Abstract
The binding of killer cell Ig-like Receptors (KIR) to their Class I MHC ligands was shown previously to be characterized by extremely rapid association and dissociation rate constants. During experiments to investigate the biochemistry of receptor–ligand binding in more detail, the kinetic parameters of the interaction were observed to alter dramatically in the presence of Zn2+ but not other divalent cations. The basis of this phenomenon is Zn2+-induced multimerization of the KIR molecules as demonstrated by BIAcore, analytical ultracentrifugation, and chemical cross-linking experiments. Zn2+-dependent multimerization of KIR may be critical for formation of the clusters of KIR and HLA-C molecules, the “natural killer (NK) cell immune synapse,” observed at the site of contact between the NK cell and target cell.
One of the major influences regulating the behavior of human natural killer (NK) cells is the interaction of killer cell Ig-related receptors (KIR) with their Class I MHC ligands. KIR ligation may either inhibit or activate the lytic machinery of the NK cell (1, 2) reflecting the diversity of these molecules in their transmembrane and intracellular portions. Inhibitory KIR have a long cytoplasmic tail that contains two copies of an immune tyrosine-based inhibitory motif (ITIM; ref. 3). Binding of an inhibitory KIR to its class I MHC ligand results in phosphorylation of the tyrosines in the ITIM units and recruitment of the protein tyrosine phosphatase SHP-1, initiating inhibition of the NK cell cytotoxic process (4). In contrast, activating KIR have a short cytoplasmic tail with a lysine residue in the transmembrane domain through which they associate with the positively signaling KARAP/DAP12 molecule that contains an immune tyrosine-based activation motif (5, 6). Ligation of a short-tailed KIR results in activation of the NK cell (2). Despite these marked differences in signaling potential, the inhibitory and activating KIR are highly homologous in their extracellular portions (7), and in every case examined thus far it seems that the activating receptors bind to HLA-C proteins with a much lower affinity than their inhibitory counterparts (8–10).
Many KIR2D molecules interact specifically with HLA-C alleles, and these receptors can be divided in two groups: receptors belonging to the NK1 group (e.g., KIR2DL1) recognize HLA-Cw2, -Cw4, -Cw6, and related alleles; and receptors belonging to the NK2 group (e.g., KIR2DL3 and KIR2DS2) recognize HLA-Cw1, -Cw3, -Cw7, and related alleles (11). Many KIR2D molecules also are able to bind zinc through a zinc-binding motif in their extracellular domains (12, 13), and Zn2+ binding has been shown to be important for KIR inhibitory function (13). However, KIR2D molecules can bind to HLA-C in a Zn2+-independent manner (13–15). Thus the role of Zn2+ in the biology of the KIR family of molecules is unclear. Another divalent cation, cobalt, has been shown recently to induce stable dimerization of soluble KIR2DL1, although as discussed by those authors the physiological significance of this observation is unclear (16).
Previous studies of the interaction between KIR2D molecules and their HLA-C ligands using soluble recombinant proteins and surface plasmon resonance technology have shown that the specificity of this interaction could be reproduced in vitro. These experiments also revealed that the association and dissociation rate constants of this interaction are very fast; the half-life of the complexes is less than one second (15, 17).
Here, experiments studying the detailed biochemistry of the KIR2D–HLA-C interaction are presented. The addition of Zn2+ to the reaction affected the kinetic parameters of this interaction in a dramatic manner. In the presence of Zn2+ but not other divalent cations such as Mg2+ or Ni2+, the association and dissociation phases of KIR2D binding to HLA-C were altered by the appearance of slowly interacting components alongside the fast interacting species. The molecular mechanism for this phenomenon has been explored also. Zn2+ binding drove KIR multimerization as demonstrated in vitro by BIAcore, analytical ultracentrifugation (AUC) and chemical cross-linking experiments.
Experimental Procedures
Materials.
Chloride and sulfate salts of the various divalent cations were purchased from Sigma. Peptides were synthesized by the Biopolymers Laboratory at Harvard Medical School (Cambridge, MA). Monoclonal antibody HP3E4 (18) was a gift of M. López-Botet (Universitat Pompeu Fabra, Barcelona, Spain). Monoclonal antibody EB6 (19) was purchased from Serotec.
Preparation of Recombinant Proteins.
Soluble KIR2D and HLA-C complexes were produced, refolded, and purified as described (9, 15). The recombinant KIR proteins used in the previous work had a histidine tag at the C terminus. For these experiments, new constructs without a histidine tag were prepared by using site-directed mutagenesis to introduce a stop codon before the sequence encoding the histidine tag using the QuikChange Mutagenesis kit (Stratagene) with the primer 5′-GGTAACCCCAGACACCTTCATTAGCACCATTAAAAGCTTGC-3′ and its complement.
Surface Plasmon Resonance.
Surface plasmon resonance experiments were done by using a BIAcore 2000 instrument (BIAcore, Piscataway, NJ) as described (15), modified by using Hepes buffered saline with 0.005% surfactant without EDTA (HBS-P, BIAcore) as the running buffer. For the experiments with divalent cations, the running buffer included from 10 to 100 μM MgCl2 or ZnCl2. The same results were obtained by using MgSO4 and ZnSO4. Analytes were injected simultaneously over the test and control (no protein immobilized) surfaces by using the multichannel flow option at a flow rate of 30 μl/min. The data-collection rate was set as high as possible. When divalent cations were included in the running buffer, washes with HBS buffer containing 3 mM EDTA were performed between injections of analyte.
Analysis of interaction kinetics was carried out by using BIAEVALUATION 3.0 software (BIAcore).
AUC.
AUC experiments were carried out by using a Beckman Coulter Optima XL-1 analytical ultracentrifuge. Proteins in the presence of 100 μM ZnCl2, 100 μM MgCl2, or no added ion were centrifuged to equilibrium in runs carried out at 5°C. These experiments were carried out at a range of concentrations and over a range of speeds varying from 12,000 to 25,000 rpm. The runs were analyzed by using ORIGIN 4.0 software (Beckman Coulter). Partial specific volume of solute was calculated from amino acid sequence and the densities were calculated by using the SEDNTERP program (20).
Chemical Cross-Linking Studies.
These experiments were carried out by using the bisimidoester dimethyl suberimidate (DMS) as the cross linker. Soluble proteins were prepared at a range of concentrations (50–200 μg/ml) in 100 mM triethanolamine (TEA; pH 8.5). To these solutions was added a 1:10 volume of freshly prepared DMS (11 mg/ml in TEA), and a T = 0 sample was collected. The reaction then was incubated at room temperature; at the indicated times, aliquots were removed and placed on ice, and the reaction was quenched by addition of 1:20 volume 1 M glycine. Proteins were precipitated by addition of an equal volume of 50% trichloroacetic acid and incubation on ice. Precipitated material was recovered by centrifugation at 4°C, washed once with ice-cold acetone/10 mM HCl, and washed again with ice-cold acetone. Then the samples were dissolved in SDS/PAGE sample buffer with protease inhibitors (leupeptin and pepstatin A) and analyzed by SDS/PAGE on 10% gels and western blotting with KIR-reactive mAbs.
Results
KIR2D Bind Zn2+.
To investigate whether a KIR–Zn2+ interaction could be detected in the BIAcore system, buffer supplemented with different divalent cations was injected over a chip containing the extracellular portions of the NK receptors KIR2DL1, KIR2DL3, and a control surface with no protein immobilized on it. Magnesium, manganese, nickel, and zinc salts were tested, but only Zn2+ showed binding to KIR over the control surface. Fig. 1 shows the binding of 100 μM Zn2+ to both KIR2DL1 and KIR2DL3, but it also was possible to detect a Zn2+–KIR interaction at 10 μm Zn2+ (data not shown). Fig. 1 illustrates two points: (i) the Zn2+–KIR interaction occurs only transiently, and (ii) the change in RU observed after injection of Zn2+ over the various KIR molecules is greater than would be expected when the Mr of Zn2+ is considered. This discrepancy may reflect an induced change in density of the KIR-coated dextran matrix (see Discussion). The interaction of Zn2+ with KIR molecules was specific, because no binding was seen when Zn2+ was injected through flow cells coated with either an mAb or peptide–HLA complexes (data not shown).
Figure 1.
Zn2+ binding to KIR2D molecules. Buffer without EDTA (containing 100 μM Zn2+) was injected over KIR2DL1 [910 resonance units (RU)] and KIR2DL3 (1066 RU). The control flow cell had no protein immobilized on it.
The Kinetics of KIR2D Binding to HLA-C Are Slower in the Presence of Zn2+ but Not Mg2+.
The effect of inclusion of divalent cations on the interaction between HLA-C and KIR2D was studied next. Because the “standard” BIAcore running buffer contains 3 mM EDTA, this new set of experiments was done in EDTA-free running buffer. Omission of EDTA from the running buffer did not affect the very fast kinetics of the interaction between the HLA-C and KIR2D molecules (data not shown). Fig. 2 shows a comparison of increasing concentrations of HLA-C injections over three different soluble KIR molecules in the presence of either Mg2+ or Zn2+. The characteristics of the interaction did not change substantially in the presence of Mg2+ ions: the recognition of HLA-C by KIR2D still had very fast association and dissociation rates, and the specificity of binding of HLA-Cw6 to KIR2DL1 and of HLA-Cw7 to KIR2DL3 and KIR2DS2 was maintained (Fig. 2 A and B). In contrast, inclusion of Zn2+ in the running buffer altered the association and dissociation phases of all of these interactions such that they became a mixture of fast and slow rate components (Fig. 2 C and D). However, Zn2+ had a stronger effect on the HLA-Cw6–KIR2DL1 interaction than on that of HLA-Cw7 with KIR2DL3 and KIR2DS2.
Figure 2.
Binding of HLA-C to KIR2DL in the presence of either injected over KIR2D receptors. Increasing concentrations (0.5 mg/ml and 1 mg/ml) of HLA-Cw6 (A and C) and HLA-Cw7 (B and D) were injected over KIR2DL1 (668 RU), KIR2DL3 (644 RU), and KIR2DS2 (608 RU) in the presence of 100 μM Mg2+ (A and B) and 100 μM Zn2+ (C and D) in the running buffer. The space in between the curves in C and D are caused by the elimination of an EDTA wash performed between injections.
Interestingly, in the presence of Zn2+, these receptors also displayed a small but highly reproducible degree of cross-reactive binding (compare the binding of KIR2DL3 and KIR2DS2 to HLA-Cw6 in Fig. 2 A and C). Moreover, inspection of the sensorgrams shows that the bulk of the cross-reactive binding had slow association/dissociation kinetics.
The reverse experiment, HLA-C molecules immobilized on the chip surface and NK receptors injected through the various flow cells, also was performed, and again slow components appeared in the association and dissociation phases of the KIR2D–HLA-C interaction in the presence of Zn2+ but not other divalent cations (data not shown).
In an attempt to characterize the altered kinetics of these interactions in the presence of Zn2+, titrations of both the amount of Zn2+ in the running buffer and the concentration of HLA-C ligand injected were done. Fig. 3 shows the interaction between HLA-Cw6 and KIR2DL1 (A–C) and between HLA-Cw7 and KIR2DL3 (D–F) in the absence of cations and in the presence of 10 μM and 100 μM of Zn2+. The general shape of the curves revealed that the observed on and off rates of the interaction between the NK receptor and the HLA-C molecule were much slower in the presence of 100 μM Zn2+ than in the presence of 10 μM Zn2+ or in the absence of the ion, but analysis of these data has proved to be extremely complex and difficult. What is clear is that HLA-C remained bound to KIR for a longer period at the end of the injection. If the slow-dissociation part of the curve is analyzed separately, then an observed koff can be derived, which although subject to error, gives estimates for the t1/2 of the KIR2DL1–HLA-Cw6 and the KIR2DL3–HLA-Cw7 complexes in the presence of 100 μM Zn2+ to be ≈ 200 and 60 sec, respectively. Similar analyses of the “reverse-experiments” gave values in good agreement with these estimates.
Figure 3.
Increasing concentrations of HLA-C injected over KIR2DL in the presence of 10μM, 100μM, or no Zn2+. (A–C) HLA-Cw6 (0.1, 0.25, 0.5, and 1 mg/ml) was injected over KIR2DL1 (910 RU). (D–F) HLA-Cw7 (0.1, 0.25, 0.5, and 1 mg/ml) was injected over KIR2DL3 (1066 RU). The control flow cell (no protein immobilized) has been subtracted from the experimental flow cells.
These experiments were done at Zn2+ concentrations of 10 and 100 μM, because serum and blood concentrations of Zn2+ are in the range of 10–120 μM, respectively. Precise estimations of the amount of available Zn2+ are difficult to make, because the majority of Zn2+ in blood is associated with albumin and other proteins (21, 22). At higher, nonphysiological concentrations of Zn2+, the slow components of the binding became even more marked (data not shown).
KIR2D Are Able to Self-Associate.
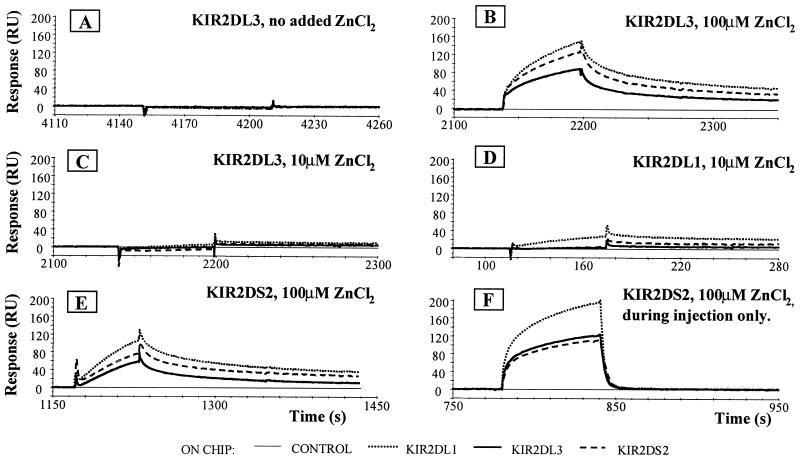
To test whether KIR2D molecules were able to interact among themselves in the presence of Zn2+, BIAcore experiments involving injections of NK receptors over NK receptors were performed. Strikingly, all of the KIR tested bound to the KIR immobilized on the BIAcore chip in the presence of Zn2+ but not in the absence of the cation.
Fig. 4A shows the binding of KIR2DL3, injected in the absence of Zn2+, over a chip comprising a control surface followed by immobilized KIR2DL1, KIR2DL3, and KIR2DS2. No binding was observed. Fig. 4B shows a repeat KIR2DL3 injection over the same chip, but this time with the presence of 100 μM Zn2+ in the running buffer. At this concentration of Zn2+, KIR2DL3 interacted with the extracellular portions of both inhibitory and activating forms of KIR2D molecules. At 10 μM Zn2+, only very weak interactions of KIR2DL3 with other KIR could be seen (Fig. 4C). In the presence of 100 μM Zn2+, KIR2DL1 behaved similarly to KIR2DL3 in that this receptor interacted with all KIR tested (data not shown). In contrast, at 10 μM Zn2+ self-association of KIR2DL1 was obvious still, although only weak interactions with KIR2DL3 or KIR2DS2 molecules were seen (Fig. 4D). These data are consistent with Fig. 2: KIR2DL1 was more susceptible to Zn2+-induced multimerization than was KIR2DL3. These experiments also revealed that KIR multimerization strictly depended on the presence of Zn2+ and was reversible. This is illustrated best by comparison of Fig. 4 E and F. As expected from previous data, KIR2DS2 was able to interact with both KIR2DL1 and KIR2DL3 as well as with itself. Further, it also can be seen that when the experiment is done with 100 μM Zn2+ in the running buffer, the dissociation of the KIR proteins from each other was slow (Fig. 4E). In contrast, when 100 μM Zn2+ was present only during the time of the injection, the complexes of KIR molecules fell apart extremely rapidly at the end of the injection as the Zn2+, which binds only transiently to the KIR, was washed away (Fig. 4F).
Figure 4.
Self-association of KIR2D in the presence of Zn2+. (A) KIR2DL3 (0.4 mg/ml) was injected over KIR2DL1 (910 RU), KIR2DL3 (1066 RU), or KIR2DS2 (1187 RU) in running buffer without any cations. (B) KIR2DL3 (0.4 mg/ml) was injected over the same surfaces as in A in the presence of 100 μM Zn2+ in the running buffer. (C) KIR2DL3 (0.4 mg/ml) was injected over the same surfaces as in A in the presence of 10 μM Zn2+ in the running buffer. (D) KIR2DL1 (0.4 mg/ml) was injected over the same surfaces as in A in the presence of 10 μM Zn2+ in the running buffer. (E) KIR2DS2 (0.4 mg/ml) was injected over KIR2DL1 (668 RU), KIR2DL3 (644 RU), and KIR2DS2 (608 RU) in the presence of 100 μM Zn2+ in the running buffer. (F) KIR2DS2 (0.4 mg/ml) was injected over the same surfaces as in E in the presence of 100 μM Zn2+, present only during the time of the injection. The difference in RU maximum is probably caused by the additive effect of Zn2+ binding to KIR. The control flow cell (no protein immobilized) has been subtracted from the experimental flow cells.
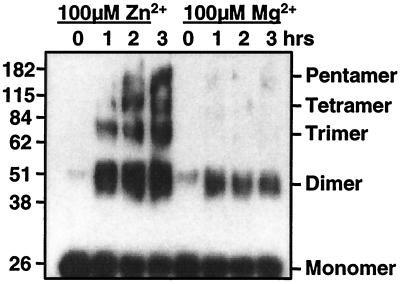
Given that KIR2D molecules are able to interact with each other in the presence of Zn2+, we attempted to determine the degree of KIR multimerization induced in the presence of Zn2+. Initial attempts to analyze the size of the KIR aggregates were made by AUC. In either plain buffer or buffer supplemented with 100 μM Mg2+, the dominant species observed was of Mr 28,000. In the presence of 100 μM Zn2+, the majority of the KIR2DL1 protein spun out of solution and accumulated at the bottom of the centrifuge tube, presumably as large protein aggregates. In contrast, when soluble KIR2DL3 protein was analyzed by AUC, a species of Mr consistent with a KIR2DL3 dimer was observed in the presence of Zn2+ but not Mg2+. Zn2+-induced aggregation of KIR also has been analyzed by chemical cross linking of the KIR proteins and SDS/PAGE after exposure to either Zn2+ or control cations. These experiments have provided evidence for the existence of a variety of KIR multimers ranging in size from dimers to, at least, pentamers. Fig. 5 shows an example of this type of experiment done with soluble KIR2DL1 protein. Even at time 0, a dimer of KIR2DL1 protein can be detected, and with time, dimer and higher Mr species of KIR multimer accumulate in the presence of Zn2+. The dimer band detected in the presence of Mg2+ can be detected also in the absence of cation (data not shown), and higher Mr species of KIR do not accumulate with time. This experiment was done over several hours in the presence of 100 μM Zn2+, but Zn2+-induced multimerization can be observed after only 10 min of incubation and with as little as 10 μM cation (data not shown). Similar data have been obtained with KIR2DL3 and KIR2DS2 proteins, confirming the BIAcore data showing that Zn2+ can induce multimerization of multiple KIR2D family members including both inhibitory and activating receptors.
Figure 5.
Cross linking of KIR2DL1 in the presence of Zn2+. Soluble KIR2DL1 proteins (molecular mass, 25 kDa) at 200 μM in triethanolamine (pH 8.5) were incubated with the cross linker in the presence of either 100 μM Zn2+ or 100 μM Mg2+ for the indicated time and then run on 10% SDS/PAGE. After electrophoresis, the KIR proteins were visualized by Western blotting with the mAbs HP3E4 and EB6.
Discussion
This paper describes the effect of Zn2+ on the interaction of KIR2D with HLA-C molecules. Zn2+ binding to KIR2D molecules induced multimerization of KIR proteins. Zn2+-induced clustering of KIR modified substantially the kinetic characteristics of KIR2D–HLA-C binding and influenced the specificity of the interaction between the receptor and its ligand. In the absence of Zn2+, this reaction is a simple first-order interaction with extremely fast association and dissociation rates (15, 17). In contrast, in the presence of Zn2+ the association and dissociation phases of the KIR2D–HLA-C interaction became a mixture of fast and slow rate components.
Initially, however, it was not obvious that the basis of this slow component of the KIR–HLA-C interaction was KIR multimerization. Evidence that Zn2+ could be inducing association of the immobilized KIR proteins came from inspection of the curves obtained after injection of Zn2+ over a chip on which various KIR proteins had been immobilized. The Mr of Zn2+ is too low for a BIAcore to detect direct binding of Zn2+ to KIR. Thus, the amplitude of the change in RU observed after injection of Zn2+ over the various KIR molecules was surprising and suggested that a change in the organization of the KIR molecules attached to the dextran matrix was being induced by Zn2+. In light of subsequent experiments, the hypothesis would be that Zn2+-induced aggregation of KIR was occurring, altering the physical properties of the dextran matrix. Self-association of proteins while immobilized on a BIAcore chip surface has been observed previously (23).
Definitive evidence for multimerization of KIR was obtained by using BIAcore, AUC (data not shown), and chemical cross-linking experiments. These data revealed that in the presence of Zn2+, KIR molecules in solution formed aggregates ranging from dimers to, at least, pentamers. Even in the absence of added Zn2+, dimers of soluble KIR proteins could be detected; however, this species must be only a very small proportion of the KIR present in the preparations used in these experiments, because the dimer species was not visible on gel-filtration analysis of the KIR proteins. Further, the standard Langmuir 1:1 binding model fits very well to the equilibrium and kinetic data obtained on analysis of the KIR2D–HLA-C interaction in the absence of Zn2+ or the presence of Mg2+ (15, 17). Perhaps KIR have a weak tendency to self-associate that is enhanced markedly in the presence of Zn2+. Certainly, the formation of large aggregates of KIR depends on the presence of Zn2+, e.g., at the concentrations of KIR used (8–32 μM), KIR–KIR association can only be detected on the BIAcore in the presence of Zn2+. Moreover, the AUC analyses indicated that in the presence of sufficient Zn2+, but not Mg2+, essentially all of a given sample of KIR2DL1 aggregated with time. This conclusion was backed up by the chemical cross-linking experiments, in which the proportion of KIR present as multimers again increased with time.
A striking feature of KIR multimerization induced by Zn2+ is that different members of the KIR2D family differ in their tendencies to self-associate in the presence of Zn2+. The interaction of KIR2DL1 with HLA-Cw6 was markedly more affected by the presence of Zn2+ than the interaction of either KIR2DL3 or KIR2DS2 with HLA-Cw7. These data correlate with the BIAcore data shown in Fig. 4, in which KIR2DL1 showed a greater tendency to associate in the presence of Zn2+ than either KIR2DL3 or KIR2DS2. The basis of this difference is unclear. The HEGVH sequence, found at the amino terminus of many KIR2D molecules and important for Zn2+ binding (13), is conserved between KIR2DL1, KIR2DL3, and KIR2DS2. Thus other residues of the KIR molecules, presumably some of the 13 dimorphic residues that vary between KIR2DL1 and KIR2DL3, must determine their susceptibility to Zn2+-induced multimerization. This effect might be mediated at either the level of Zn2+ binding to the receptor or receptor–receptor association.
Another feature of note is that Zn2+ not only induced self-association of KIR but also induced association between inhibitory KIR2D molecules of different HLA-C specificities, e.g., KIR2DL1 with KIR2DL3, and association of inhibitory KIR with activating KIR, e.g., KIR2DL1 with KIR2DS2. The functional consequences of these phenomena remain to be investigated, but it is worth noting that the majority of the human population is heterozygous at HLA-C. An ability to integrate the action of NK receptors with different HLA-C specificities might well be important for KIR function in vivo.
Having established that Zn2+ can induce multimerization of KIR in vitro, is there any evidence that Zn2+ is important for KIR function in vivo? KIR molecules with mutations in the putatively Zn2+-binding HEGVH motif were shown to be impaired in their ability to deliver inhibitory signals but equally as competent as wild-type KIR to bind HLA-C (13). Similarly, treatment of NK cells with the divalent cation chelator 1,10-phenanthroline reduced both KIR2D-mediated inhibition of target-cell lysis (12, 13) and clustering of HLA-C at the NK cell–target cell interface induced by the presence of inhibitory KIR molecules (24), although these data are subject to the caution that treatment of cells with an inhibitor such as 1,10-phenanthroline affects multiple pathways, not just the KIR2D–HLA-C interaction. These data suggested that Zn2+ mediated some protein–protein interaction important for the inhibitory function of KIR. The data presented in this paper strongly suggest that this interaction is Zn2+-dependent multimerization of KIR proteins. Moreover, these data demonstrate that Zn2+ binding to KIR2D molecules can induce multimerization of both inhibitory and activating KIR proteins independent of their HLA-C specificity, and indeed in the absence of their class I MHC ligand.
How might KIR aggregation in the presence of Zn2+ affect KIR2D function? It has not proved possible to analyze precisely the altered kinetics of KIR2D binding to HLA-C, because these curves are composed of multiple interactions in which the valency and concentration of the KIR reactants are unknowable. That said, comparison of experiments done with Zn2+ present in the running buffer with those in which Zn2+ is only present during the time of injection (data not shown) confirms that the slow component of the dissociation is caused by the effect of Zn2+ (presumably the interaction of KIR multimers with HLA-C), whereas the fast component of the dissociation represents the interaction of the KIR monomer with HLA-C. Thus it is obvious that the half-life of the complex formed between HLA-C and clustered KIR2D molecules would be much longer than that of the complex formed between the monomeric species. This extra stability of the interaction might well be important for a productive inhibitory or activating interaction. It has been shown for T cells that sustained engagement of the T cell antigen receptor with peptide–MHC complex is critical for T cell activation (25).
It is interesting to note also that KIR clustering can induce cross-reactive binding of HLA-C by KIR. Inspection of the BIAcore sensorgrams shows that the bulk of the cross-reactive binding had slow association/dissociation kinetics; presumably; this component is ligand binding by KIR multimers. Given the high degree of homology between KIR, it is perhaps not surprising that a degree of cross reactivity is revealed when the strength of the receptor–ligand interaction is boosted by receptor clustering. Perhaps receptor-clustering and avidity effects are the basis of some of the cross-reactive binding of HLA-C by KIR that has been reported (10, 26).
Finally, it is of some interest to compare the roles of metal ions in KIR function with their role in integrin function. Zn2+ binding induces KIR aggregation, which increases the strength of the receptor–ligand interaction by increasing receptor valency. Metal cation binding by integrins also changes the affinity of receptor–ligand binding but probably by inducing a conformational change of the receptor and not by inducing receptor clustering (27, 28), although the avidity of integrin–ligand binding also can be affected by receptor clustering (28).
Acknowledgments
We thank M. López-Botet for the gift of antibodies. We also thank Dr. P. A. van der Merwe for helpful discussion on the BIAcore experiments, Professor Jean Thomas for advice on chemical cross linking of proteins, and Dr. Peter Leadley for access to the analytical ultracentrifuge. This work was supported by grants from the Wellcome Trust to H.T.R. and the National Institutes of Health (CA-47554) to J.L.S.
Abbreviations
- NK
natural killer
- KIR
killer cell Ig-related receptors
- AUC
analytical ultracentrifugation
- RU
resonance units
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041618298.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041618298
References
- 1.Moretta A, Vitale M, Bottino C, Orengo A M, Morelli L, Augugliaro R, Barbaresi M, Ciccone E, Moretta L. J Exp Med. 1993;178:597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, Bottino C, Moretta L. J Exp Med. 1995;182:875–884. doi: 10.1084/jem.182.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vivier E, Daeron M. Immunol Today. 1997;18:286–291. doi: 10.1016/s0167-5699(97)80025-4. [DOI] [PubMed] [Google Scholar]
- 4.Renard V, Cambiaggi A, Vely F, Blery M, Olcese L, Olivero S, Bouchet M, Vivier E. Immunol Rev. 1997;155:205–221. doi: 10.1111/j.1600-065x.1997.tb00953.x. [DOI] [PubMed] [Google Scholar]
- 5.Olcese L, Cambiaggi A, Semenzato G, Bottino C, Moretta A, Vivier E. J Immunol. 1997;158:5083–5086. [PubMed] [Google Scholar]
- 6.Lanier L L, Corliss B C, Wu J, Leong C, Phillips J H. Nature (London) 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 7.Biassoni R, Cantoni C, Falco M, Verdiani S, Bottino C, Vitale M, Conte R, Poggi A, Moretta A, Moretta L. J Exp Med. 1996;184:645–650. doi: 10.1084/jem.183.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 9.Valés-Gómez M, Reyburn H T, Erskine R A, Strominger J L. Proc Natl Acad Sci USA. 1998;95:14326–14331. doi: 10.1073/pnas.95.24.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter C C, Gumperz J E, Parham P, Long E O, Wagtmann N. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 11.Colonna M, Borsellino G, Falco M, Ferrara G B, Strominger J L. Proc Natl Acad Sci USA. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajagopalan S, Winter C C, Wagtmann N, Long E O. J Immunol. 1995;155:4143–4146. [PubMed] [Google Scholar]
- 13.Rajagopalan S, Long E O. J Immunol. 1998;161:1299–1305. [PubMed] [Google Scholar]
- 14.Fan Q O R, Garboczi D N, Winter C C, Wagtmann N, Long E O, Wiley D C. Proc Natl Acad Sci USA. 1996;93:7178–7183. doi: 10.1073/pnas.93.14.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valés-Gómez M, Reyburn H T, Mandelboim M, Strominger J L. Immunity. 1998;9:337–344. doi: 10.1016/s1074-7613(00)80616-0. , and erratum (1998) 9, 892. [DOI] [PubMed] [Google Scholar]
- 16.Fan Q R, Long E O, Wiley D C. J Biol Chem. 2000;275:23700–23706. doi: 10.1074/jbc.M003318200. [DOI] [PubMed] [Google Scholar]
- 17.Maenaka K, Juji T, Nakayama T, Wyer J R, Gao G F, Maenaka T, Zaccai N R, Kikuchi A, Yabe T, Tokunaga K, et al. J Biol Chem. 1999;274:28329–28334. doi: 10.1074/jbc.274.40.28329. [DOI] [PubMed] [Google Scholar]
- 18.Melero I, Salmeron A, Balboa M A, Aramburu J, López-Botet M. J Immunol. 1994;152:1662–1673. [PubMed] [Google Scholar]
- 19.Moretta A, Bottino C, Pende D, Tripodi G, Tambussi G, Viale O, Orengo A, Barbaresi M, Merli A, Ciccone E, et al. J Exp Med. 1990;172:1589–1598. doi: 10.1084/jem.172.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laue T M, Stafford W F., III Annu Rev Biophys Biomol Struct. 1999;28:75–100. doi: 10.1146/annurev.biophys.28.1.75. [DOI] [PubMed] [Google Scholar]
- 21.Harrison T R. Harrison's Principles of Internal Medicine. New York: McGraw–Hill; 1987. [Google Scholar]
- 22.Wellinghausen N, Rink L. J Leukocyte Biol. 1998;64:571–577. doi: 10.1002/jlb.64.5.571. [DOI] [PubMed] [Google Scholar]
- 23.Mavaddat N, Mason D W, Atkinson P D, Evans E J, Gilbert R J, Stuart D I, Fennelly J A, Barclay A N, Davis S J, Brown M H. J Biol Chem. 2000;275:28100–28109. doi: 10.1074/jbc.M004117200. [DOI] [PubMed] [Google Scholar]
- 24.Davis D M, Chiu I, Fassett M, Cohen G B, Mandelboim O, Strominger J L. Proc Natl Acad Sci USA. 1999;96:15062–15067. doi: 10.1073/pnas.96.26.15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valitutti S, Dessing M, Aktories K, Gallati H, Lanzavecchia A. J Exp Med. 1995;181:577–584. doi: 10.1084/jem.181.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajagopalan S, Long E O. J Exp Med. 1997;185:1523–1528. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart M, Hogg N. J Cell Biochem. 1996;61:554–561. doi: 10.1002/(sici)1097-4644(19960616)61:4<554::aid-jcb8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 28.Stewart M P, McDowall A, Hogg N. J Cell Biol. 1998;140:699–707. doi: 10.1083/jcb.140.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]