Abstract
The pore and gate regions of voltage-gated cation channels have been often targeted with drugs acting as channel modulators. In contrast, the voltage-sensing domain (VSD) was practically not exploited for therapeutic purposes, although it is the target of various toxins. We recently designed unique diphenylamine carboxylates that are powerful Kv7.2 voltage-gated K+ channel openers or blockers. Here we show that a unique Kv7.2 channel opener, NH29, acts as a nontoxin gating modifier. NH29 increases Kv7.2 currents, thereby producing a hyperpolarizing shift of the activation curve and slowing both activation and deactivation kinetics. In neurons, the opener depresses evoked spike discharges. NH29 dampens hippocampal glutamate and GABA release, thereby inhibiting excitatory and inhibitory postsynaptic currents. Mutagenesis and modeling data suggest that in Kv7.2, NH29 docks to the external groove formed by the interface of helices S1, S2, and S4 in a way that stabilizes the interaction between two conserved charged residues in S2 and S4, known to interact electrostatically, in the open state of Kv channels. Results indicate that NH29 may operate via a voltage-sensor trapping mechanism similar to that suggested for scorpion and sea-anemone toxins. Reflecting the promiscuous nature of the VSD, NH29 is also a potent blocker of TRPV1 channels, a feature similar to that of tarantula toxins. Our data provide a structural framework for designing unique gating-modifiers targeted to the VSD of voltage-gated cation channels and used for the treatment of hyperexcitability disorders.
Keywords: M-current, ion channel, KCNQ, opener
Voltage-sensitive cation channels play crucial roles in brain and cardiac excitability. These channels are endowed with two main transmembrane modules, a voltage-sensing domain (VSD) and a pore domain. Mutations of ion channel genes in humans lead to severe inherited neurological, cardiovascular, or metabolic disorders, called “channelopathies” (1). So far, the medicinal toolbox has focused on the pore domain and its gate in an attempt to cure ion channel-related dysfunctions by channel blockers or openers (2).
In contrast, the VSD of voltage-gated cation channels was virtually not exploited for therapeutic purposes. VSDs are found in voltage-dependent cation channels and other voltage-regulated proteins (3). In voltage-gated cation channels, the linker S4–S5 of the VSD serves as an electromechanical coupling device, which opens the channel pore. VSDs have also been recently characterized in voltage-regulated proteins that lack associated ion channel pores (4–6). A voltage-sensitive phosphatase, Ci-VSP, has a VSD that is coupled to a phosphatase domain (4). In the human voltage-activated proton channel (Hv1), the VSD itself functions as a proton channel (5–7).
Crystallographic studies of voltage-gated K+ channels (Kv) have described the VSD architecture in its open-state conformation. It forms a module of four membrane-spanning segments (S1–S4) with the S3b helix and the charge-bearing S4 helix forming a helix-turn-helix structure, termed the “paddle motif,” which is buried in the membrane and moves at the protein–lipid interface (8, 9). Recent data indicate that the VSD paddle motif can drive channel opening when transplanted from the archaebacterial Kv channel or VSD proteins (Hv1 and Ci-VSP) into eukaryotic Kv channels (3, 10). Thus, VSD paddle motifs are modular and transferable structures, whose functions are conserved in voltage-sensing proteins (3).
In our quest to target neuronal M-type K+ channels, encoded by Kv7.2/Kv7.3 subunit assembly, we recently designed unique diphenylamine carboxylates that are powerful Kv7.2 channel openers or blockers (11). M-channels generate subthreshold, noninactivating voltage-gated K+ currents that play an important role in controlling neuronal excitability (12, 13). Here we show that the unique opener compound, NH29, acts as a nontoxin gating modifier, which robustly increases Kv7.2 K+ currents. Experimental and modeling data suggest that in the Kv7.2 channel open state, NH29 docks to the external surface of the VSD at the interface of helices S1, S2, and S4, where it stabilizes the interaction between two conserved residues in S2 and S4, known to interact electrostatically in Kv channels. Intriguingly, NH29 is also a potent blocker of TRPV1 channels, a feature similar to that described for tarantula toxins (14, 15). Results indicate that NH29 may operate via a voltage-sensor trapping mechanism similar to that depicted for various gating-modifier toxins (16). Thus, the VSD represents a unique and important pharmacological target for the treatment of major hyperexcitability disorders, like epilepsy, migraine, or neuropathic pain.
Results
Compound NH29 Increases Kv7.2 Currents and Inhibits Neuronal-Spike Discharge and Neurotransmitter Release.
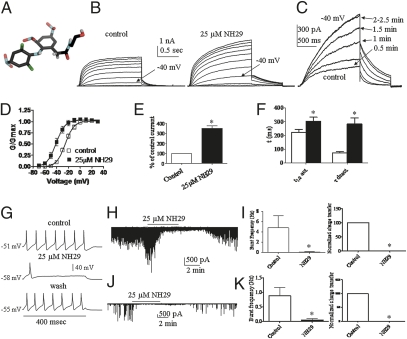
We recently designed unique derivatives of diphenylamine carboxylate, which act as potent M-channel openers or blockers (11). Along this series, we characterized the pharmacological properties and the target site of the Kv7.2 channel opener NH29. The carboxylate group of the diphenylamine diclofenac was derivatized to an amide functionality with ethanolamine, and the aromatic rings were substituted with nitro groups, yielding compound NH29 (Fig. 1A). NH29 was initially examined on WT Kv7.2 homomeric channels expressed in CHO cells. External application of NH29 rapidly increases Kv7.2 K+ currents at threshold potentials. At −40 mV, NH29 (25 μM) enhances by 3.5 ± 0.3–fold Kv7.2 current amplitude (n = 15, P < 0.01) (Fig.1 C and E). However, at positive potentials, the effects of NH29 become much weaker (Fig. 1B) and little change in maximal conductance is observed. The overall increase in WT Kv7.2 current amplitude mainly arises from a hyperpolarizing shift of the activation curve. At 25 μM, NH29 causes a left-shift of −15.5 mV from V50 = −25.4 ± 0.5 mV to V50 = −40.9 ± 0.6 mV (n = 15, P < 0.01) (Fig. 1D and SI Appendix, Table S1), although at 100 μM the shift is −31 mV. In addition, NH29 slows down the kinetics of Kv7.2 activation and more noticeably those of deactivation (Fig. 1F and SI Appendix, Table S1). The NH29-induced slowing of deactivation (4-fold) can be accounted for by only 33% the negative shift in the voltage dependence of activation (1.3-fold slowing for a 15-mV shift), suggesting that NH29 affects Kv7.2 gating by multiple mechanisms.
Fig. 1.
NH29 increases Kv7.2 currents and inhibits neuronal-spike discharge and neurotransmitter release. (A) Chemical structure of NH29. The atom color codes are gray, black, green, red, and blue for hydrogen, carbon, chloride, oxygen, and nitrogen, respectively. (B) Representative Kv7.2 whole-cell currents, in transfected CHO cells, in the absence and presence of 25 μM NH29. Cells, held at −90 mV, were stepped from −70 mV to +30 mV in 10-mV increments and repolarized at −60 mV. (C) Representative trace of a cell, stepped at −40 mV every 30 s in the absence (control) and presence of 25 μM NH29. (D) The normalized conductance was plotted as a function of the test voltages for cells in the absence (open squares) and presence (filled squares) of 25 μM NH29 (n = 15; *P < 0.01). (E) Potentiation of WT Kv7.2 currents recorded at −40 mV in the presence of NH29 expressed as percentage of control in the absence of opener (n = 15; *P < 0.01). (F) Kinetics of Kv7.2 were measured in the absence (open bars) and presence (filled bars) of 25 μM NH29 (n = 15; *P < 0.01). Activation kinetics was evaluated at −40 mV by t1/2, the time at which half of the current amplitude developed. Deactivation kinetics was measured at −60 mV and fitted by one exponential function. (G) Representative rat DRG spiking discharge, evoked by a squared depolarizing current pulse (100 pA for 400 ms) before (control), during exposure to 25 μM NH29 and after washout (wash). Representative traces of spontaneous EPSCs (H) and IPSCs (J) recorded before, during exposure to 25 μM NH29 and after washout. Effects of NH29 on burst frequency and charge transfer of spontaneous EPSCs (I) and IPSCs (K) (n = 4; *P < 0.05).
Next, we examined the selectivity of NH29 on other Kv7 channels (SI Appendix, Fig. S1). NH29 increases the current amplitude of heteromeric Kv7.2/Kv7.3 channels with a similar potency compared with homomeric Kv7.2 channels with an EC50 = 14 ± 2 μM (n = 5). In contrast, NH29 is virtually ineffective on homomeric Kv7.3 channels (SI Appendix, Fig. S1). NH29 weakly increases homomeric Kv7.4 currents (1.3-fold at −40 mV) (SI Appendix, Fig. S1) and does not affect homomeric Kv7.1 and heteromeric Kv7.1/KCNE1 channels (SI Appendix, Fig. S1). NH29 appears to act on Kv7.2 both in the closed and open states. Indeed, when NH29 is first applied for 4 min at −90 mV, a potential where Kv7.2 channels are in the closed state, the opener increases by 3.4 ± 0.3–fold the currents at the first step depolarization to −40 mV, indicating a closed-state interaction (SI Appendix, Fig. S2 and SI Text). In addition, when Kv7.2 channels are first opened by depolarization, subsequent application of NH29 significantly increases the currents by 2.8 ± 0.3–fold, suggesting an open-state interaction (SI Appendix, Fig. S2 and SI Text).
We further characterized the properties of NH29 on central and peripheral neurons. NH29 (25 μM) hyperpolarizes the resting membrane potential (from −56.0 ± 2.0 mV to −65.0 ± 2.2 mV, n = 9; P < 0.01) and reversibly reduces the number of evoked spikes of dorsal root ganglion (DRG) neurons (from 9.0 ± 0.8 to 0.9 ± 0.1, n = 12; P < 0.001) (Fig. 1G). NH29 powerfully depresses synaptic transmission (Fig. 1 H–K); it inhibits glutamate and GABA release, as shown from the depression of network-driven excitatory and inhibitory postsynaptic currents (EPSCs and IPSCs), which were recorded from primary cultures of hippocampal neurons (17). This inhibition is reflected by the reduced charged transfer and frequency of postsynaptic currents (Fig. 1 H–K).
NH29 Does Not Act at the Target Site of Retigabine.
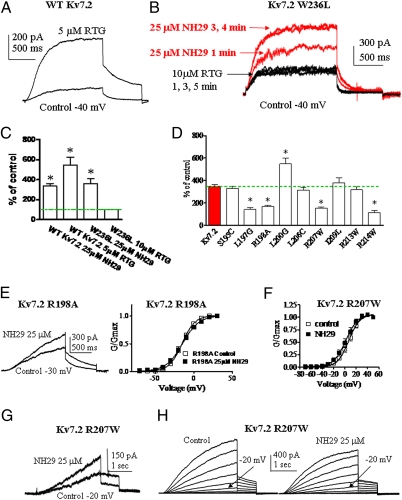
We searched for the Kv7.2 residues involved in NH29 interaction. We previously showed that a parent template of NH29, meclofenamate, exhibits an additive effect with the M-channel opener retigabine (RTG), suggesting that the two drugs act via different mechanisms (18). Thus, we checked whether NH29 interacts with the target site of RTG, which involves a crucial tryptophan residue in S5 (19–21). The Kv7.2 W236L mutant leads to currents insensitive to RTG (20, 21). Although RTG (5 μM) increases by 5.5-fold the current of WT Kv7.2 (at −40 mV), it does not affect that of the Kv7.2 W236L mutant (Fig. 2 A−C). Interestingly, NH29 (25 μM) markedly enhances the current amplitude of this RTG-insensitive mutant W236L (by 3.6-fold at −40 mV) (Fig. 2 B and C) mainly by causing a left-shift of the activation curve from V50 = −26.6 ± 1.7 mV to V50 = −38.9 ± 1.6 mV (n = 4, P < 0.01). These data suggest that NH29 does not act at the target site of RTG in the pore region.
Fig. 2.
Sensitivity of pore and S4 residues to NH29 modulation. (A) Representative trace of a cell expressing WT Kv7.2, stepped at −40 mV in the absence (control) and presence of 5 μM RTG. (B) Representative trace of a cell expressing mutant Kv7.2 W236L, stepped at −40 mV in the absence (control) and presence of 10 μM RTG and subsequently exposed to 25 μM NH29. (C) Potentiation of WT Kv7.2 and mutant W236L currents recorded at −40 mV in the presence of NH29 or RTG expressed as percentage of control (n = 5–8; *P < 0.01). (D) Potentiation of the current produced by 25 μM NH29 on WT Kv7.2 (red bar) and the different S4 mutants, expressed as percent of control determined in the absence of the opener (n = 6–15; *P < 0.01). (E) Representative trace of a cell expressing mutant Kv7.2 R198A, stepped at −20 mV in the absence (control) and presence of 25 μM NH29 (Left) and normalized conductance (Right) in the absence (open squares) and presence (filled squares) of 25μM NH29 (n = 8). (F) Normalized conductance of mutant Kv7.2 R207W in the absence (open squares) and presence (filled squares) of 25 μM NH29 (n = 8). (G) Representative trace of a cell expressing mutant Kv7.2 R207W, stepped at −20 mV in the absence (control) and presence of 25 μM NH29. (H) Representative Kv7.2 R207W currents recorded as in Fig. 1B, in the absence and presence of 25 μM NH29.
Residues Involved in Kv7.2 Modulation by NH29.
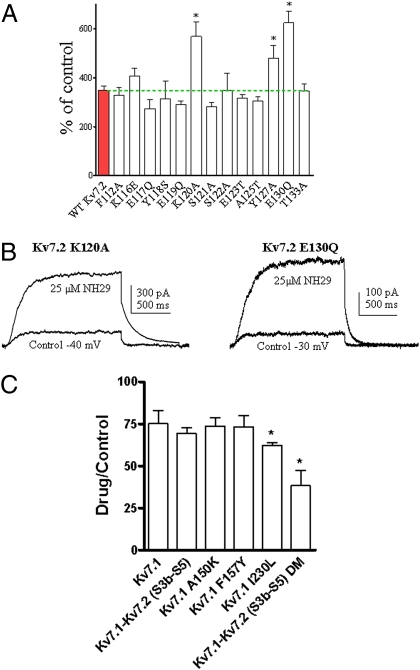
Next, we turned to the VSD to explore potential residues involved in Kv7.2 modulation by NH29. We mutated residues of helices S4 and S2 and of loop S1–S2 and characterized the biophysical properties of each mutant in the absence and presence of NH29 (25 μM) (SI Appendix, Tables S1 and S2). Most Kv7.2 residues were mutated to amino acids of the homologous position in Kv7.1. Some bulky residues were mutated to small-size amino acid (glycine), and a few S4 residues were mutated to naturally occurring epilepsy mutants. We quantified the sensitivity of each mutant channel to NH29 by measuring its ability to increase the current amplitude at a voltage yielding a normalized open probability of about 0.1 (Figs. 2 D, E, and G and 3). In helix S4, mutant L200G demonstrates stronger stimulation by NH29 compared with WT Kv7.2 (5.5 ± 0.5–fold vs. 3.5 ± 0.3–fold, n = 7–15, P < 0.05), but mutants L197G, R198A, and two epilepsy-associated mutations, R207W and R214W (22), exhibit significantly weaker stimulation by 25 μM NH29 (1.4-, 1.6-, 1.5-, and 1.1-fold, respectively; n = 8–15, P < 0.001) (Fig. 2 D–H). Notably, NH29 produces a very weak left-shift, if any, of the activation curve of R198A and R207W mutant channels compared with WT Kv7.2 (e.g., ΔV50 = 0.6 ± 2.6 mV and ΔV50 = −5.3 ± 0.9 mV for R198A and R207W, respectively, compared with ΔV50 = −15.5 ± 0.5 mV for WT Kv7.2, n = 8–15, P < 0.01) (Figs. 1D and 2 E and F). In addition, in R198A and R207W mutants the kinetics of channel activation and deactivation are not appreciably affected by NH29, compared with WT Kv7.2 (Fig. 2 E, G, and H and SI Appendix, Table S1). Noticeably, the S4 mutants, which are less sensitive to NH29, are highly responsive to other M-channel openers, RTG (5 μM), and zinc pyrithione (10 μM ZnPy) (23). RTG and ZnPy vigorously increase (4- to 6-fold) the current amplitude of R198A and R207W mutants and of WT Kv7.2 (SI Appendix, Figs. S3 and S4). Mutant K120A in linker S1–S2 and mutant Y127A in helix S2 show significant larger stimulation by NH29 compared with WT Kv7.2 (5.7- and 4.8-fold increase, respectively, versus 3.5-fold increase for WT; n = 8–15, P < 0.01) (Fig. 3A and SI Appendix, Table S2).
Fig. 3.
Sensitivity of Kv7.2 S2 residues and of Kv7.1 chimeras to NH29 modulation. (A) Potentiation of the currents produced by 25 μM NH29 on WT Kv7.2 (red bar) and on different S1–S2 linker and S2 mutants, expressed as percent of control determined in the absence of the opener (n = 4–15; *P < 0.01). (B) Representative trace of cells expressing mutant Kv7.2 K120A (Left) or mutant Kv7.2 E130Q (Right) in the absence (control) and presence of 25 μM NH29. (C) Effect of NH29 on WT Kv7.1, Kv7.1 mutants and Kv7.1 chimeras expressed in CHO cells. The effect of 25 μM NH29 was checked at 0 mV on Kv7.1 (WT), Kv7.1 A150K, Kv7.1 F157Y, Kv7.1 I230L, Kv7.1–Kv7.2 (S3b–S5), and Kv7.1–Kv7.2 (S3b–S5) DM (Kv7.1 A150K, Kv7.1 F157Y). The effect of NH29 was expressed as the ratio of the current amplitude in the presence and the absence of the drug. n = 3–8; *P < 0.01.
With these experimental constraints, we attempted to dock NH29 onto a 3D-model structure of Kv7.2 in an open conformation. The NH29 docking site is an external, water-exposed crevice within the VSD interface of helices S1 to S4 (Fig. 4A). Accordingly, we expected Y127 and E130 in helix S2, and R207 in helix S4, to affect the stability of the Kv7.2–NH29 complex, (Fig. 4C and Discussion). To further validate this docking model, we checked the effects of NH29 on mutant E130Q in helix S2. Although NH29 induces similar left-shift of the activation curve of mutant E130Q compared with WT Kv7.2, it produces significantly larger current-amplitude stimulation compared with WT Kv7.2 (6.2-fold versus 3.5-fold, respectively, n = 8–15, P < 0.01) (Fig. 3 A and B and SI Appendix, Table S2), suggesting that the opener may also affect the channel conductance of these mutants. In an attempt to explore the molecular basis for Kv7.2 specificity versus Kv7.1, for which NH29 does not act as an opener, we transferred into the Kv7.1 channel some relevant nonconserved Kv7.2 residues (K120, Y127, and L200). In addition, we swapped onto Kv7.1 the paddle motif of Kv7.2, which includes the S4–S5 linker (Kv7.1–Kv7.2 S3b–S5). NH29 (25 μM) did not significantly affect the chimera Kv7.1–Kv7.2 S3b–S5 and the single Kv7.1 mutants A150K and F157Y, but inhibited by 38% the current amplitude of mutant Kv7.1 I230L (n = 3–8, P < 0.01) (Fig. 3C). NH29 (25 μM) produced a strong inhibition (62%) of the chimera Kv7.1–Kv7.2 S3b–S5 bearing the double mutation (DM) A150K and F157Y (Kv7.1–Kv7.2 S3b–S5 DM) (n = 3–8, P < 0.01) (Fig. 3C). We attempted to generate chimeras where the entire VSD of Kv7.2 was swapped by that of Kv7.1 (Kv7.2–Kv7.1 S1–S4) and reciprocally (Kv7.1–Kv7.2 S1–S4). Unfortunately, these chimeras did not express functional channels to probe the impact of NH29.
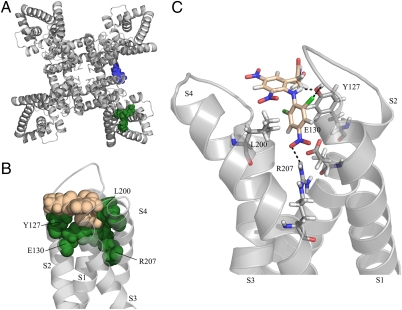
Fig. 4.
Docking of NH29 onto a model structure of Kv7.2 in an open state. (A) Top view of the model structure showing on one subunit the S5 residue W236 (blue) involved in RTG interaction and on another subunit the VSD residues (green) (Y127A, E130, L200, and R207) forming the binding pocket of NH29. (B) Front view of one VSD showing the bulky residues (green-filled spheres) sandwiching one NH29 molecule (salmon-filled spheres). (C) Side view of VSD residues (sticks) engaged in hydrogen bonding with one NH29 molecule (stick).
To investigate the selectivity of NH29 for the VSD of Kv7.2 subunits within the M-channel heteromer complex Kv7.2/Kv7.3, we coexpressed the Kv7.2 S4 mutants R198A and R207W with WT Kv7.3 (SI Appendix, Fig. S5). Results indicate that despite coexpression with WT Kv7.3, the Kv7.2 S4 mutations dominate within the complex and lead to channels, which display significantly weaker stimulation by NH29 compared with WT Kv7.2/Kv7.3 (SI Appendix, Fig. S5 and Table S3). These Kv7.2 S4 mutations were then introduced at homologous positions in helix S4 of Kv7.3 subunits (Kv7.3 R227A and Kv7.3 R236W, respectively) and were coexpressed with WT Kv7.2. NH29 potently increases the current amplitude of the latter channel complexes and produces significant left-shift of their activation curves similarly to WT Kv7.2/Kv7.3 (SI Appendix, Fig. S5 and Table S3). These data suggest that NH29 mainly acts on the VSD of Kv7.2, but barely affect that of Kv7.3.
Discussion
The results of this study suggest that the unique Kv7.2 channel opener, NH29, acts as a gating modifier. NH29 increases Kv7.2 current amplitude by affecting channel gating, thereby producing a hyperpolarizing shift of the activation curve. In addition, NH29 significantly reduces both activation and deactivation rates.
So far, RTG is the most extensively characterized Kv7 channel opener. RTG docks to a pore hydrophobic pocket formed by the cytoplasmic ends of helices S5 and S6 (Fig. 4A) (19–21). Our data clearly indicate that NH29 does not interact with the RTG site on Kv7.2, as it can potently stimulate the RTG-insensitive mutant W236L and left-shifts its voltage-dependence of activation.
Among the residues tested in S4, two mutants, L197G, R198A, and two epilepsy mutations, R207W and R214W (22), exhibit significantly weaker stimulation by NH29 compared with WT Kv7.2. In contrast, mutants L200G in S4, K120A in loop S1–S2, E130Q, and Y127A in S2 show significantly higher activation by NH29. The S4 mutants that are much less sensitive to NH29 (e.g., R198A and R207W) are potently activated by M-channel openers acting at the pore region, like RTG and ZnPy, suggesting that these molecules interact with a different target from NH29 on Kv7.2 subunits. Thus, the results suggest an NH29 interaction with the VSD of Kv7.2 subunits. Docking data indicate that NH29 interacts with the external groove formed by the interface of helices S1, S2, and S4 in a pocket formed by the bulky residues K120 in loop S1–S2, Y127 and E130 in helix S2, and L200 and R207 in helix S4 (Fig. 4 A and B). The docking model of NH29 predicts that a nitro group of one aromatic ring is engaged in hydrogen bonding with the guanidinium of R207 in S4 and with the carboxylate of E130 in S2 (Fig. 4C). In addition, the carbonyl oxygen of the NH29 amide function also forms hydrogen bonds with the hydroxyl of Y127 in S2. In this orientation, the hydroxyl group of NH29 lies close to the aromatic ring of Y127, consistent with the formation of a hydrogen bond with the π-electrons of Y127. Remarkably, in the open state the guanidinium of R207 and the carboxylate of E130 are sufficiently close to form a salt bridge. The model suggests that in the presence of NH29, this interaction could be stabilized, which favors the activated VSD conformation (Fig. 4C). The mutation R207W destabilizes the interaction with NH29, and in E130Q the nitro group of NH29 can still interact via H-bonding with the glutamine side chain and the guanidinium of R207, and even act as a surrogate of E130. In the closed state, the S4 helix rotates and moves downward, leading R207 to interact with E140 (3, 24). We speculate that NH29 stabilizes the Kv7.2 channel open state by preventing this movement. In the docking open-state model, the S4 residue R198, whose mutant A198 displays significant weaker stimulation by NH29, does not face NH29; instead, it is located at 180° opposite to the molecule (SI Appendix, Fig. S6). We suggest that R198 interacts with NH29 in the closed state, in line with our electrophysiological data (SI Appendix, SI Text and Figs. S2, S7, S8, S9, S10, S11, S12, and S13).
Despite the overall agreement between the computationally suggested binding mode of NH29 and the mutagenesis data, one should keep in mind the inherent limitations of small molecule docking protocols. Such procedures involve searching for ligand binding sites in a query protein and the prediction of the preferred positioning of the ligand in the sites, and a scoring function to estimate its binding affinity. In the present study, in the absence of a high-resolution structure, a homology model of human Kv7.2 was used, increasing the uncertainty of the procedure. Although the protein backbone is expected to be of relatively high accuracy, the side chains’ orientation is based on rotamer libraries and is, therefore, presumably imprecise. The LigandFit method that we used (25), akin to most docking protocols, treats proteins as rigid bodies to make the search process computationally feasible; thus, side-chain movements were not included. Moreover, any other possible protein rearrangements taking place upon ligand binding were left out as well. To evaluate ligand positions obtained with a search algorithm, scoring functions were exploited. Scoring functions use approximations and simplifications to moderate their complexity with a consequent cost in terms of accuracy. We combined the results of three different scoring functions to reduce the risk. Clearly, further mutagenesis data will be necessary to increase the structural constraints and refine the docking model.
Additional residues in the Kv7.2 VSD are likely involved in NH29 interaction and need to be further identified. Our results with the chimera Kv7.1–Kv7.2 S3b–S5 indicate that swapping helices S3 and S4 is not enough to recover the opener effect of NH29. In line with the docking data, the whole VSD (S1–S4) is apparently necessary to retrieve the opener action of NH29. Unfortunately, the lack of functionality of fully swapped VSD chimeras Kv7.2–Kv7.1 S1–S4 and Kv7.1–Kv7.2 S1–S4 prevented us to confirm the necessity of the whole VSD for supporting NH29 opener action. This lack of functionality of fully swapped VSD chimeras was already emphasized by Alabi et al. (10). Nevertheless, it is interesting to notice that the chimera Kv7.1–Kv7.2 S3b–S5 DM converts the opener effect of NH29 into a strong inhibition, supporting the view that this region is important for NH29 interaction. This notion is reminiscent to the finding that a point mutation (S512Y), in the S1–S2 loop converts the VSD targeted TRPV1 antagonist iodoresiniferatoxin into an intrinsic agonist (26). Our data are in line with a very recent study suggesting that the Kv7.2/3 channel opener ICA-27243 may interact with the VSD (27).
Specific residues, found to be critical for the opener action, are also key positions in structural interactions between helices S2 and S4. In Kv7.2, charged residues E130 in S2 and R207 in S4 are homologous to E283 and R371, respectively, in Shaker channel, where they were shown to interact electrostatically (28). E283 and R371 interact in the activated conformation of the VSD (28). Our data suggest that NH29 resides in the S2–S4 interface and contacts both R207 and E130 in the channel open state. Interestingly, combining charge-reversal mutations in Shaker, a previous study (28) showed that the double mutant, E283R+R371E, left-shifts the channel conductance-voltage relation and slows down the kinetics of activation and deactivation. These features are very similar to those exerted by NH29 on WT Kv7.2. We suggest that NH29 stabilizes the interaction between E130 and R207, thereby increasing the relative stability of the activated state as well as the heights of energetic barriers for transitions into and out of the open state.
The mechanism of NH29 activation of the Kv7.2 VSD may be similar to that of gating-modifier toxins. The VSD of voltage-gated Na+ channels is the target of polypeptide toxins from scorpions and sea anemones. These toxins bind to the S3–S4 loop at the outer end of S4 segments of Na+ channels and impede their movements during gating (16). Hence, α-scorpion and sea-anemone toxins specifically block inactivation by holding the S4 of domain IV in its inward position, and β-scorpion toxins enhance activation of Na+ channels by holding the S4 of domain II in its outward position (16). Similarly, hanatoxins from tarantula venom bind to the S3–S4 loop of Kv2.1 K+ channels and inhibit their activity by right-shifting the voltage dependence of activation (15). Interestingly, the vanillotoxin VaTx1 from tarantula venom, not only inhibits Kv2.1 in a similar manner to hanatoxin but also activates TRPV1 channels (14, 15). Along this line, our preliminary data intriguingly show that in addition to activating Kv7.2 K+ channels, NH29 is also a potent blocker of TRPV1 channels with an IC50 of 4.2 μM (SI Appendix, Fig. S14). NH29 changes the rather linear TRPV1 current-voltage relation obtained with capsaicin to an outwardly rectifying shape (SI Appendix, Fig. S14). A voltage-sensor trapping mechanism was suggested to be the molecular basis of action of polypeptide toxins that alter the voltage dependence of channel gating (16). Such mechanism may also operate for NH29.
In conclusion, the modular nature of the VSD and its rather unexploited targeting by drugs makes this strategic domain an important pharmacological target. Our data provide a framework for designing nontoxin gating-modifiers of voltage-gated cation channels to treat hyperexcitability disorders.
Materials and Methods
Details concerning the synthesis of NH29 have been previously described as compound 6 in Peretz et al. (11).
Cell Cultures and Electrophysiology.
CHO and primary hippocampal and DRG neuronal cultures were performed as described (17). Whole-cell patch-clamp recordings in CHO cells, in DRGs and in hippocampal neurons, were carried out as previously described (17). All data were expressed as mean ± SEM. Statistically significant differences were assessed by unpaired t test (two-tail) assuming equal variances for comparing parameters of WT Kv7.2 with those of Kv7.2 mutants, and by paired t test (two-tail) for comparing parameters in the same cell without and with the opener.
Structural Modeling and Docking.
We used comparative (homology) modeling techniques to produce an approximate 3D-model structure of the human Kv7.2 transmembrane domain (residues 92–323). The possible templates were the X-ray structure of the rat Kv1.2 channel (PDB entry 2A79) (9) and its modified chimeric version containing the rat Kv2.1 paddle (PDB entry 2R9R) (29). However, the query human Kv7.2 channel shares a very low sequence identity of about 22% with these structures, which complicates the modeling. Thus, we chose a model-structure of the human Kv7.1 channel (30), sharing about 60% sequence identity with our query, as a template instead. To increase the accuracy, the pair-wise alignment between the sequences of the query and target (SwissProt entries KCNQ2_HUMAN and KCNQ1_HUMAN, respectively) was derived from a multiple alignment of 128 sequences. These sequences were collected as KCNQ2_HUMAN homologs from the CleanUniProt database (31). Redundant (> 99% sequence identity) and fragmented sequences were discarded. The full-length sequences of these proteins were then aligned using the MAFFT program (32). We next constructed the 3D-model structure of the human Kv7.2 channel based on the obtained pair-wise alignment using NEST (33), with default parameters. We used the Plop program (34) to further optimize the model-structure (i.e., to eliminate steric clashes and improve side-chains packing in the protein core). The refined model structure was used for substrate docking, using the LigandFit tool (25) from the DiscoveryStudio 2.0 software (Accelrys). Forty-one docking cavities were detected using the LigandFit site search utility. Of these, only one was in line with the mutagenesis data and was selected as the seed binding site. The cavity was then expanded with the Discovery Studio site editing tool to a distance of 1.5 Ǻ in all directions and edited manually to remove points protruding far outside the main cavity. To improve the search in conformational space, the binding pocket was partitioned into three regions. The docking of the NH29 ligand into each of the three regions was conducted using the Dreiding force field (35) and putative NH29/channel complexes were selected. These complexes were minimized using the Smart Minimizer algorithm and evaluated with the DockScore (25), LigScore (36), and PMF04 (37) modules of the Discovery Studio software.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Jentsch (Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin) for the human clones of Kv7.2 and Kv7.3. This work is supported by the Ministry of Science and Technology (Tachtiot) and by the Deutsch-Israelische Projektkooperation fund (Deutsche Forschungsgemeinschaft) (to B.A.) and the European Commission FP6 program through the Marie-Curie Action: BIOCONTROL, Contract Marie Curie Research Training Network–33439 (to Y.G. and N.B.-T.)
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0911294107/-/DCSupplemental.
References
- 1.Ashcroft FM. Ion Channels and Diseases. Oxford: Academic Press; 2000. [Google Scholar]
- 2.Camerino DC, Tricarico D, Desaphy JF. Ion channel pharmacology. Neurotherapeutics. 2007;4(2):184–198. doi: 10.1016/j.nurt.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 5.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 7.Lee SY, Letts JA, MacKinnon R. Functional reconstitution of purified human Hv1 H+ channels. J Mol Biol. 2009;387:1055–1060. doi: 10.1016/j.jmb.2009.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 9.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 10.Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–375. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peretz A, et al. A tale of switched functions: From cyclooxygenase inhibition to M-channel modulation in new diphenylamine derivatives. PLoS ONE. 2007;2:e1332. doi: 10.1371/journal.pone.0001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmas P, Brown DA. Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nat Rev Neurosci. 2005;6:850–862. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- 13.Jentsch TJ. Neuronal KCNQ potassium channels: Physiology and role in diseases. Nat Rev Neurosci. 2000;1:21–30. doi: 10.1038/35036198. [DOI] [PubMed] [Google Scholar]
- 14.Siemens J, et al. Spider toxins activate the capsaicin receptor to produce inflammatory pain. Nature. 2006;444:208–212. doi: 10.1038/nature05285. [DOI] [PubMed] [Google Scholar]
- 15.Swartz KJ. Tarantula toxins interacting with voltage sensors in potassium channels. Toxicon. 2007;49:213–230. doi: 10.1016/j.toxicon.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Catterall WA, et al. Voltage-gated ion channels and gating modifier toxins. Toxicon. 2007;49(2):124–141. doi: 10.1016/j.toxicon.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 17.Peretz A, et al. Pre- and postsynaptic activation of M-channels by a novel opener dampens neuronal firing and transmitter release. J Neurophysiol. 2007;97:283–295. doi: 10.1152/jn.00634.2006. [DOI] [PubMed] [Google Scholar]
- 18.Peretz A, et al. Meclofenamic acid and diclofenac, novel templates of KCNQ2/Q3 potassium channel openers, depress cortical neuron activity and exhibit anticonvulsant properties. Mol Pharmacol. 2005;67:1053–1066. doi: 10.1124/mol.104.007112. [DOI] [PubMed] [Google Scholar]
- 19.Lange W, et al. Refinement of the binding site and mode of action of the anticonvulsant Retigabine on KCNQ K+ channels. Mol Pharmacol. 2009;75:272–280. doi: 10.1124/mol.108.052282. [DOI] [PubMed] [Google Scholar]
- 20.Schenzer A, et al. Molecular determinants of KCNQ (Kv7) K+ channel sensitivity to the anticonvulsant retigabine. J Neurosci. 2005;25:5051–5060. doi: 10.1523/JNEUROSCI.0128-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wuttke TV, Seebohm G, Bail S, Maljevic S, Lerche H. The new anticonvulsant retigabine favors voltage-dependent opening of the Kv7.2 (KCNQ2) channel by binding to its activation gate. Mol Pharmacol. 2005;67:1009–1017. doi: 10.1124/mol.104.010793. [DOI] [PubMed] [Google Scholar]
- 22.Dedek K, et al. Myokymia and neonatal epilepsy caused by a mutation in the voltage sensor of the KCNQ2 K+ channel. Proc Natl Acad Sci USA. 2001;98:12272–12277. doi: 10.1073/pnas.211431298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong Q, Sun H, Li M. Zinc pyrithione-mediated activation of voltage-gated KCNQ potassium channels rescues epileptogenic mutants. Nat Chem Biol. 2007;3:287–296. doi: 10.1038/nchembio874. [DOI] [PubMed] [Google Scholar]
- 24.Miceli F, et al. Gating consequences of charge neutralization of arginine residues in the S4 segment of K(v)7.2, an epilepsy-linked K+ channel subunit. Biophys J. 2008;95:2254–2264. doi: 10.1529/biophysj.107.128371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatachalam CM, Jiang X, Oldfield T, Waldman M. LigandFit: A novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph Model. 2003;21:289–307. doi: 10.1016/s1093-3263(02)00164-x. [DOI] [PubMed] [Google Scholar]
- 26.Johnson DM, et al. Functional mapping of the transient receptor potential vanilloid 1 intracellular binding site. Mol Pharmacol. 2006;70:1005–1012. doi: 10.1124/mol.106.023945. [DOI] [PubMed] [Google Scholar]
- 27.Padilla K, Wickenden AD, Gerlach AC, McCormack K. The KCNQ2/3 selective channel opener ICA-27243 binds to a novel voltage-sensor domain site. Neurosci Lett. 2009;465(2):138–142. doi: 10.1016/j.neulet.2009.08.071. [DOI] [PubMed] [Google Scholar]
- 28.Tiwari-Woodruff SK, Lin MA, Schulteis CT, Papazian DM. Voltage-dependent structural interactions in the Shaker K(+) channel. J Gen Physiol. 2000;115(2):123–138. doi: 10.1085/jgp.115.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 30.Smith JA, Vanoye CG, George AL, Jr, Meiler J, Sanders CR. Structural models for the KCNQ1 voltage-gated potassium channel. Biochemistry. 2007;46:14141–14152. doi: 10.1021/bi701597s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: Pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009;37(Database issue):D323–D327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrey D, et al. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53(Suppl 6):430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 34.Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 35.Mayo SL, Olafson BD, Goddard WA. Dreiding: A generic force-field for molecular simulations. J Phys Chem. 1990;94:8897–8909. [Google Scholar]
- 36.Krammer A, Kirchhoff PD, Jiang X, Venkatachalam CM, Waldman M. LigScore: A novel scoring function for predicting binding affinities. J Mol Graph Model. 2005;23:395–407. doi: 10.1016/j.jmgm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Muegge I. PMF scoring revisited. J Med Chem. 2006;49:5895–5902. doi: 10.1021/jm050038s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.