Abstract
It is known from paleontology studies that two premolars have been lost during mouse evolution. During mouse mandible development, two bud-like structures transiently form that may represent rudimentary precursors of the lost premolars. However, the interpretation of these structures and their significance for mouse molar development are highly controversial because of a lack of molecular data. Here, we searched for typical tooth signaling centers in these two bud-like structures, and followed their fate using molecular markers, 3D reconstructions, and lineage tracing in vitro. Transient signaling centers were indeed found to be located at the tips of both the anterior and posterior rudimentary buds. These centers expressed a similar set of molecular markers as the “primary enamel knot” (pEK), the signaling center of the first molar (M1). These two transient signaling centers were sequentially patterned before and anterior to the M1 pEK. We also determined the dynamics of the M1 pEK, which, slightly later during development, spread up to the field formerly occupied by the posterior transient signaling center. It can be concluded that two rudimentary tooth buds initiate the sequential development of the mouse molars and these have previously been mistaken for early stages of M1 development. Although neither rudiment progresses to form an adult tooth, the posterior one merges with the adjacent M1, which may explain the anterior enlargement of the M1 during mouse family evolution. This study highlights how rudiments of lost structures can stay integrated and participate in morphogenesis of functional organs and help in understanding their evolution, as Darwin suspected long ago.
Keywords: rudiment, signaling center, tooth evolution, SHH, molar development
Adult vestiges and embryonic rudiments are defined as traces of ancestral structures lost during evolution (1, 2). These evolutionary remnants are present in many species, including humans, and have fascinated scientists since Aristotle and have been at the heart of evolutionary thought since Darwin (3), who first suggested that they can help to reveal ancestry. These residual structures may represent key intermediates in morphological innovation (2). Indeed, once a body part loses functionality following a change in lifestyle, it will degenerate in a neutral manner, and this neutral evolution makes it prone to assume new functions. To our knowledge, however, very few examples have been described yet (1, 4).
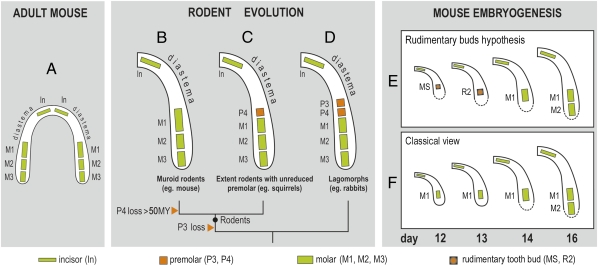
If they are not identified, rudiments can obscure the developmental studies of prospective functional structures. However, consideration of the evolutionary history of species can help to draw attention to the possible presence of rudiments. For example, mouse dentition is strongly reduced compared with other mammals. In each jaw quadrant, one incisor is separated from three molars by a gap (diastema), where incisor, canine, and premolar teeth are present in other species (Fig. 1A). The common ancestor of lagomorphs (e.g., rabbits) and early rodents in the mouse lineage had premolars, and these were maintained in some extant rodent or lagomorph families (Fig. 1 C and D), where they develop earlier than and anterior to molars (5). Despite the lack of premolars in the adult mouse, there is morphological evidence for bud-like structures that develop earlier than and anterior to the upper and lower first molar (M1), and which have been interpreted as rudimentary (vestigial) premolar buds (6, 7). In the lower jaw these buds (called MS and R2) develop transiently at embryonic day (ED) 12 and 13, respectively, and then they regress (Fig. 1E). The M1 becomes a distinct structure posteriorly by day 14, and the R2 is thought to be integrated into it (8, 9).
Fig. 1.
Reduction of the lower cheek teeth during mouse evolution and their pattern during mouse ontogeny. (A) Dentition in the adult mouse is considerably reduced. Each jaw quadrant comprises only one incisor (In) and three molars (M1, M2, M3); a large toothless diastema occurs at the place of missing canine and premolar teeth. (B–D) Evolution of the mouse lower molars from a common ancestor of lagomorphs and rodents. (D) Two premolars (called P3 and P4) were lost in the mouse lineage (B). (E and F) The two current interpretations of mouse lower molar development. The “rudimentary buds hypothesis” of mouse lower molar row development (E) has a basis in descriptive morphological studies and evolutionary data: Two rudimentary premolar buds (MS and R2) are the first tooth primordia, which sequentially develop in the cheek region of mandible, before and in front of molars. These buds’ progression is stopped by apoptosis. (F) In the classic view, the first molar (M1) is the first tooth primordium that appears in the cheek region of the mandible. Afterward, the other molars (M2, M3) are sequentially added.
However, the existence of the two premolar rudimentary buds has been poorly recognized in the literature, mainly because of a lack of molecular data correlated with their development. As a consequence, the morphological changes, molecular events, and tooth-specific signaling centers evident at ED 12 and 13 are generally thought to reflect M1 development (Fig. 1F) (e.g., ref. 10, http://bite-it.helsinki.fi/). Interestingly, a supernumerary tooth occurs in front of the M1 in some mouse mutants (11–23), and this has been considered as an extra molar (19, 20, 23) or related to the lost premolar (13–16, 18, 21, 22). In the latter case, some authors explicitly proposed the continued development of a rudimentary premolar (diastemal) bud as a possible origin for this supernumerary tooth in mutants (8, 12–16). However, the “rudimentary bud hypothesis” (Fig. 1E) has not yet been experimentally tested or validated on the basis of molecular data.
To clarify whether the prominent bud-like structures detected in WT mouse mandible at ED 12 and 13 reflect the rudimentary tooth primordia (Fig. 1E) or the M1 anlage (Fig. 1F), we set out to test these two concepts by correlating morphological and molecular aspects of tooth development in the mouse embryonic mandible. By studying the dynamics of molecular markers of tooth development and performing lineage tracing during early cheek tooth formation, we validated the vestigial bud hypothesis. Our studies provide a unique understanding of molar row development and first molar morphogenesis. They also provide a clear example of participation by a rudiment in development and evolution of a functional organ.
Results and Discussion
Three Shh Signaling Centers Are Sequentially Patterned in the Cheek Region of Mandible and Colocalized, Respectively, with MS, R2, and M1 Tooth Buds.
To investigate the dynamics of tooth development in the cheek region of embryonic mandible, we first assessed Shh expression, which is widely recognized as a marker of early tooth development in mammals (24), and also in fish (25), at closely spaced intervals using Shh-EGFP mice (26), in which EGFP is inserted at the Shh locus.
To avoid heterogeneity of data resulting from the intra- and interlitter variability in developmental progress of embryos harvested at a similar day of pregnancy, both age- and weight-staging criteria (27) were used: the age of embryos counted in number of ED postcoitum was further refined using wet body weight in milligrams (for details, see Materials and Methods). Body weight has been shown to be a reliable indicator of small stage differences, which allows for fine-ranking embryos of similar age, and enables a detailed series of progressive stages of tooth development (27).
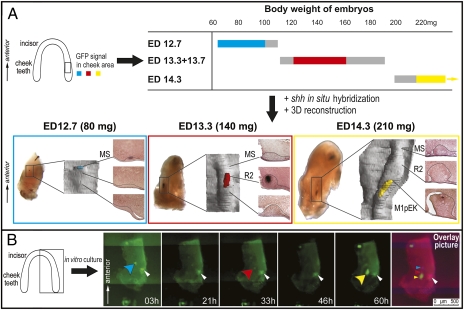
Because of the detailed staging, three distinct periods of bright GFP expression were revealed in the cheek region of the developing mandible. These periods of bright signal alternated with periods of a weak or indistinct signal (Fig. 2A). Time-lapse microscopy of mandibles cultured in vitro (Fig. 2B, Fig. S1, and Movie S1) and Shh in situ hybridization of dissected mandibles (Fig. 2A and Fig. S2) confirmed this dynamic change in pattern of expression.
Fig. 2.
Three Shh signaling centers are sequentially patterned in the cheek region of the mandible. (A) The time-table shows in colors (blue, red, yellow) the presence of a distinct signal patch in the cheek region of mandible of the Shh-EGFP mice according to their age (ED) and body weight (mg). Gray represents the mandibles with weak or indistinct signal. Blue, red, and yellow boxes represent the mandibles with the signal patch in the cheek region at ED 12.7, 13.3 + 13.7, and 14.3, respectively. The mandibles hybridized with Shh antisense probe were sectioned frontally to show dental epithelium (in a rectangle), which was reconstructed in 3D (Shh signal in color according to the time-table). The Shh signaling centers correspond to the respective morphological structures MS, R2, and M1 bud. (B) Time-lapse microscopy beginning at ED 12.7 (90 mg), culture time in picture corner. Blue, red, or yellow arrowhead designates three successive EGFP patches; the white arrowhead designates a necrotic zone. An overlay of the pictures at 0, 31, and 60 h shows three distinct EGFP signals.
The distinct domains of strong Shh expression became progressively more posterior in the mandible over time (Fig. S1 and Movie S1). This pattern was consistent with the notion that these domains corresponded to sequentially developing MS, R2, and M1. We next analyzed sections through Shh-hybridized mandibles, followed by 3D reconstructions of dental epithelium. The Shh expression was indeed found at the tip of the morphological structures called MS, R2, or M1 bud, at ED 12.7, 13.3, or 14.3, respectively (Fig. 2A). Of note, two signals (a small anterior and a larger posterior Shh domain) occasionally coexisted in ED 13.3 embryos (Fig. S2). Such a double signal has already been reported in day 13 mouse embryos (19, 22): the anterior weak signal has been attributed to an extra (possibly vestigial) tooth and the posterior, larger signal to the developing M1. According to the dynamics of Shh expression reported here, we propose that both these Shh spots in fact mark rudimentary signaling centers: one disappearing in the MS rudiment (weak anterior signal) and another newly formed in the R2 rudiment (strong posterior signal) (Fig. S2). In contrast, the M1 signaling center (primary enamel knot) appears 1 d later and even more posteriorly (Fig. 2).
The finding of a series of three Shh-signaling centers, which colocalized with three successive buds (MS, R2, and M1, respectively) (Figs. 1E and 2A), strongly contrasts with the classical interpretation of Shh expression in the cheek region of mouse mandible, which attributes the Shh expression at ED 12 to 14 to successive stages of M1 development (Fig. 1F).
Rudimentary Buds Have Their Own Signaling Center Resembling the M1 Signaling Center, Known as the “Primary Enamel Knot.”
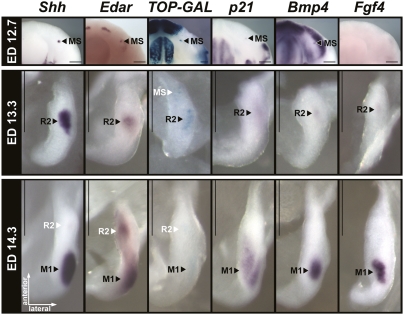
An important consequence of the colocalization of the Shh expression at ED 12 and 13 to MS and R2, respectively, was that Shh expression in M1 was first found at the late-bud stage of M1 epithelium at day 14 (more exactly at ED 14.3 in the present study). In M2, which contrary to M1 cannot be confused with a rudimentary structure, Shh expression is also not found before a late-bud stage, consistent with the present interpretation of M1 (Fig. S3). This Shh expression coincided with onset of formation of the molar primary enamel knot (pEK), a recognizable morphological structure (28) with signaling center properties (24, 29). The Shh expression in molars just preceded the transition to the cap stage of tooth development and was maintained during that stage. Two days and 1 d before, the respective MS and R2 buds also had their own Shh signaling center (Fig. 2 and Fig. S1), but failed to maintain it. This observation suggested that MS and R2 initiate a pEK but fail to maintain it. In agreement with this view, a rudimentary enamel knot was described in R2 [in terms of cell arrangement, presence of apoptosis, and Shh expression (8, 15)]. We therefore looked for other markers (Fig. 3) that are typical of the cap stage M1 pEK signaling center, such as activation of the Wnt pathway reported by TOPGAL transgene activity and expression of Edar, p21, Bmp4, and Fgf4 (29–31). Most of these markers could indeed be detected in both MS and R2 bud, although at lower levels than in M1 bud (Fig. 3). Only Fgf4 could not be detected in MS bud, which may reflect its growth arrest at an earlier developmental stage compared with R2.
Fig. 3.
MS and R2 display typical pEK markers. Localization of several pEK markers in whole mandibles (ED 12.7, 80–90 mg) or dissociated dental epithelium from the lower cheek tooth region (ED 13.3, 130–140 mg; and ED 14.3, 220–230mg), as revealed by in situ hybridization with Shh, Edar, p21, Fgf4, and Bmp4 antisense probes of CD1 embryos or by X-gal staining of TOPGAL embryos carrying a Wnt pathway reporter transgene (41). The residual signal is indicated by white letters. (Scale bars, 250 μm.)
These findings demonstrate that a series of three signaling centers, which colocalized with the morphological primordia MS, R2, and M1, are sequentially patterned in the budding epithelium of the cheek tooth-forming region at early stages. All three signaling centers displayed common molecular markers. This finding suggests that similar gene networks are reiteratively deployed in the development of MS, R2, and M1, and further strengthens the view that MS and R2 are rudiments of teeth (Fig. 3).
M1 Shh Signaling Spreads Anteriorly up to the Adjacent R2 Bud, Which Is Integrated into M1.
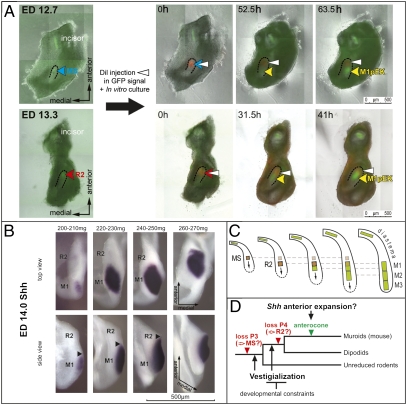
Previous morphological study has suggested that the R2 bud is incorporated in the M1 cap (9). This suggestion prompted us to trace the fate of MS and R2 buds (Fig. 4A and Movies S2 and S3). For that purpose, the MS or R2 bud of Shh-EGFP mandibles were marked at the signaling center with DiI (at ED 12.7 or 13.3, respectively), and then traced by time-lapse microscopy in vitro (Fig. 4A and Movies S2 and S3). In both cases, the label was localized anteriorly to the rising EGFP signal of the M1 pEK (Fig. 4A, 52.5-h culture of the labeled MS, or 31.5-h culture of the labeled R2). These results confirmed that the signaling center of MS, R2, and M1 are topographically distinct structures.
Fig. 4.
Sequential patterning of the cheek teeth in mouse mandible. (A) Time-lapse microscopy pictures of Shh-EGFP mandibles cultured after DiI microinjection at ED 12.7, 90 mg (in MS) or at ED 13.3, 145 mg (in R2), (culture time in picture corner). Note the pEK of the M1 appears posteriorly to the DiI label. The R2 label is later overlapped by the anteriorly extending M1pEK. (B) A series of ED 14.0 dissociated dental epithelia hybridized with Shh antisense probe document the secondary anterior extension of the M1 Shh-signaling at the bud-cap transition. (C) A model of integration of the premolar rudiments in patterning of mouse molars. (Arrows) The influence of a tooth primordium on the newly rising one. (D) A two-step working model of mouse tooth row evolution.
However, when R2-labeled mandibles were cultured longer, the EGFP signal of M1 pEK spread anteriorly to overlap with the R2 label (Fig. 4A, 41-h culture of the labeled R2). This result implied that the Shh expression related to the M1 pEK was initiated in a distinct region posterior to the R2, but later extended anteriorly to include the R2 domain. This striking spreading was confirmed by harvesting embryos in a very narrow time window during bud-to-cap transition (Fig. 4B).
Thus, the M1 Shh signaling center (pEK) is initiated in a field posterior to R2. It is secondarily repatterned in a broader field encompassing R2, which is thus integrated into M1. These data suggest a possible mechanism by which a large signaling center can be formed during ontogeny. Such a large signaling center could in turn explain the markedly long anterior part (anteroconid) of the M1 in adult mouse mandible when compared with the M2 (Fig. S3A), as well as the shorter M1 of those rodents that retained a lower premolar.
Concluding Remarks.
In the classic view of mouse odontogenesis, all events in the posterior part of the mandible of day 12 to 14 embryos are considered to exclusively represent the early stages of M1 formation (Fig. 1F). Instead, we demonstrated that three distinct signaling centers are sequentially patterned along the antero-posterior jaw axis. These three centers are associated with morphological structures previously shown to be transiently discernable in the dental epithelium (6, 9): the anterior rudimentary bud (MS), posterior rudimentary bud (R2), and M1 in day 12, 13, and 14 embryos, respectively (Fig. 1E). Our results point to a previously unexplored view in which dental epithelial budding, as well as appearance of the signaling centers, progresses sequentially along the antero-posterior axis of the mandible at embryonic days 12 to 14, as later occurs with M2 and M3 development. The two earliest signaling centers (in day 12 and 13 embryos) are transitory and respectively belong to the rudimentary MS and R2 primordia, whose development is arrested at the bud stage. Only the third signaling center, which appears at day 14, corresponds to the M1 pEK. The M1 pEK, as represented by Shh expression, later spreads anteriorly as far as the field of the R2 primordium, which becomes integrated into M1. We propose that the rapid sequential development of the cheek teeth, together with the later fusion between R2 and M1, have led to a misinterpretation of mouse molar morphogenesis. The integration of R2 into the anterior part of the M1 (Fig. 4) documents that a tooth, here the large mouse first molar, can arise from the fusion of several tooth primordia, here the R2 bud and the M1 bud. Although this developmental mechanism (concrescence or connation of tooth primordia) has been proposed long before on the basis of descriptive embryological studies (32–34), it has not been generally accepted (for details, see refs. 8 and 35).
This study raises several questions regarding the initiation of the whole tooth row in the cheek region (including MS, R2, M1, M2, and M3) (Fig. 4C). A main issue is how and when the most anteriorly located rudimentary bud MS is patterned. The present data suggest that MS signaling center at ED 12.3 express key genes similar to the pEK, which is found in the M1 and M2 from a late-bud stage (at ED 14.3 and 16.0, respectively) (Figs. 2A and 3, and Fig. S3). Therefore, earlier events (ED 10.5–12.0) should be responsible for the positioning of the MS, which might be the first of a cascade that patterns the whole tooth row. Such a cascade has already been suggested for the patterning of M1, M2, and M3 (10, 36, 37), and rudimentary buds could act in it, independently of their later fate. This result would be reminiscent of the situation known in fish, in which a pioneer tooth often sets the stage for the development of an entire row (38). Shh is known to be expressed as a spot in the tooth-forming region from end of ED 11 and 12 (39), which has been referred to as a “placode” (19, 22). This early expression is thought to play a role in the positioning of the tooth-forming region along the mouse embryonic jaw (40). Future work should focus on how this expression may relate to the signaling center, shown here in MS bud at ED 12.7 (Fig. 2), and how it is initiated and controlled. The proposed patterning model of the cheek teeth row (Fig. 4C) can be tested in mammals where premolars and molars are sequentially patterned, or in genetically altered mice with an extra tooth in front of the M1, such as K14-Eda mice (19, 22) or Sprouty mutant mice (12).
Our study also provides a unique framework for comparing the rudimentary tooth buds (which regress) with the M1 bud (which forms a functional tooth) to find the genes involved in this fine tuning, from both a developmental and evolutionary point of view. Such comparisons should take into account qualitative as well as quantitative and temporal aspects of gene regulations and may be helpful for tooth engineering and regeneration.
From an evolutionary perspective, the rudimentary tooth buds in mouse molar development illustrate how rudiments can be maintained. At least three nonmutually exclusive mechanisms may explain why the two rudimentary tooth buds have been maintained over approximately 50 million years. First, gene networks acting in the rudimentary buds could to some extent be preserved because they are fundamental for odontogenesis, being shared with developing molars that were maintained. Indeed, the present data showed that several genes are expressed in both rudimentary and M1 buds (Fig. 3). Second, the sequential development of the cheek tooth row is likely to be an integrated system, in which the development of molars and rudimentary buds is not independent. Recent articles have shown that during the first, second, and third molar sequential development, each developing molar exerts an inhibitory influence on the next developing molar (36, 37). The present study suggests that this model should be extended to include the rudimentary buds that presumably represent premolars, meaning that the premolar rudiments would have an influence on patterning of molars (Fig. 4C). Moreover, the rudiments may function as a barrier between the developing molars and inhibitory influences found in the diastema [such as Gas1, an antagonist of the Shh pathway (40)]. The third mechanism more specifically concerns the R2 bud. The present data demonstrated that R2 is incorporated into M1 during development, when M1 pEK elongates anteriorly to encompass R2. This phenomenon may be related to the formation of a prolonged anterior part (the anteroconid) in the mouse M1, especially as this part arose, evolutionarily speaking, following the loss of premolars (Fig. 4D) (7). This hypothesis can be tested in several manners. In mice, one can compare M1 with M2 development, because M2 looks like the M1 deprived of anteroconid in WT mice (Fig. S3A), but can develop a small anteroconid in some “boosted” conditions (22). Furthermore, M1 in genetically altered mice with an extra tooth in the premolar position should be examined in more detail, as some of these mice have the extra tooth and a well-developed M1 anteroconid (12, 19, 22), but others have an extra tooth but reduced anteroconid (13, 16, 17). This difference has been proposed to result from differential participation of the rudiments in supernumerary tooth formation: MS or R2 could be involved where the anteroconid is maintained or reduced, respectively (16, 35). Finally, a comparison with other species (e.g., rodent species that retained a premolar) will be very helpful to better understand the specificities of mouse first molar development.
We propose a two-step model for mouse molar row evolution in which sharing of genetic networks and developmental integration might have slowed down rudiment regression, but still allows a certain degree of freedom, thus facilitating morphological innovation of the first muroid molar. Work at the intersection of developmental biology and paleontology will help to test this model by further focusing on development and evolution of the anterior part of M1.
Materials and Methods
Mouse Breeding.
The CD1 mice were purchased from the Charles River. C57BL/6 mouse strain carrying fusion protein Shh-EGFP and Cre recombinase from endogenous Shh locus (B6.Cg-Shhtm1(EGFP/cre)Cjt/J), first introduced by Harfe (26), were purchased from The Jackson Laboratory. The transgenic mice were genotyped using Jackson's Lab Protocol for fluorescent proteins. TOPGAL mice [Tg(Fos-lacZ)34Efu] carry three LEF1/TCF1 binding sites fused to a minimal c-fos promoter driving lacZ expression (41). For experiments, CD1 males, Shh-EGFP males or TOPGAL males were crossed with WT CD1 females. All of the animals' treatment satisfied the requirements of the Institutional Review Board of the Institute of Experimental Medicine, Academy of Sciences of the Czech Republic, Prague, Czech Republic, and of the Institut de Génomique Fonctionnelle de Lyon, Université de Lyon, Lyon, France.
Embryo Harvesting and Staging.
Mice were mated overnight and vaginal plugs were detected in the next morning, noon being indicated as ED 0.5. Pregnant females were killed by a cervical dislocation at 3:00 to 4:00 PM on ED 12 and 13 (stages ED 12.7 and 13.7, respectively), or at 7 to 8 AM on ED 13 and 14 (stages ED 13.3 or 14.3, respectively). These two harvesting terms allowed getting a broader spectrum of developmental stages of the mouse embryos during the period under observation.
An important variability in stages of morphological development may exist not only among litters but even within litters harvested at a similar day of pregnancy (ED) (e.g., ref. 42). The wet body weight allows further sorting of the embryos of a specific chronological age, being a very good indicator of developmental progress of tooth development (27). Therefore, each embryo was weighed after a drop of a fluid on its surface was removed by gentle dabbing on a dry Petri dish. For example, in Fig.2, the embryos harvested at 3:00 to 4:00 PM at ED 12 (when counted according to the day of detection of vaginal plug = day 0), were said to be ED 12.7 embryos (according to the present counting, when the midnight before morning detection = ED 0.0), and were further ranked according to their body weight from 65 to 110 mg.
Then, the Shh-EGFP embryos were sorted according to the presence/absence of a green fluorescence in their tails under inverted fluorescence microscope Leica AF6000. For further experiments, only the EGFP-positive embryos were used. The lower jaw arch was dissected and split at a midline. The left half was always used for Shh whole mount in situ hybridization, and the right one for detection of Shh-EGFP during in vitro culture (time-laps microscopy, microinjection).
Mandible Epithelium Dissociations.
Mandibles were dissected in Hank's medium and treated with Dispase II (Roche) 10 mg/mL at 37 °C for 1 to 2 h, depending on embryonic stage. Epithelium was carefully removed and fixed overnight in PFA 4%.
Whole-Mount in Situ Hybridization and X-Gal Staining.
Embryonic mandibles or dissociated epithelia were fixed in 4% PFA solution over night at 4 °C and In situ hybridization was done according to a standard protocol. DIG RNA probes were transcribed in vitro from plasmids described elsewhere: Shh (43), Edar (44), Fgf4 (29), and Bmp4 (45), except for p21, which was made from a PCR fragment amplified with primers GAGCAAAGTGTGCCGTTGTCTCT and ACCAATCTGCGCTTTGGAGTGATA and cloned in Topo-pCRII (Invitrogen). TOPGAL embryonic mandibles or dissociated epithelia were fixed in 4% PFA for 15 min only and stained with X-gal according to a standard protocol. The samples were documented by a Leica MZ6 stereomicroscope with a Leica DC480 digital camera or on a Zeiss LUMAR stereomicroscope with a CCD CoolSNAP camera (PLATIM).
Cryosections and 3D Reconstruction.
The hybridized jaw halves were embedded in a series of graded solutions of sucrose (Sigma) diluted in PBS (pH 7.4). Next, the specimens were embedded in OCT Tissue Tek (Sakura) diluted 1:1 with 20% sucrose and frozen in isopentan (Sigma) cooled on dry ice to −60 °C, and sectioned on a cryostat microtome Mikrom HM 560 (Mikrom) in 10-μm sections. The sections were postfixed in 4% formaldehyde and counterstained by Nuclear fast red (Fluka), dehydrated, and mounted in Neomount (Merck).
Computer-Aided 3D Reconstruction.
Contours of the dental and adjacent oral epithelium were drawn from consecutive sections using a Leica DMLB microscope equipped with a drawing chamber at a magnification of 320×. The digitalization of the serial drawings and the correlation of successive images have been previously described (46). The generation of 3D pictures was made using VG Studio Max 2.0 software (VG Studio Max).
In Vitro Culture.
Dentition explants were dissected under sterile conditions from mandibles of ED 12.7 and 13.3 embryos, and cultured in an agarose semisolide medium for 60 h. The semisolid culture medium was prepared according to Hu et al. (47).
Time Lapse Microscopy.
Mandibles from heterozygous Shh-EGFP embryos at ED 12.7 (80–90 mg) were cultured in vitro. The time lapse experiments were made on the inverted fluorescence microscope Leica AF6000 with transparent incubator associated with humidifier, and CO2- (5%) and temperature- (37.5 °C) regulating units. Experiments were driven and evaluated by LeicaAF software with a well-plate acquisition software module (Leica). For the overlay of time-laps pictures, three pictures taken respectively at 0, 31, and 60 h were first converted to artifical colors (respectively, blue, red, and green). They were put together by using original Leica software, and adjusted according to the shape of dental epithelium and the autofluorescence of necrotic zone.
DiI Labeling.
Upon dissection (at ED 12.7 or 13.3) and accommodation of an explant in a semisolid medium, DiI microinjection was targeted into EGFP fluorescence area selected under green fluorescence cube. DiI was kept as a stock solution (solution of 0.25 μg/μL DiI in 100% DMSO). Before use, the stock solution was dissolved 1:1 by 50% glycerol in aqua pro injectione. Micromanipulator (Narishige) and microinjector (Narishige) were connected with the Leica microscope working station described above. Microinjection itself was performed with a capillary needle (Narishige) with 5 μm in diameter.
Supplementary Material
Acknowledgments
We thank B. Hu for sharing his skills with in vitro molar culture, D. Lyons (University of California, San Francisco, CA) for kindly providing the epithelium dissociation protocol, A. McMahon (Harvard University, Cambridge, MA), S. Vincent (Institut de Génomique Fonctionnelle de Lyon, University of Lyon, Lyon, France), I. Thesleff (Institute of Biotechnology, University of Helsinki, Helsinki, Finland), D. Duprez (Université Pierre et Marie Curie, Paris, France), and M. Vieux-Rochas (Evolution des Régulations Endocriniennes, Muséum National d'Histoire Naturelle, Paris, France) for kindly providing probe plasmids, L. Krasny, H. Šanderová (Institute of Microbiology Academy of Sciences of the Czech Republic) and R. Sedláček with his colleagues (Institute of Molecular Genetics, Academy of Sciences of the Czech Republic) for providing technical support; the Plateau d'Imagerie (IFR128) staff and B. Rokytova for imaging assistance; the Plateau de Biologie Expérimentale de la Souris (IFR128) staff for animal care and breeding; and F. Spoutil for helpful discussion and critical reading of the manuscript. This work was supported by the Grant Agency of the Czech Republic (304/07/0223), Ministry of Education, Youth, and Sports of the Czech Republic (MSM0021620843), the Center National pour la Recherche Scientifique, the Ecole Normale Supérieure de Lyon, the Région Rhône-Alpes, the Fondation pour la Recherche Médicale, the “Fondation Singer-Polignac,” the Agriculture and Natural Resources program “Quenottes,” and a grant for supporting project for Strategic Research of Nihon Univiversity School of Dentistry at Matsudo from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT), 2008–2012.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002784107/-/DCSupplemental.
References
- 1.Hall BK. Descent with modification: the unity underlying homology and homoplasy as seen through an analysis of development and evolution. Biol Rev Camb Philos Soc. 2003;78:409–433. doi: 10.1017/s1464793102006097. [DOI] [PubMed] [Google Scholar]
- 2.Müller GB. Vestigial organs and structures. In: Pagel M, editor. Encyclopedia of Evolution. Vol. 2. New York: Oxford University Press; 2002. pp. 1131–1132. [Google Scholar]
- 3.Darwin C. On the Origin of Species. A Facsimile of the First Edition. Cambridge, Massachusetts: Harvard University Press; 1859. Sixteenth printing, 2000. [Google Scholar]
- 4.Walker-Larsen J, Harder LD. Vestigial organs as opportunities for functional innovation: The example of the Penstemon staminode. Evolution. 2001;55:477–487. doi: 10.1554/0014-3820(2001)055[0477:voaoff]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 5.Luckett W, Hartenberger J-L. The Order Rodentia: Major Questions on Their Evolutionary Origin, Relationships and Suprafamilial Systematics. New York: Plenum press; 1985. [Google Scholar]
- 6.Peterková R, Peterka M, Viriot L, Lesot H. Dentition development and budding morphogenesis. J Craniofac Genet Dev Biol. 2000;20:158–172. [PubMed] [Google Scholar]
- 7.Viriot L, Peterková R, Peterka M, Lesot H. Evolutionary implications of the occurrence of two vestigial tooth germs during early odontogenesis in the mouse lower jaw. Connect Tissue Res. 2002;43:129–133. doi: 10.1080/03008200290001168. [DOI] [PubMed] [Google Scholar]
- 8.Peterková R, Peterka M, Viriot L, Lesot H. Development of the vestigial tooth primordia as part of mouse odontogenesis. Connect Tissue Res. 2002;43:120–128. doi: 10.1080/03008200290000745. [DOI] [PubMed] [Google Scholar]
- 9.Viriot L, et al. The presence of rudimentary odontogenic structures in the mouse embryonic mandible requires reinterpretation of developmental control of first lower molar histomorphogenesis. Int J Dev Biol. 2000;44:233–240. [PubMed] [Google Scholar]
- 10.Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- 11.Grüneberg H. Genes and genotypes affecting the teeth of the mouse. J Embryol Exp Morphol. 1965;14:137–159. [PubMed] [Google Scholar]
- 12.Klein OD, et al. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohazama A, et al. Primary cilia regulate Shh activity in the control of molar tooth number. Development. 2009;136:897–903. doi: 10.1242/dev.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohazama A, et al. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS ONE. 2008;3:e4092. doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterkova R, et al. Revitalization of a diastemal tooth primordium in Spry2 null mice results from increased proliferation and decreased apoptosis. J Exp Zoolog B Mol Dev Evol. 2009;312B:292–308. doi: 10.1002/jez.b.21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterková R, Lesot H, Viriot L, Peterka M. The supernumerary cheek tooth in tabby/EDA mice—a reminiscence of the premolar in mouse ancestors. Arch Oral Biol. 2005;50:219–225. doi: 10.1016/j.archoralbio.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Charles C, et al. Distinct impacts of Eda and Edar loss of function on the mouse dentition. PLoS One. 2009;4:e4985. doi: 10.1371/journal.pone.0004985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristenová-Cermáková P, Peterka M, Lisi S, Lesot H, Peterková R. Postnatal lower jaw dentition in different phenotypes of tabby mice. Connect Tissue Res. 2002;43:283–288. doi: 10.1080/03008200290000727. [DOI] [PubMed] [Google Scholar]
- 19.Mustonen T, et al. Ectodysplasin A1 promotes placodal cell fate during early morphogenesis of ectodermal appendages. Development. 2004;131:4907–4919. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- 20.Pispa J, et al. Tooth patterning and enamel formation can be manipulated by misexpression of TNF receptor Edar. Dev Dyn. 2004;231:432–440. doi: 10.1002/dvdy.20138. [DOI] [PubMed] [Google Scholar]
- 21.Tucker AS, Headon DJ, Courtney JM, Overbeek P, Sharpe PT. The activation level of the TNF family receptor, Edar, determines cusp number and tooth number during tooth development. Dev Biol. 2004;268:185–194. doi: 10.1016/j.ydbio.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Kangas AT, Evans AR, Thesleff I, Jernvall J. Nonindependence of mammalian dental characters. Nature. 2004;432:211–214. doi: 10.1038/nature02927. [DOI] [PubMed] [Google Scholar]
- 23.Kassai Y, et al. Regulation of mammalian tooth cusp patterning by ectodin. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- 24.Thesleff I, Keränen S, Jernvall J. Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation. Adv Dent Res. 2001;15:14–18. doi: 10.1177/08959374010150010401. [DOI] [PubMed] [Google Scholar]
- 25.Fraser GJ, Graham A, Smith MM. Conserved deployment of genes during odontogenesis across osteichthyans. Proc Biol Sci. 2004;271:2311–2317. doi: 10.1098/rspb.2004.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Peterka M, Lesot H, Peterková R. Body weight in mouse embryos specifies staging of tooth development. Connect Tissue Res. 2002;43:186–190. doi: 10.1080/03008200290000673. [DOI] [PubMed] [Google Scholar]
- 28.Butler PM. The ontogeny of molar teeth. Biol Rev Camb Philos Soc. 1956;31:30–70. [Google Scholar]
- 29.Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: Non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463–469. [PubMed] [Google Scholar]
- 30.Liu F, et al. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev Biol. 2008;313:210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucker AS, et al. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development. 2000;127:4691–4700. doi: 10.1242/dev.127.21.4691. [DOI] [PubMed] [Google Scholar]
- 32.Adloff P. Die Entwicklung des Zahnsystems der Säugetiere und des Menschen. 1916. (Berlin, Verlag von Hermann Meusser) [Google Scholar]
- 33.Kükenthal W Űber den Ursprung und die Entwickelung der Säugertierzähne (About the origin and evolution of mammalian teeth) Jenaer Zeitsch Naturwiss. 1892;26:469–489. [Google Scholar]
- 34.Röse C Über die Entstehung und Formveränderungen des menschlichen Molaren (About the formation and shape changes of human molars) Anatomischer Anzeiger. 1892;7:393–421. [Google Scholar]
- 35.Peterkova R, Lesot H, Peterka M. Phylogenetic memory of developing mammalian dentition. J Exp Zoolog B Mol Dev Evol. 2006;306:234–250. doi: 10.1002/jez.b.21093. [DOI] [PubMed] [Google Scholar]
- 36.Kavanagh KD, Evans AR, Jernvall J. Predicting evolutionary patterns of mammalian teeth from development. Nature. 2007;449:427–432. doi: 10.1038/nature06153. [DOI] [PubMed] [Google Scholar]
- 37.Renvoisé E, et al. Evolution of mammal tooth patterns: New insights from a developmental prediction model. Evolution. 2009;63:1327–1340. doi: 10.1111/j.1558-5646.2009.00639.x. [DOI] [PubMed] [Google Scholar]
- 38.Huysseune A, Witten PE. Developmental mechanisms underlying tooth patterning in continuously replacing osteichthyan dentitions. J Exp Zoolog B Mol Dev Evol. 2006;306:204–215. doi: 10.1002/jez.b.21091. [DOI] [PubMed] [Google Scholar]
- 39.Dassule HR, McMahon AP. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev Biol. 1998;202:215–227. doi: 10.1006/dbio.1998.8992. [DOI] [PubMed] [Google Scholar]
- 40.Cobourne MT, Miletich I, Sharpe PT. Restriction of sonic hedgehog signalling during early tooth development. Development. 2004;131:2875–2885. doi: 10.1242/dev.01163. [DOI] [PubMed] [Google Scholar]
- 41.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 42.Miyake T, Cameron AM, Hall BK. Detailed staging of inbred C57BL/6 mice between Theiler's [1972] stages 18 and 21 (11–13 days of gestation) based on craniofacial development. J Craniofac Genet Dev Biol. 1996;16:1–31. [PubMed] [Google Scholar]
- 43.Echelard Y, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 44.Laurikkala J, et al. TNF signaling via the ligand-receptor pair ectodysplasin and edar controls the function of epithelial signaling centers and is regulated by Wnt and activin during tooth organogenesis. Dev Biol. 2001;229:443–455. doi: 10.1006/dbio.2000.9955. [DOI] [PubMed] [Google Scholar]
- 45.Merlo GR, et al. The Dlx5 homeobox gene is essential for vestibular morphogenesis in the mouse embryo through a BMP4-mediated pathway. Dev Biol. 2002;248:157–169. doi: 10.1006/dbio.2002.0713. [DOI] [PubMed] [Google Scholar]
- 46.Lesot H, et al. Mouse molar morphogenesis revisited by three-dimensional reconstruction. II. Spatial distribution of mitoses and apoptosis in cap to bell staged first and second upper molar teeth. Int J Dev Biol. 1996;40:1017–1031. [PubMed] [Google Scholar]
- 47.Hu B, Nadiri A, Bopp-Küchler S, Perrin-Schmitt F, Lesot H. Dental epithelial histomorphogenesis in vitro. J Dent Res. 2005;84:521–525. doi: 10.1177/154405910508400607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.