic and claudin-low tumors and FOXC1 expression marks basal-like tumors and is a predictor of poor clinical outcome. (A) Data were extracted for EMT-related genes and samples were ordered by intrinsic subtype as in Herschkowitz et al. (22). The twelve metaplastic tumors from Hennessey et al. (23) were also included. (B) Patients from the NKI and UNC datasets were divided into high- and low-FOXC1 expressers and their survival was compared. The P value was generated using the χ2 test of equality.
Correlation of Expression of an EMT-Inducer with Patient Survival.
We anticipated that tumors expressing the EMT-associated genes would exhibit a poorer survival than tumors not expressing the EMT-associated genes. For this reason, we performed clinical prediction analyses on the breast tumors in the Netherlands Cancer Institute (NKI) and University of North Carolina (UNC) databases (49, 51), using the EMT core signature. We found, however, that expression by a tumor of the EMT core signature failed to predict patient survival.
We therefore pursued an alternative hypothesis: that expression of individual genes capable of inducing an EMT, rather than multigene signatures, could predict patient survival. Although the expression of Gsc, Snail, Twist, or TGF-β1 genes did not predict clinical outcome, high expression of the FOXC1 TF was indeed a powerful predictor of poor clinical outcome. Thus, those patients whose tumors exhibited high levels of FOXC1 gene expression showed a significantly poorer survival outcome (Fig. 5B). Although, FOXC1 was not induced by Gsc, Snail, Twist, or TGF-β1, expression of FOXC1 in immortalized, but nontumorigenic, human mammary epithelial MCF12A cells has been shown to induce an EMT (8, 13).
Recent reports have linked expression of specific sets of genes to resistance to chemotherapy in breast cancer patients (52, 53). Using an M.D. Anderson Cancer Center study of response to neoadjuvant chemotherapy with paclitaxel, 5-fluorouracil, doxorubicin, and cyclophosphamide (54), we asked if tumor gene-expression profiles from patients that had a pathological complete response (pCR) correlated with the EMT core signature. We found that gene-expression profiles from patient tumors with pCR showed a negative correlation to the EMT core signature, whereas GESs from tumors from patients without pCR showed a slightly positive correlation to the EMT Core Signature (Fig. S6).
Discussion
The EMT core signature that we have identified was generated by comparing the gene-expression changes that occurred by overexpressing either Gsc, Snail, Twist, or TGF-β1, or by reducing levels of E-cadherin. As we found, this signature is enriched for genes containing the Zeb1 transcription factor-binding site near their transcription start sites (Table S2), and is most similar to GES from claudin-low and metaplastic breast cancers, as well as cancers without pCR.
Of note, a recent study demonstrated that the GES obtained from normal mammary epithelial stem cells is most similar to the claudin-low signature (55). This finding is consistent with the present findings that the expression changes associated with the EMT-inducing TFs correlate most closely with the claudin-low and metaplastic breast cancers and with our earlier demonstration that passage through an EMT results in the acquisition of stem-cell characteristics (6).
Analysis of mRNA expression levels in cells overexpressing EMT-inducers reveals that Twist, Snail, and TGF-β1 each up-regulate expression of Foxc2, Zeb1, and Zeb2 (Fig. 2C). Furthermore, we demonstrate that Snail and Twist generate the most closely related GESs, and expression of TGF-β1, Gsc, and knockdown of E-cadherin generate quite distinct, less closely related signatures. Further exploration of the differences between these various GESs will likely yield insights into the mechanisms required to activate the EMT program and those involved in maintaining the resulting mesenchymal/stem cell state.
Surprisingly, we were unable to observe any correlation between expression of genes in the EMT core signature and a poorer survival outcome among breast cancer patients, despite the observation that the EMT core signature correlates with metaplastic tumors, which are themselves linked with poorer patient survival (56). Interestingly, a recent report by Creighton et al. found that the GES derived from breast tumor-initiating cells was also enriched in claudin-low tumors, but likewise did not serve as a useful prognostic marker for clinical progression (53). However, they found that cell populations that survive conventional chemotherapeutic treatment are enriched for cells with EMT-associated mesenchymal and stem cell-associated tumor-initiating features (53). Additionally, an association between EMT and chemotherapy resistance, rather than survival, is suggested by Farmer et al., who show that a stroma-related GES predicts shorter relapse-free survival among patients who received chemotherapy, but not among untreated breast cancer patients (52). These findings are consistent with our data indicating that gene expression profiles of tumors from patients that responded to chemotherapy correlate negatively with the EMT core signature.
The use of total mRNA isolated from entire tumors for annotation of the expression datasets may preclude detection of cells that have undergone EMT, as only a small proportion of the neoplastic cells in each tumor may exhibit an EMT phenotype. Poor-prognosis tumors may therefore harbor an insufficient number of cells with a mesenchymal phenotype, so that there is no detectable effect on the gene-expression profile of the tumor as a whole. Accordingly, it may be necessary to collect tumor cells at the invasive edges of tumor cell islands and analyze these for the expression of the EMT-associated genes to gauge the true malignant potential of the tumor as a whole.
Among the group of EMT-inducing genes studied here, FOXC1 expression most closely correlates with a poorer survival of the breast cancer patients in the NKI and UNC datasets (Fig. 5B). FOXC1 was highly expressed in metaplastic and basal-like breast cancer subtypes (Fig. 5A), for which highly effective treatments are not currently available. The closely related FOXC2 TF is known to play a key role in inducing an EMT, promoting metastasis and to be associated with aggressive basal-like breast cancers as studied using immunohistochemistry methods (14). However, expression of FOXC2 could not be correlated with survival in the cited microarray-based stratification of breast cancer patients, because the arrays used did not contain suitable FOXC2 probes. Given the consistently high levels of FOXC1 in basal-like and metaplastic breast cancer subtypes, the high levels of Zeb2 in claudin-low breast cancers, and the contribution of FOXC2 to basal-like breast cancer (14), these genes or the pathways that regulate these genes would seem to represent potential targets for the development of novel anticancer therapeutics.
Methods
RT-PCR.
RNA was prepared from cultured cells by TRIzol extraction (Invitrogen). Complementary DNA was synthesized using Moloney Murine Leukemia virus reverse transcriptase (Invitrogen). Relative quantification values were calculated using the ddCt method (57) and values were plotted with SD using GraphPad Prism v5.0 (GraphPad Software, Inc.).
Cell Culture.
Immortalized HMLE were grown as previously described (58). MCF-7, SUM1315, and MDA-MB 231 cells were maintained according to ATCC instructions. The SUM159 cell line used for the study was developed from pleural effusions of breast cancer patients (59). SUM159 cells were cultured in F-12 Hams (Gibco) supplemented with 5% FBS (Tissue Culture Biologicals), 5 μg/mL of insulin, and 1 μg/mL hydrocortisone at 37 °C in 5% CO2.
Microarray Data Analysis and Deposition.
Microarray data for HMLE shCDH1 and vector control were extracted from the Gene Expression Omnibus (GEO) database under the accession GSE9691. Microarray data for HMLE Gsc, Snail, Twist, TGF-β1, and vector control has been deposited in GEO under accession number GSE9691.
To compare gene expression among different EMT inducers, a heat map was generated using genes with at least a 2-fold change in at least one condition included in the clustering. To compare individual EMT signatures to breast cancer patient gene-expression data, a heat map showing correlations between gene-expression profiles of the EMT inducers and the gene-expression profiles of tumors from breast cancer patients in the UNC cohort was created. Pearson correlation coefficients were calculated between the gene-expression profiles of all EMT inducer-patient pairs.
Significance Analysis of Microarrays.
The list of up- and down-regulated EMT genes was derived using Significance Analysis of Microarrays and cut off at the top 1,155 probes, 544 up and 611 down. The genes in the dataset were then extracted and averaged in each sample (up and down separately). ANOVA was performed and boxplot graphs were plotted with gene expression using GraphPad Prism v5.0 (GraphPad Software, Inc.). The one-way ANOVA for each plot was P < 0.0001.
Survival Analysis.
Patients from the NKI and UNC datasets were divided into high- and low-FOXC1 expressers and their survival was compared. The P value was generated using the χ2 test of equality using the survival package in R (www.r-project.org).
Supplementary Material
Acknowledgments
We thank the members of the Mani laboratory for helpful discussions and Biogen for the SM16 (TGF-β inhibitor). Additional help with data analysis was provided by Dr. Keith Beggarly and Nianxiang Zhang. This work was supported in part by the Broad Institute, Komen post-doctoral Fellowship PDF0707744 (to. J.I.H.), KG091219 (to B.G.H), National Institutes of Health Grant R01CA125109 (to P.T.R.), a pilot grant from the Dan L. Duncan Cancer Center (to J.M.R. and S.A.M.), a Research Trust award from M. D. Anderson Cancer Center, the V Foundations V scholar award, and Cancer Center Support Grant CA016672 (to S.A.M.), and National Institutes of Health Grant RO1 CA78461, the Breast Cancer Research Foundation, and Ludwig Center for Molecular Oncology at the Koch Institute (to R.A.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004900107/-/DCSupplemental.
References
- 1.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Frixen UH, et al. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113(1):173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabbah M, et al. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Updat. 2008;11(4-5):123–151. doi: 10.1016/j.drup.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morel AP, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 9.Hartwell KA, et al. The Spemann organizer gene, Goosecoid, promotes tumor metastasis. Proc Natl Acad Sci USA. 2006;103:18969–18974. doi: 10.1073/pnas.0608636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 11.Batlle E, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 12.Cano A, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 13.Bloushtain-Qimron N, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci USA. 2008;105:14076–14081. doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mani SA, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104:10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aigner K, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 17.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 18.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onder TT, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 25.Comijn J, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 26.Vandewalle C, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savagner P, Yamada KM, Thiery JP. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derycke LD, Bracke ME. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int J Dev Biol. 2004;48:463–476. doi: 10.1387/ijdb.041793ld. [DOI] [PubMed] [Google Scholar]
- 30.Burdsal CA, Damsky CH, Pedersen RA. The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development. 1993;118:829–844. doi: 10.1242/dev.118.3.829. [DOI] [PubMed] [Google Scholar]
- 31.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154(1):8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 32.Yu M, et al. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev. 2009;23:1737–1742. doi: 10.1101/gad.1809309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bracken CP, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 34.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: The molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki E, et al. A novel small-molecule inhibitor of transforming growth factor beta type I receptor kinase (SM16) inhibits murine mesothelioma tumor growth in vivo and prevents tumor recurrence after surgical resection. Cancer Res. 2007;67:2351–2359. doi: 10.1158/0008-5472.CAN-06-2389. [DOI] [PubMed] [Google Scholar]
- 36.Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. Dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 1992;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- 37.Vega S, et al. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitali R, et al. Slug (SNAI2) down-regulation by RNA interference facilitates apoptosis and inhibits invasive growth in neuroblastoma preclinical models. Clin Cancer Res. 2008;14:4622–4630. doi: 10.1158/1078-0432.CCR-07-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue A, et al. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer Cell. 2002;2:279–288. doi: 10.1016/s1535-6108(02)00155-1. [DOI] [PubMed] [Google Scholar]
- 40.Roy HK, et al. Down-regulation of SNAIL suppresses MIN mouse tumorigenesis: Modulation of apoptosis, proliferation, and fractal dimension. Mol Cancer Ther. 2004;3:1159–1165. [PubMed] [Google Scholar]
- 41.Sayan AE, et al. SIP1 protein protects cells from DNA damage-induced apoptosis and has independent prognostic value in bladder cancer. Proc Natl Acad Sci USA. 2009;106:14884–14889. doi: 10.1073/pnas.0902042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eswaramoorthy R, et al. DDR1 regulates the stabilization of cell surface E-cadherin and E-cadherin-mediated cell aggregation. J Cell Physiol. 2010;224:387–397. doi: 10.1002/jcp.22134. [DOI] [PubMed] [Google Scholar]
- 43.Wang CZ, Yeh YC, Tang MJ. DDR1/E-cadherin complex regulates the activation of DDR1 and cell spreading. Am J Physiol Cell Physiol. 2009;297:C419–C429. doi: 10.1152/ajpcell.00101.2009. [DOI] [PubMed] [Google Scholar]
- 44.Maeyama M, et al. Switching in discoid domain receptor expressions in SLUG-induced epithelial-mesenchymal transition. Cancer. 2008;113:2823–2831. doi: 10.1002/cncr.23900. [DOI] [PubMed] [Google Scholar]
- 45.Blick T, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. J Mammary Gland Biol Neoplasia. 2010;15:235–252. doi: 10.1007/s10911-010-9175-z. [DOI] [PubMed] [Google Scholar]
- 46.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 48.Nogai H, et al. Follistatin antagonizes transforming growth factor-beta3-induced epithelial-mesenchymal transition in vitro: Implications for murine palatal development supported by microarray analysis. Differentiation. 2008;76:404–416. doi: 10.1111/j.1432-0436.2007.00223.x. [DOI] [PubMed] [Google Scholar]
- 49.Hoadley KA, et al. EGFR associated expression profiles vary with breast tumor subtype. BMC Genomics. 2007;8:258. doi: 10.1186/1471-2164-8-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van de Vijver M. Gene-expression profiling and the future of adjuvant therapy. Oncologist. 2005;10(Suppl 2):30–34. doi: 10.1634/theoncologist.10-90002-30. [DOI] [PubMed] [Google Scholar]
- 52.Farmer P, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15(1):68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 53.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hess KR, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 55.Lim E, et al. kConFab Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 56.Luini A, et al. Metaplastic carcinoma of the breast, an unusual disease with worse prognosis: The experience of the European Institute of Oncology and review of the literature. Breast Cancer Res Treat. 2007;101:349–353. doi: 10.1007/s10549-006-9301-1. [DOI] [PubMed] [Google Scholar]
- 57.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 58.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15(1):50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ethier SP. Human breast cancer cell lines as models of growth regulation and disease progression. J Mammary Gland Biol Neoplasia. 1996;1(1):111–121. doi: 10.1007/BF02096306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.