Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that have important roles in the regulation of gene expression. The roles of individual miRNAs in controlling vertebrate eye development remain, however, largely unexplored. Here, we show that a single miRNA, miR-204, regulates multiple aspects of eye development in the medaka fish (Oryzias latipes). Morpholino-mediated ablation of miR-204 expression resulted in an eye phenotype characterized by microphthalmia, abnormal lens formation, and altered dorsoventral (D-V) patterning of the retina, which is associated with optic fissure coloboma. Using a variety of in vivo and in vitro approaches, we identified the transcription factor Meis2 as one of the main targets of miR-204 function. We show that, together with altered regulation of the Pax6 pathway, the abnormally elevated levels of Meis2 resulting from miR-204 inactivation are largely responsible for the observed phenotype. These data provide an example of how a specific miRNA can regulate multiple events in eye formation; at the same time, they uncover an as yet unreported function of Meis2 in the specification of D-V patterning of the retina.
Keywords: microRNA, eye development, medaka fish
Development of the vertebrate eye takes place through a series of morphogenetic events that are controlled by molecular networks in which transcription factors and signaling pathways have major roles (1). Specific components of these networks are reiteratively exploited in space and time to pattern eye tissues and to control the subsequent cellular programs, such as cell proliferation, differentiation, migration, and programmed cell death (2, 3). Most of these developmental processes are critically sensitive to gene dose, and variations in the normal levels of regulatory proteins appear to result in a variety of eye anomalies (4, 5). Posttranscriptional regulatory mechanisms maintain the appropriate levels of expression of these proteins and enable rapid changes in the cellular proteome; thus, they have fundamental roles in the development of the nervous system (6).
MicroRNAs (miRNAs) are a class of 20- to 25-nucleotide noncoding RNA molecules that mediate a newly recognized level of posttranscriptional control of gene expression. Indeed, miRNAs can impair either mRNA translation or stability by binding through imperfect base pairing to specific sites in the 3′-UTR of target mRNAs (7). Recently, many miRNAs have been shown to be required for vertebrate developmental processes, such as cell fate determination and patterning as well as cell death and proliferation (8).
A number of miRNAs show restricted expression patterns in the developing lens, retinal pigment epithelium (RPE), neural retina, and other ocular tissues, which suggests their potential relevance to eye development and function (9, 10). However, the precise roles of individual miRNAs and their specific influences on given mRNA targets that are important for vertebrate eye development remain unclear.
In the present study, we show that miR-204, which is highly expressed in the presumptive RPE, lens, ciliary body, and neural retina (11, 12), is required for correct lens and optic cup development. Using a variety of gain- and loss-of-function approaches in the medaka fish [Oryzias latipes (ol)], we demonstrate that miR-204–mediated modulation of the Meis2 gene dose has a significant impact on regulation of the genetic pathways controlling eye morphogenesis and differentiation.
Results
miR-204 Knockdown Causes Lens Abnormalities, Microphthalmia, and Eye Coloboma.
We found that during early medaka development [stage (St) 23], ol-miR-204 was expressed in the lens placode and in the presumptive RPE, with an apparent dorsalhigh-to-ventrallow gradient (Fig. S1A). At later stages, ol-miR-204 expression was also detected in the ciliary marginal zone, ciliary body, and presumed migratory neural crest cells (Fig. 1 A–A′). This expression pattern suggested that miR-204 might modulate different aspects of eye development.
Fig. 1.
miR-204 directly targets Meis2. (A–C′) Frontal sections of St24 WT medaka embryos hybridized in both single (A–B′) and double (C and C′) whole-mount RNA ISH with probes against olMeis2 (red) and miR-204 (blue). miR-204 (A and A′) and olMeis2 (blue) (B–B′) colocalize in the lens placode and ciliary marginal zone (C–C′). Boxed areas are magnified in A′–C′. (D) Predicted target site of miR-204 within the 3′-UTR of the Meis2 gene in different species, showing conserved nucleotides (red) and nonconserved nucleotides (black). The blue line represents the sequence against which Meis2-TPmiR-204 Mo was designed. (E) Relative Luc activities in H36CE cells as fold differences in the Luc/Renilla ratios normalized to the value of Luc reporter constructs. miR-204 addition significantly decreases Luc activity of the construct containing 3′-UTR of MEIS2 when compared with controls. ***P < 0.0001 (t tests). Three point mutations in the predicted miR-204 target site in Meis2 inhibit this effect (no significant variation when compared with the thymidine kinase (TK)-Luc control). Densitometric analysis (F) of Western blotting (G) shows reduction of Meis2 protein levels in the presence of miR-204 duplexes and increase after miR-204 depletion when compared with cel-miR-67 control transfections. **P < 0.001; ***P < 0.0001 (t tests). 1, inhibitor hsa-miR-204; 2, inhibitor cel-miR-67; 3, mimic hsa-miR-204; 4, mimic cel-miR-67. Relative levels of the Meis2 protein measured 48 h after transfection of H36CE cells. (H–M) Frontal sections of St24 control (H and K), miR-204–overexpressing (I and L), and Mo-miR-204–injected (J and M) embryos treated for whole-mount RNA ISH with an olMeis2 probe (H–J) or immunostained with an anti-Meis2 antibody (green) (K–M). Sections are counterstained with propidium iodide (PI, red). Both olMeis2 mRNA and protein are down-regulated in lens placode and retina of miR-204–overexpressing embryos (I and L) but up-regulated in miR-204 morphants (J and M). (Scale bars: 20 μm.).
To investigate this further, we interfered with miR-204 processing and activity using a multiblocking morpholino (Mo)-based knockdown approach (13). To this end, we designed two Mos, Mo-miR-204- and Mo-miR-204-2, against the two ol-miR-204-1 and ol-miR-204-2 precursor sequences present in the medaka genome, which give rise to an identical mature miR-204 sequence. Embryos injected with either of these two Mos at the one-cell stage were morphologically indistinguishable from control embryos up to the optic-vesicle stage. In contrast, from St24 onward, an aberrant eye phenotype was clearly visible in most of the Mo-miR-204–injected embryos (65 ± 5% of 3,000 injected embryos). Growth of the eye cup was significantly impaired and culminated in evident microphthalmia at St40 (Fig. S2 A and D). In 90% of the microphthalmic embryos, lens development was also impaired. Specifically, at St24, the monolayer of lens epithelial cells was positioned in the dorsal region of the lens vesicle instead of lining its distal surface in morphants (Fig. 2 A–F, Fig. S3 H, H’, J, and J’, and Fig. S4 A, B, G, and H). Furthermore, the primary fiber cells that are located in the center of the lens vesicle were misplaced and disorganized, whereas those of the control embryos had begun to elongate to form the organized concentrical layers (Fig. 2 A–F and Fig. S4 A, B, G, and H). This altered cellular organization was also evident at later stages, when abnormal ventral-distal herniations of the lens were also evident (Fig. S2E). Finally, a significant number of the microphthalmic morphant embryos (30%) were characterized by a ventral coloboma, through failure of optic fissure closure (Fig. S2 C and F).
Fig. 2.
Interference with miR-204 expression modifies lens cell differentiation via Meis2 targeting. Frontal sections of St24 control (A–C), Mo-miR-204 (D–F), miR-204 (G–I), Mo-Meis2/Mo-miR-204 (J–L), Meis2-TPmiR-204 (M–O), and Meis2-TPmiR-204/miR-204 (P–R)–injected medaka embryos processed for whole-mount RNA ISH with probes specific for olMeis2 (A, D, G, J, M, and P), olPax6 (B, E, H, K, N, and Q), and olα-ACrystallin (C, F, I, L, O, and R). Expression of olMeis2 (D and M) and olPax6 (E and N) is up-regulated in lens of miR-204 and Meis2-TPmiR-204 morphant embryos, whereas that of olα-ACrystallin is increased in the lens placode and ectopically expressed in the epithelial lens monolayer (F and O, yellow arrowheads). Lens epithelial (D–F and J–L, red arrowheads) and primary fiber (D–F and J–L, black arrowheads) cells are displaced. (J) MOs act at the translational level; thus, Mo-Meis2/Mo-miR-204 coinjection does not rescue olMeis2 mRNA expression. miR-204 gain-of-function has opposite effects in lens gene expression, without affecting lens epithelial monolayer (G–I, red arrowheads) and the primary fibers (G–I, black arrowheads). Mo-Meis2/Mo-miR-204 and Meis2-TPmiR-204/miR-204 coinjections restore correct expression of lens differentiation markers (J–L and P–R). Mo-Meis2/Mo-miR-204 coinjections do not rescue cell displacement (J–L, red and black arrowheads). Broken lines mark boundaries between the lens epithelial monolayer and the primary fiber cells. (Scale bars: 20 μm.)
We did not observe any qualitative and quantitative phenotypic differences following the injection of Mo-miR-204-1 or Mo-miR-204-2; hence, subsequent studies were performed only with Mo-miR-204-1. The Mo blocking efficiency and the specificity of the eye phenotype were verified through a series of experiments described in SI Text (Fig. S1 and Tables S1 and S2). These included RNA in situ hybridization (ISH) using the miR-204-locked nucleic acid antisense probe and injections of 6-bp–mismatched Mo (mm-Mo-miR-204), which gave no aberrant phenotype at any concentration (Table S2). To rule out off-targeting Mo effects, we coinjected Mo-miR-204-1 and a Mo against p53 (Mo-p53). We did not observe any modifications of the phenotype, which further confirmed the specificity of Mo-miR-204 targeting (14) (SI Text and Fig. S2 V–X).
Meis2 Gene Is a Direct Target of miR-204.
Given the specificity of this miR-204 loss-of-function phenotype, we searched for its potential mRNA targets using the recently developed host-gene oppositely correlated target (HOCTAR) tool, which integrates expression profiling and sequence-based miRNA target recognition software (15). Among the predicted miR-204 targets, the homeobox transcription factors Meis1 and Meis2 appeared particularly attractive to explain the observed eye defects because they had been previously shown to regulate vertebrate eye development by modulating Pax6 expression (16). Furthermore, the predicted target site of miR-204 within the 3′-UTR of Meis2 (but not of Meis1) was highly conserved across all vertebrate species analyzed, including medaka (Fig. 1D). To validate this prediction, we cloned the 3′-UTR of the human MEIS2 gene containing the miR-204 target site downstream of the coding region of the Luciferase (Luc) reporter gene, and tested the ability of miR-204 to affect reporter expression in vitro. The presence of the MEIS2 3′-UTR sequence specifically inhibited Luc activity in response to miR-204 (Fig. 1E). In addition, point mutations in the miR-204 binding site of the MEIS2 3′-UTR abolished this repression, indicating that miR-204 directly and specifically targets MEIS2 (Fig. 1E). In agreement with these observations, the levels of MEIS2 protein in H36CE human lens epithelial cells were decreased in the presence of miR-204 duplexes and elevated on miR-204 inhibition (Fig. 1 F and G). The specificity of this repression was confirmed by additional controls described in SI Text (i.e., a Luc construct containing the 3′-UTR of PAX6 and miR-182, an unrelated miRNA expressed in the eye) (Fig. S3 K–N).
The miR-204 targeting of Meis2 was also confirmed in vivo. miR-204 and Meis2 showed overlapping expression patterns in the lens and in the peripheral optic cup (Fig. 1 A–C′). Moreover, injections of miR-204 duplexes resulted in a decrease in endogenous Meis2 mRNA and protein levels, whereas injections of Mo-miR-204 led to an increase in the optic cup of medaka embryos (Fig. 1 H–M). The expansion of the Meis2 domain in the miR-204 morphants was not caused by a generalized delay of eye development because (i) Meis2 was correctly expressed at early stages of lens and retinal development in the morphants (Fig. S3 G–J′) and (ii) Ath5, an early marker of retinal ganglion cell differentiation, was expressed normally at St26, when the retina begins to differentiate (Fig. S3 O and P). Altogether, these data strongly indicate that Meis2 is a bona fide miR-204 target.
miR-204 Controls Lens Differentiation by Targeting Meis2 and Modulating the Pax6 Transcriptional Pathway.
We next sought to determine whether the miR-204 morphant phenotype was indeed related to abnormal activation of olMeis2 expression. Meis2 has been reported to regulate Pax6 activity in the lens in a direct and positive way (16). Pax6, in turn, controls the expression of genes involved in lens differentiation, including Sox2, Prox1, and α-ACrystallin (17, 18). With the exception of Meis2, none of these genes are predicted to contain miR-204 target sites. We thus reasoned that if miR-204 directly controls expression of Meis2 in vivo, the levels of expression of all these genes should be increased in the miR-204 morphant as a consequence of alterations in Meis2 expression.
Indeed, RNA ISH demonstrated that in the morphant optic cup, olPax6, olSox2, olProx1, and olα-ACrystallin showed up-regulated expression and/or were misexpressed in both lens ectoderm and primary lens fiber cells when compared with control embryos (Fig. 2 A–F and Fig. S4 A, B, G, and H). To demonstrate a direct interaction between miR-204 and Meis2 further, and to dissect out the role of this interaction in lens development, we increased the levels of miR-204 by injecting miR-204 duplexes. From St20 onward, duplex-injected embryos were severely microphthalmic, with small lenses (Fig. S2 J–L), which strongly resembled the phenotype reported for Meis2.2 zebrafish morphant embryos (19). Consistent with the hypothesis that olMeis2 is an important miR-204 target, expression of the olMeis2, olPax6, olSox2, olProx1, and olα-ACrystallin genes was significantly reduced in the lens placode of all the duplex-injected embryos (Fig. 2 G–I and Fig. S4 C and I), as confirmed by quantitative real-time qRT (PCR) (Fig. S4M). Of note, inhibition of miR-204 activity by Mo-miR-204 injections was accompanied by an opposite trend in the relative transcript levels (Fig. S4N). Interestingly, both the lens epithelial monolayer and the primary fiber cells were normally localized in duplex-injected embryos (Fig. 2 G–I and Fig. S4 C and I).
If most of the changes in lens differentiation marker expression caused by miR-204 knockdown are mediated by Meis2, coinjection of a Mo against olMeis2 (Mo-Meis2) should reestablish normal expression levels in miR-204 morphants. Consistent with this, Mo-Meis2 injection was sufficient to rescue the normal expression of lens differentiation markers in a substantial proportion of miR-204 morphants (Fig. 2 J–L, Fig. S4 D and J, and Table S2), although defects in epithelial and lens fiber cell organization were not rescued (Fig. 2 J–L and Fig. S4 D and J). This suggests that miR-204 regulates additional, and as yet unidentified, genes that are important for correct lens development.
To obtain additional support for the importance of miR-204–mediated regulation of olMeis2, we disrupted the interaction of miR-204 with its target site in olMeis2 3′-UTR by injecting a Meis2 “target protector” (20) Mo (Meis2-TPmiR-204) in WT embryos (SI Text and Fig. S3 A–F). A significant percentage of Meis2-TPmiR-204–injected embryos were morphologically indistinguishable from miR-204 morphants and characterized by similar defects in the lens, optic cup size, and optic fissure coloboma (Fig. S2 G–I and Table S2). Moreover, protection of the miR-204 target site within Meis2 mRNA resulted in expansion of the lens placode expression domains of olMeis2, olPax6, and olSox2 and mislocalization of olProx1 and olα-ACrystallin, the mRNAs of which were ectopically detected in the epithelial cell monolayer (Fig. 2 M–O and Fig. S4 E and K). Unlike our observations in miR-204 morphant embryos, these changes were not associated with defects in epithelial and lens fiber cell organization (Fig. 2 M–O and Fig. S4 E and K), further supporting the possibility that these alterations are mediated by other mRNA targets. Finally, protection of Meis2 targeting was sufficient to rescue the correct expression of lens differentiation markers in miR-204–overexpressing embryos (Fig. 2 P–R and Fig. S4 F and L).
Altogether, these data indicate that miR-204 controls lens cell differentiation by modulating the expression of lens placode differentiation genes via the Meis2/Pax6 pathway.
miR-204 Has an Active Role in Establishment of Dorsoventral Polarity of the Optic Cup.
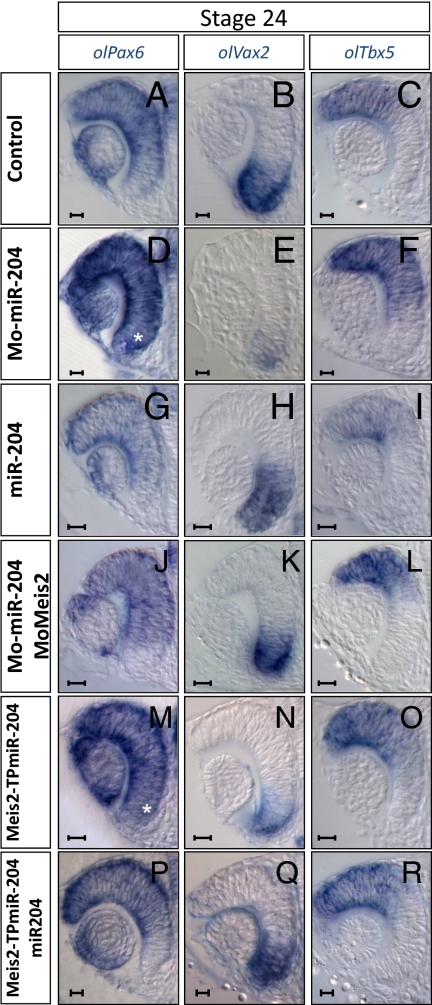
Defects in optic fissure closure were observed in both the Mo-miR-204 and Meis2-TPmiR-204 morphants (Fig. S2 D–I). Colobomas have been frequently described as a consequence of impaired dorsoventral (D-V) polarity of the optic cup (21). As observed in the lens, this defect was associated with concomitant up-regulation of olMeis2 (Fig. 1 H, J, K, and M and Fig. S5B) and olPax6 expression. In particular, olPax6 expression extended to the ventral retina, where it is normally expressed at relatively low levels (Fig. 3 A, D, and M). Previous studies have shown that expansion of Pax6 expression in the ventral retina results in alterations in D-V polarity of the optic cup (22, 23). Therefore, we asked whether expression of D-V optic cup markers was modified in Mo-miR-204 and Meis2-TPmiR-204 morphants. In all the injected embryos, the expression domain of the ventral marker olVax2 and the ventral expression domain of olPax2 (24) were reduced or absent (Fig. 3 B, E, and N and Fig. S5 E and H), whereas the expression domains of the dorsal markers olBmp4 and olTbx5 were ventrally expanded (Fig. 3 C, F, and O and Fig. S5 K and N). A reciprocal molecular phenotype was observed after miR-204 overexpression: The olBmp4 and olTbx5 domains were largely reduced, whereas those of olPax2 and olVax2 were dorsally expanded (Fig. 3 H and I and Fig. S5 F and L). Furthermore, coinjection of Mo-Meis2 with Mo-miR-204 (Fig. S2 M–O, Fig. S5A, and Table S2) or that of miR-204 with the Meis2-TPmiR-204 target protector (Fig. S2 S–U, Fig. S5C, and Table S2) fully rescued the coloboma phenotype and the normal levels of D-V markers and Pax6 expression (Fig. 3 J–L, P–R and Fig. S5 G, I, M, and O).
Fig. 3.
miR-204 is required for establishment of D-V polarity of the optic cup. Frontal vibratome sections of control (A–C), Mo-miR-204 (D–F), miR-204 (G–I), Mo-Meis2/Mo-miR-204 (J–L), Meis2-TPmiR-204 (M–O), and Meis2-TPmiR-204/miR-204 (P–R)–injected embryos processed for whole-mount RNA ISH with probes specific for olPax6 (A, D, G, J, M, and P), olVax2 (B, E, H, K, N, and Q), and olTbx5 (C, F, I, L, O, and R). olPax6 is up-regulated and ectopically expressed in ventral retina of morphant embryos (D and M, asterisk). Expression of ventral gene olVax2 is reduced, whereas that of dorsal marker olTbx5 is expanded ventrally in morphant embryos (E, F, N, and O) when compared with control embryos (B and C). miR-204–overexpressing embryos show reciprocal alterations in Pax6 and D-V marker expression (G–I). Defects in D-V optic cup polarity are completely rescued by coinjection of Mo-miR-204 with Mo-Meis2 (J–L) and Meis2-TPmiR-204 with miR-204 (P–R). (Scale bars: 20 μm.)
Altogether, these data demonstrate that miR-204–mediated control of the Meis2/Pax6 pathway contributes to D-V patterning of the optic cup. Moreover, our findings reveal a previously unidentified role for Meis2 in this morphogenetic event.
Discussion
MiRNAs appear to function as “master regulators” of key molecular pathways through their ability to fine-tune gene dose (25); consequently, they have basic roles in vertebrate organogenesis and pathogenesis. The general importance of this class of molecule in eye development is supported by effects observed in mice after genetic inactivation of Dicer, a key enzyme of miRNA biogenesis (26). However, there is little information available on individual miRNAs that contribute to the correct development and function of the eye. Examples exist with miR-24a, miR-124, and miR-26a for the regulation of apoptotic pathways, retinogenesis, and circadian rhythms of mRNAs in the retina, respectively (27–29). Here, we showed that a single miRNA can regulate multiple aspects of eye development. Indeed, miR-204 is required for lens differentiation, optic cup development, and optic fissure closure.
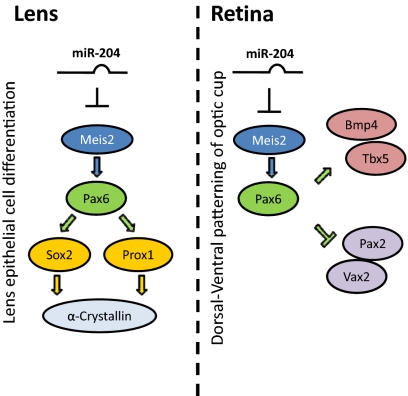
Starting from in silico predictions and using a variety of in vitro and in vivo experimental approaches, we have shown that Meis2 is a bona fide target of miR-204 activity. Our data also show that the specific miR-204–mediated regulation of Meis2 modulates the function of the Pax6 transcriptional network. These data indicate that miR-204 is an essential component of the Meis2/Pax6 molecular pathway, and hence is an important element of the molecular network that regulates eye development in vertebrates (Fig. 4). This regulatory cascade is strongly supported by a number of observations. Meis2 directly activates Pax6 expression during lens and retina development in the zebrafish, chick, and mouse (16, 19, 30). In medaka fish embryos, expression levels of olMeis2 and, concomitantly, those of olPax6 and its downstream targets are up-regulated on miR-204 knock-down or inhibition of the miR-204 interaction with its specific target site in the Meis2 3′-UTR. Furthermore, the phenotypic alterations observed in miR-204 morphants strongly resembled those reported for Meis2 or Pax6 gain-of-function models (4, 16, 19, 30), which include aberrant lens differentiation and microphthalmia. Conversely, miR-204 over-expression led to a significant down-regulation of Meis2 activity, with phenotypic consequences similar to those observed after Meis2 loss-of-function, in which a reduction of Pax6 gene dose was also observed (16, 19, 30). Finally, concomitant knock-down of olMeis2 and miR-204 and protection of the olMeis2 target site in miR-204–overexpressing embryos, significantly rescued expression of olPax6, olSox2, olProx1, and olα-ACrystallin in the lens, which is consistent with a pathway in which miR-204 controls lens differentiation through regulation of olMeis2 levels (Fig. 4).
Fig. 4.
Model for function of miR-204 in lens and retina development. miR-204 controls expression of the Meis2 gene, contributing to regulation of the Meis2/Pax6 pathway in both lens and retina development.
During lens development, the progeny of epithelial cells migrate into the transitional zone and elongate and differentiate into lens fiber cells (18, 31). Timely differentiation and correct migration of lens fibers are crucial for continuous addition of fiber mass and formation of a correctly organized lens. The miR-204–mediated control of olMeis2 appears to modulate gene expression programs that control these events because its over-expression or inactivation produces significant changes in the expression of lens differentiation markers. miR-204 also appears to control lens morphogenesis, because lens epithelial cells were abnormally positioned in Mo-204 morphants. However, this miR-204 activity is very likely to be Meis2-independent, because protection of Meis2 mRNA did not alter epithelial and lens fiber cell organization. Thus, miR-204 might ensure correct control of lens morphogenesis by targeting other genes involved (e.g., in the control of cell polarity, cell-cell signaling, tissue polarity, or cell migration). This last possibility is particularly attractive, because cells with elevated miR-204 levels are highly mobile and have invasive properties (32, 33).
The miR-204 contributed to other aspects of eye morphogenesis, as expected by its specific distribution in ocular tissues other than the lens. The requirement for miR-204 in establishment of D-V polarity in the optic cup and in optic fissure closure can also be explained by its control of Meis2 expression. Indeed, the defects in D-V polarity and optic fissure closure in the miR-204 and Meis2-TPmiR-204 morphants were rescued by coinjection with Mo-Meis2 or miR-204, respectively. This is an as yet unreported aspect of Meis2 function that is probably mediated by the observed ectopic ventral expression of Pax6, the overexpression of which correlates with significant down-regulation of ventral determinant genes, which leads to the formation of optic fissure coloboma (22, 23). However, the relatively low frequency of optic fissure defects in miR-204 morphants suggests that miR-204 might not serve as an “on-off” switch for the genetic program required for correct optic fissure closure.
In miR-204 morphants, eye formation was initiated and progressed normally up to the optic cup stage; thereafter, it did not advance correctly, leading to microphthalmia. This phenotype might be a consequence of alterations in apoptosis and/or cell proliferation, because previous studies have shown that members of the Meis gene family can directly regulate these events. However, we found that the concomitant knock-down of Meis2 and miR-204 was insufficient to restore normal eye size (Fig. S2), possibly because Mo-Meis2 reduces the levels of Meis2 expression well below normal levels. Indeed, a microphthalmic phenotype has been reported for loss-of-function Meis mutants in the chick and zebrafish (19, 29). Alternatively, the miR-204–mediated regulation of eye size involves other, as yet unidentified, transcriptional pathways.
Although miRNAs have the potential to regulate the expression of hundreds of genes, we have shown that the specific miR-204-Meis2 interaction has multiple consequences in eye development. Of note, this action is mediated by a single miR-204 target site within the 3′-UTR of the Meis2 gene, as demonstrated by the specific target protector assay. Previous reports have proposed that the presence of multiple target sites for the same miRNA within the 3′-UTR of a given mRNA is a strong indication of the “strength” and biological relevance of these interactions (34). However, in agreement with our data, it has also been reported that point mutations in a single miRNA recognition site have a pathogenetic role in human genetic diseases (35). Thus, the mode of action of miRNAs and their relevance in the control of basic biological processes may be more complex than initially envisaged.
In conclusion, we have begun to unravel the function of miR-204 during eye development, and we have demonstrated that this is largely mediated by Meis2 targeting. As shown by our data, miR-204 may have additional target genes in the eye. It will be of the utmost importance to identify these and to determine whether alterations in miR-204 expression contribute to the pathogenesis of eye malformations in humans.
Materials and Methods
Medaka Stocks.
Samples of the Cab strain of WT medaka fish were kept and staged as described (36).
Computational Analysis.
Prediction of miRNA targets was performed using the Host gene Opposite Correlated TARgets (HOCTAR) tool (http://hoctar.tigem.it) (15).
Mo and miR-204 Duplex Injections.
Mos (Gene Tools, LLC) were designed and injected into fertilized one-cell embryos, as detailed in Table S1. The specificity and inhibitory efficiency of each Mo were determined as described (14). Optimal Mo concentrations (Tables S1 and S2) were determined on the basis of morphological criteria. miRIDIAN (Dharmacon) miRNA Mimics for miR-204 were injected at a final concentration of 4 μM. Embryos injected with mm-Mo-miR-204 were used as controls.
Whole-Mount ISH.
Whole-mount RNA ISH was performed, photographed, and sectioned as described (37). Digoxigenin-labeled antisense and sense riboprobes for olMeis2, olPax6, olα-ACrystallin, olSox2, olProx1, olBmp4, olTbx5.2, olVax2, and olPax2 were used. The miRCURY detection miR-204 probe (Exiqon) was used according to Karali et al. (11).
Western Blotting.
Immunoblotting was performed as described (38), with a rabbit polyclonal antibody against Meis2 (1:1,000) or an anti-β-tubulin monoclonal antibody (1:1,000; Sigma).
Immunolabeling.
Medaka embryos were cryostat-sectioned, and immunochemistry was performed as described (39) using an anti-phospho-histone H3 monoclonal antibody (1:100; Cell Signaling Technology) and an anti-Meis2 rabbit polyclonal antiserum. Alexa-488–conjugated goat anti-rabbit or anti-mouse (1:1,000; Invitrogen) IgGs were used as secondary antibodies. Alternatively, a peroxidase-conjugated anti-rabbit antibody (1:200; Vector Laboratories) was used, followed by diaminobenzidine staining, as described previously (40).
Cell Transfection, qRT-PCR, and Luc Assays.
The H36CE human lens epithelial cell line was grown as described (41). Cell transfections and qRT-PCR experiments were performed as described (15). Cells were transfected with either 50 nM miRIDIAN miRNA Mimics or 80 nM miRIDIAN miRNA Inhibitor (Dharmacon). Plasmids containing the 3′-UTR of the human MEIS2 gene and psiUx plasmid constructs containing the hsa-premiR-204 sequence were used in Luc assays, as described previously (15). Each assay was performed in duplicate, and all the results are shown as means ± SD of at least three independent assays. The primer sequences used to amplify each transcript are shown in Table S1.
Supplementary Material
Acknowledgments
We are grateful to M. Torres (Centro Nacional de investigaciones Cardiovasculares, Madrid) for providing the Meis2 antibody. We thank M. Studer and G. Diez-Roux (Telethon Institute of Genetics and Medicine, Naples) for critical reading of the manuscript. We thank F. Salierno, M. Pirozzi (Telethon Institute of Genetics and Medicine), and the Institute of Genetics and Biophysics Open Laboratory for technical support. This study was supported by the Italian Telethon Foundation, European Union Grants LSHG-CT-2005-512036 and PERG03-GA-2008-231068 (to S.B.) and Spanish Ministerio de Ciencia e Innovación Grant BFU2007-61774 (to P.B.). R.M.F holds an I3P-Consejo Superior de Investigaciones Cientificas Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914785107/-/DCSupplemental.
References
- 1.Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: Lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 3.Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- 4.Schedl A, et al. Influence of PAX6 gene dosage on development: Overexpression causes severe eye abnormalities. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- 5.Taranova OV, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15:410–415. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Huang KM, Dentchev T, Stambolian D. MiRNA expression in the eye. Mamm Genome. 2008;19:510–516. doi: 10.1007/s00335-008-9127-8. [DOI] [PubMed] [Google Scholar]
- 10.Xu S. microRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res. 2009;28:87–116. doi: 10.1016/j.preteyeres.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- 12.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 13.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen JS, Smith JC. Controlling morpholino experiments: Don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 15.Gennarino VA, et al. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009;19:481–490. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Friedman A, Heaney S, Purcell P, Maas RL. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev. 2002;16:2097–2107. doi: 10.1101/gad.1007602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, et al. Regulation of alphaA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 2006;25:2107–2118. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang RA. Pathways regulating lens induction in the mouse. Int J Dev Biol. 2004;48:783–791. doi: 10.1387/ijdb.041903rl. [DOI] [PubMed] [Google Scholar]
- 19.Bessa J, et al. meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development. 2008;135:799–803. doi: 10.1242/dev.011932. [DOI] [PubMed] [Google Scholar]
- 20.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 21.Adler R, Canto-Soler MV. Molecular mechanisms of optic vesicle development: Complexities, ambiguities and controversies. Dev Biol. 2007;305:1–13. doi: 10.1016/j.ydbio.2007.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz M, et al. Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. Development. 2000;127:4325–4334. doi: 10.1242/dev.127.20.4325. [DOI] [PubMed] [Google Scholar]
- 23.Leconte L, Lecoin L, Martin P, Saule S. Pax6 interacts with cVax and Tbx5 to establish the dorsoventral boundary of the developing eye. J Biol Chem. 2004;279:47272–47277. doi: 10.1074/jbc.M406624200. [DOI] [PubMed] [Google Scholar]
- 24.Koster R, Stick R, Loosli F, Wittbrodt J. Medaka spalt acts as a target gene of hedgehog signaling. Development. 1997;124:3147–3156. doi: 10.1242/dev.124.16.3147. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Damiani D, et al. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L, Ko ML, Ko GY. Rhythmic expression of microRNA-26a regulates the L-type voltage-gated calcium channel alpha1C subunit in chicken cone photoreceptors. J Biol Chem. 2009;284:25791–25803. doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker JC, Harland RM. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev. 2009;23:1046–1051. doi: 10.1101/gad.1777709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu R, et al. The role of miR-124a in early development of the Xenopus eye. Mech Dev. 2009;126:804–816. doi: 10.1016/j.mod.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heine P, Dohle E, Bumsted-O'Brien K, Engelkamp D, Schulte D. Evidence for an evolutionary conserved role of homothorax/Meis1/2 during vertebrate retina development. Development. 2008;135:805–811. doi: 10.1242/dev.012088. [DOI] [PubMed] [Google Scholar]
- 31.Tholozan FM, Quinlan RA. Lens cells: More than meets the eye. Int J Biochem Cell Biol. 2007;39:1754–1759. doi: 10.1016/j.biocel.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 32.Findlay VJ, Turner DP, Moussa O, Watson DK. MicroRNA-mediated inhibition of prostate-derived Ets factor messenger RNA translation affects prostate-derived Ets factor regulatory networks in human breast cancer. Cancer Res. 2008;68:8499–8506. doi: 10.1158/0008-5472.CAN-08-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLOS Comput Biol. 2010;6:e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 35.Meola N, Gennarino VA, Banfi S. microRNAs and genetic diseases. Pathogenetics. 2009 doi: 10.1186/1755-8417-2-7. 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 2004;121:605–618. doi: 10.1016/j.mod.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 37.Conte I, Bovolenta P. Comprehensive characterization of the cis-regulatory code responsible for the spatio-temporal expression of olSix3.2 in the developing medaka forebrain. Genome Biol. 2007;8:R137.1–R137.17. doi: 10.1186/gb-2007-8-7-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercader N, Tanaka EM, Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132:4131–4142. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- 39.Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del Bene F, Tessmar-Raible K, Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- 41.Porter RM, Hutcheson AM, Rugg EL, Quinlan RA, Lane EB. cDNA cloning, expression, and assembly characteristics of mouse keratin 16. J Biol Chem. 1998;273:32265–32272. doi: 10.1074/jbc.273.48.32265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.