Abstract
Cancer-specific mutations in the iSH2 (inter-SH2) and nSH2 (N-terminal SH2) domains of p85α, the regulatory subunit of phosphatidylinositide 3-kinase (PI3K), show gain of function. They induce oncogenic cellular transformation, stimulate cellular proliferation, and enhance PI3K signaling. Quantitative determinations of oncogenic activity reveal large differences between individual mutants of p85α. The mutant proteins are still able to bind to the catalytic subunits p110α and p110β. Studies with isoform-specific inhibitors of p110 suggest that expression of p85 mutants in fibroblasts leads exclusively to an activation of p110α, and p110α is the sole mediator of p85 mutant-induced oncogenic transformation. The characteristics of the p85 mutants are in agreement with the hypothesis that the mutations weaken an inhibitory interaction between p85α and p110α while preserving the stabilizing interaction between p85α iSH2 and the adapter-binding domain of p110α.
Keywords: oncogenic transformation, target of rapamycin
The phosphoinositide 3-kinase (PI3K) signaling pathway is deregulated in most human cancers by differential gene expression, amplification, or mutation. Of particular interest are mutations that occur in the catalytic subunit p110α of class I PI3K, because they confer a strong gain of function upon the enzyme, resulting in enhanced catalytic activity, constitutive signaling, and oncogenicity in vitro and in vivo (1–8). There have also been early reports of cancer-specific mutations in p85α, a regulatory subunit of class I PI3K (9–14). Such mutations gained high significance by recent comprehensive genomic analyses of glioblastomas (15, 16). Approximately 9% of these tumors harbor a mutation in p85α. The mutations cluster in the inter-SH2 (iSH2) domain of p85α, involving residues that interact with the C2 domain of the catalytic subunit p110α (15, 17). The iSH2–C2 domain interaction has an inhibitory effect on enzyme activity, and the mutations in the iSH2 domain of p85α could weaken this interaction and release the inhibition of PI3K activity (15, 17–19). A similar mechanism has been proposed for the gain-of-function mutations in the helical domain of p110α that alleviate an inhibitory interaction with the N-terminal SH2 domain (nSH2) of p85α (20).
We have studied mutations in p85α (referred to as p85). Most of these were identified in a genomic characterization of glioblastoma (15) and map to the iSH2 domain of p85; one was an engineered mutation that maps to the nSH2 domain of p85. These mutations show oncogenic potency in cell culture and elevated levels of downstream signaling and operate through the p110α isoform of the catalytic subunit of class I PI3K. Our observations extend recent studies of the p85α mutants using different cell systems (17, 19) by providing quantitative data on the oncogenic potency of the mutations and by presenting evidence that suggests a unique role of p110α for the p85 mutation-induced gain of function in PI3K activity.
Results
Cancer-Derived Mutations of p85 Induce Oncogenic Transformation and Increase Cell Proliferation.
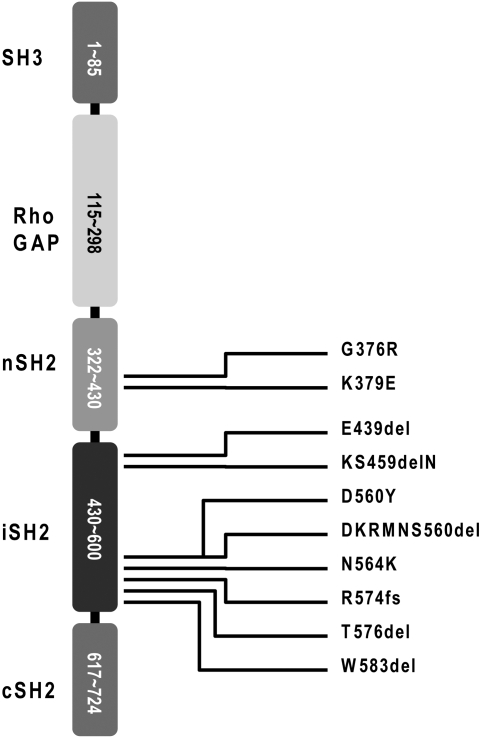
Fig. 1 lists recently identified p85 mutations and their map positions in the p85 sequence. The changes caused by the mutations in the protein sequence are summarized in Fig. S1. Most of the mutations are located in the iSH2 domain of p85. With the exception of the K379E mutation, they were first seen in human glioblastoma (15). To date, K379E has not been detected in human cancers; it is an engineered mutation designed to weaken the interaction between the nSH2 domain of p85 and the helical domain of p110α involving p110α residue E545 by disrupting an inhibitory salt bridge (20).
Fig. 1.
Domain organization of p85 and map positions of the mutants tested. SH3, Src homology domain 3; Rho GAP, GTPase activating protein domain for the Rho GTPase; nSH2, N-terminal Src homology domain 2; iSH2, inter-Src homology domain 2; cSH2, C-terminal Src homology domain 2.
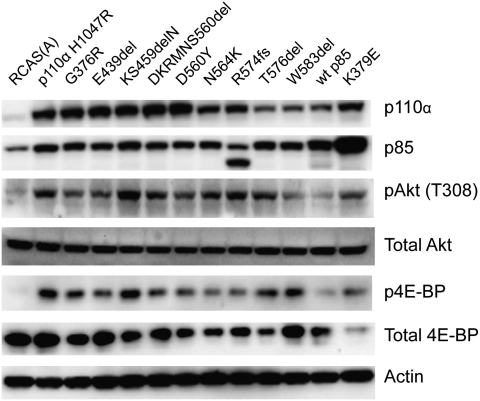
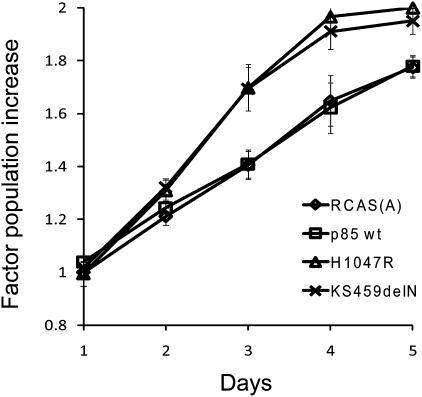
The mutant p85 proteins were expressed in chicken embryo fibroblasts (CEF) with the replication-competent avian sarcoma retroviral vector (RCAS) (21, 22), and expression was verified by Western blotting (Fig. 2). The vector-mediated expression of exogenous p85 resulted in elevated levels of endogenous p110α. After approximately 2 wk of incubation, foci of transformed cells appeared in the mutant-transfected cultures (Fig. 3A) but not on plates transfected with WT p85. The mutant p85 proteins showed different efficiencies of transformation (EOT), as defined by the number of foci induced per microgram of transfected DNA (Fig. 3B). Two of the p85 deletion mutants, KS459delN and DKRMNS560del, displayed a particularly high EOT, comparable to that of the H1047R mutant of p110α, which was used as a positive control. The nSH2 mutant, K379E, also belongs to this highly transforming category. R574fs and T576del transformed CEF with an intermediate efficiency, and the EOT of the remaining mutants was an order of magnitude lower than that of the highly transforming mutants. These differences in EOT were maintained when the cell cultures were cotransfected with WT human p110α and therefore probably reflect inherent properties of the p85 mutants. These data suggest that cancer-derived mutants of p85 have oncogenic activity, which probably reflects a mutation-mediated gain of function in the catalytic subunit. The transforming mutants of p85 also conferred increased replicative ability to the host cells. Fig. 4 documents this enhanced proliferation for the highly transforming mutant KS459delN. This enhancement was identical to that induced by the H1047R mutant of p110α. The same elevated cellular growth rates were found with the K379E mutant. Mutants R574fs, T576del, and DKRMNS560del induced an intermediate enhancement of cell growth that roughly corresponded to their intermediate efficiency of oncogenic transformation. Overexpression of WT p85 or of empty RCAS vector did not produce a detectable effect on the growth rates of CEF.
Fig. 2.
Western blots of mutant-infected and control CEF. Cells were lysed in Nonidet P-40 lysis buffer, and the lysates were clarified by centrifugation. For each lysate, 40 μg of protein were separated by SDS/PAGE and transferred to Immobilon P membranes (Millipore). Processing of the membranes is described in Materials and Methods. The R574fs mutation results in a truncated p85 protein, smaller than the endogenous WT p85.
Fig. 3.
(A) Representative focus assays of p85 mutants and controls on CEF. (B) Efficiencies of cellular transformation (number of foci per microgram of DNA) for individual p85 mutants and control constructs.
Fig. 4.
Cellular proliferation of CEF transformed by the p85 mutant KS459delN compared with control CEF transfected with RCAS(A), WT p85, and the H1047R mutant of p110α.
Mutations in p85 Induce Elevated Levels of Downstream Signaling.
As a regulatory subunit of PI3K, p85 signals in conjunction with the catalytic subunit p110 through the phosphorylation of phosphoinositide 4,5-bisphosphate, generating phosphoinositide 3,4,5-trisphosphate. The trisphosphate recruits the serine-threonine kinase Akt (cellular homolog of the Akt8 murine leukemia viral oncogene) and its activating kinase PDK1 (phosphoinositide-dependent kinase 1). Akt then initiates a cascade of downstream phosphorylations that activate TOR (target of rapamycin), S6K (p70 S6-kinase), and 4E-BP1 (eukaryotic initiation factor 4E binding protein 1). We have examined the activating phosphorylation of Akt and of 4E-BP1 as indicators of the PI3K signaling pathway (Fig. 2). CEF transfected with the mutant constructs were analyzed by Western blot. Transfection with the H1047R mutant of p110α served as positive control, and transfection with empty RCAS vector or WT p85 was used as negative control. All p85 mutants stimulated phosphorylation of Akt and of 4E-BP1. The strong differences in potency seen in the cell transformation assay were not evident in the levels of Akt or 4E-BP1 phosphorylation. These data support the conclusion that the mutations in p85 induce a gain of enzymatic function of PI3K. They also suggest, as has been observed previously, that potency in cell transformation is not always correlated with signaling levels measured by the phosphorylation of Akt and other downstream targets (23, 24).
Mutant p85 Proteins Still Bind to the Catalytic Subunits p110α and p110β.
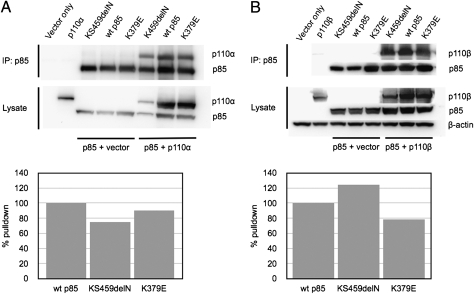
The p85 mutations cluster in regions of the protein that interact with the C2 and the helical domains of the catalytic subunit p110α (15, 25–27). These interactions mediate an inhibition of the catalytic activity of the enzyme, hence the mutant phenotypes could result from a weakening of p85–p110 binding. We therefore determined the ability of the p85 mutants to bind to p110α and p110β, the two ubiquitously expressed isoforms of p110. FLAG-tagged p85 constructs were coexpressed with human p110α or p110β in 293T cells using the pCAGGS vector (28). Coimmunoprecipitation and Western blots of cell lysates were performed as outlined in the legend to Fig. 5. All p85 mutants retained the ability to interact with the p110α and p110β isoforms of the catalytic subunit, and there was no significant reduction in this binding activity.
Fig. 5.
(A) Coimmunoprecipitation of p110α with WT p85 and with p85 mutants. (Upper) p110α is included in an immunoprecipitate of WT and of mutant p85. (Lower) Graphs show that under the conditions of the pull-down, the two mutants still bind efficiently to p110α. (B) Coimmunoprecipitation of p110β with WT p85 and with p85 mutants. Upper and Lower as in A.
Phenotypic Effects of p85 Mutations Are Mediated Exclusively by p110α.
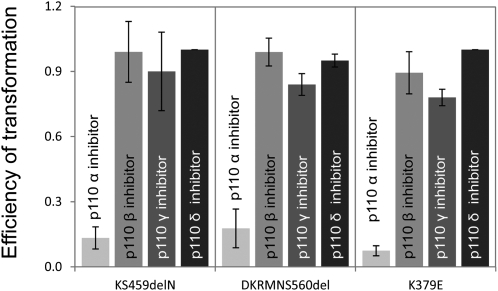
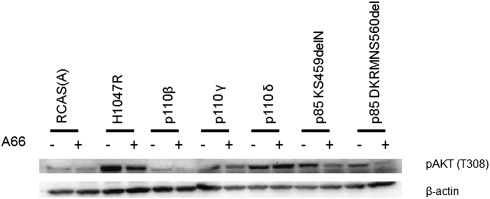
The ability of the p85 mutants to bind to p110α as well as p110β raises the question of which catalytic isoform is the most prevalent in mediating p85 mutant-induced oncogenic transformation. To answer this question, we studied the effects of p110 isoform-specific inhibitors on the formation of transformed cell foci by p85 mutants in CEF (Fig. 6). The p110α-specific inhibitor A66 (0.7 μM) induced a 75–80% reduction in focus formation by the highly transforming iSH2 mutants KS459delN, DKRMNS560del, and K379E. The p110β-specific inhibitor TGX221 (250 nM) did not interfere with focus formation by any of the p85 mutants but effectively and specifically inhibited transformation induced by p110β (Fig. S2). Neither p110γ- nor p110δ-specific inhibitors (AS-604850, 5 μM, and IC87114, 5 μM, respectively) had an effect on focus formation induced by the p85 mutants. The pan-PI3K inhibitors LY294002 (10 μM), ZK-93 (5 μM), PIK90 (500 nM), and NVP-BEZ-235 (100 nM) (29–31) also abolished focus formation by all of the p85 mutants, including K379E. We also studied the effects of the inhibitors on signaling induced by the p85 mutants (Fig. 7 and Fig. S3). The p110α-specific inhibitor A66 reduced phosphorylation of Akt on T308 by all p85 mutants (Fig. 7). The p110α-specific inhibitor also affected signaling induced by overexpressed p110β and p110γ. This activity probably reflects a dependence of p110β and p110γ signaling on p110α; it is not seen in oncogenic transformation (Fig. S2). The p110β- and p110δ-specific inhibitors showed the expected p110 isoform specificity in reducing signaling and failed to affect signaling by the p85 mutants (Fig. S3). The p110γ-specific inhibitor was not tested in signaling because p110γ does not interact with p85. These observations suggest that the predominant and possibly exclusive catalytic partner for the p85 mutants is p110α. The mutants induce a gain of function in p110α but apparently not in p110β. Rapamycin abolishes focus formation by all p85 mutants, suggesting that p85-induced cellular transformation uses the canonical PI3K signaling pathway in which TOR occupies a central branching position (Fig. S4).
Fig. 6.
Effect of isoform-specific PI3K inhibitors on focus formation in CEF induced by selected p85 mutants expressed by the RCAS retroviral vector. The following inhibitors were used in this experiment: p110α inhibitor A66 (700 nM), p110β inhibitor TGX-221 (250 nM), p110γ inhibitor AS-604850 (5 μM), and p110δ inhibitor IC87114 (5 μM). The p110α-specific inhibitor A66 reduced transformation by three potent p85 mutants. Isoform-specific inhibitors directed against p110β, p110γ, and p110δ had no significant effect on p85-induced focus formation. Controls documenting the specificity of the inhibitors are shown in Fig. S2. The plot shows relative efficiencies of transformation (ratio of focus count in the presence of inhibitor to the focus count in the absence of inhibitor).
Fig. 7.
Effect of the p110α-specific inhibitor A66 on mutant signaling. Signaling to Akt by KS459delN and DKRMNS560del was reduced by the inhibitor.
Discussion
The data presented in this article document gains of function in mutants of p85, a regulatory subunit of PI3K. Expression of the mutant p85 proteins in CEF induces oncogenic cellular transformation and increased proliferation. The p85 mutant-expressing cells show enhanced signaling through the PI3K pathway, as evidenced by the phosphorylation of Akt and 4E-BP. Expression of mutant or WT exogenous p85 in CEF also induces elevated levels of p110α by stabilizing the catalytic subunit (17, 29). The observations are in agreement with published studies on p85 mutants that used different cellular systems (17, 19). Going beyond these previous studies, we demonstrate oncogenic activity in a comprehensive collection of mutated p85, derived from glioblastoma. Most of these mutants have not been analyzed previously. Each of them can act as sole transforming agent in cultured primary cells. We document large quantitative differences in oncogenic potency of the p85 mutants; however, these differences are not reflected in signaling activity. The most potent mutants, KS459delN and DKRMNS560del, show specific transforming activity (foci per microgram of DNA) similar to the highly oncogenic p110α mutant H1047. Our experiments assessing the effects of PI3K-isoform-specific inhibitors on p85-mediated oncogenic transformation and on signaling further suggest a preferential partnering between p85 mutants and p110α in mediating the mutation-induced gain of function.
The iSH2 domain of p85 interacts with the adapter-binding and the C2 domains of p110α (18, 25–27, 32). The interaction between p85 and the adapter-binding domain stabilizes p110α. The interaction between p85 iSH2 and the C2 domain inhibits the enzymatic activity of p110α. The mutations in the iSH2 domain of p85 affect primarily the residues that interact with the C2 domain of p110α and weaken this inhibitory interaction. The result is a gain of function in PI3K activity. The engineered mutation K379E in the nSH2 domain of p85 affects the residue that is involved in an electrostatic interaction with E545 of p110α. The K379E mutation disrupts this interaction by substituting a negatively for a positively charged amino acid.
Our data show substantial differences in the oncogenic transforming efficiencies of the p85 mutants. Structural considerations offer some possible explanations for these differences in mutant potency. The highly oncogenic mutants of p85 show deletions of several amino acids or of a single amino acid in conjunction with mutation of the adjacent residue. The two most potent mutations, KS459delN and DKRMNS560del, are located on equivalent positions of two alternate helices in the iSH2 domain and could mark the inhibitory interaction surface. These mutations probably disrupt the α-helical structure of the iSH2 domain. Indeed, the secondary structure prediction program, NetSurfP, indicates a strong α-helical tendency for the two helices in the iSH2 domain of p85. Introduction of either KS459delN or DKRMNS560del significantly lowers the α-helical propensity, possibly prematurely ending the α helix. Breaking the α helix would disturb the positioning of the nSH2 and cSH2 (C-terminal SH2) domains. In addition, both of these mutations are in close proximity to the C2 domain of p110α, and disruption of the α helix could disrupt interactions with the C2 domain, further increasing catalytic activity as shown in a recent study (19). These two mutations along with the C420R mutation of p110α probably represent one of the mechanisms for aberrant activation of PI3K. They also delineate a region responsible for the inhibitory action of p85.
Among the less-oncogenic p85 proteins are those carrying the D560Y or the N564K mutation. The low oncogenic activity of D560Y is surprising, because D560 is one of the key p85 residues interacting with the C2 domain of p110α (18). However, unlike the potent mutations in this region of p85, neither D560Y nor N564K destabilize the α helix or change the length of the iSH2 domain as single mutants. The small structural consequences of these mutations may explain their weak transforming activity.
The other mutations occur toward the N- and C-terminal regions of the iSH2 domain. In the case of E439del, the shortening of the loop may influence the range of possible nSH2 conformations. Although the nSH2 domain itself is rigid, the flexible linker allows the nSH2 domain of WT p85 to sweep a significant amount of space (33). For the mutations on the C terminus of the iSH2 domain, possible mechanisms are speculative (20). This region is disordered in the structure of PI3K (18). However, the region is ordered in the structures of H1047R and the structure of the iSH2 in complex with the adapter-binding domain of p110α (20, 34). The mutations all occur in the long α helix of the iSH2 domain and likely destabilize its conformation and possibly its interaction with the disordered loop of the C2 domain. The role of the cSH2 domain remains unresolved, because it has been shown to not be required for the inhibition of PI3K activity by p85 (35–37). The oncogenicity of the p85 mutants probably confers a selective advantage to the cell that is commensurate with the strength of the oncogenic signal. Tumors carrying potently transforming mutants would then be expected to occur at higher frequencies than tumors carrying weakly transforming mutants. At present, there is insufficient genomic information to examine this suggestion, but for the mutations in p110α such a correlation between oncogenic potency and frequency of occurrence is observed (http://www.sanger.ac.uk/genetics/CGP/cosmic/).
The p85 mutants transform cells and generate downstream signals by binding and disinhibiting the catalytic subunit p110. We have used small-molecule inhibitors of p110 to identify the isoform that mediates the phenotypic changes induced by the p85 mutants. These data show that p110α is necessary and sufficient in mediating oncogenic transformation and signaling to Akt. Inhibition of p110β, p110γ, or p110δ has no effect on mutant activity. p110γ and p110δ can also be eliminated as potential partners, because they are not expressed at detectable levels in fibroblasts. We speculate that the exclusive role of p110α in mediating p85 mutant effects may reflect differences between p110α and p110β in their interaction with p85. The high sensitivity of p85 mutant-induced oncogenic transformation to rapamycin primarily reflects the fact that TOR is an essential component of the PI3K signaling pathway. However, p85 has been reported to bind to TOR directly with its cSH2 domain (38). Whether this interaction is rapamycin sensitive and whether it contributes to the oncogenic activity of the p85 mutants remains to be determined.
The results described in this communication are in agreement with the hypothesis that the gain-of-function mutations in p85 destabilize the inhibitory interaction between p85 and p110, resulting in a relief of p110 inhibition (17–19). At the same time, these mutants retain the ability to bind to p110, probably by the interaction with the adapter-binding domain, thus stabilizing p110. Our data suggest differences in the interaction of p85 with p110α vs. p110β. The exact nature of these differences and their consequences for PI3K function remain to be determined.
Materials and Methods
Plasmid Construction.
The construction of the pBSFI vector encoding p110α H1047R, p110β, p110γ, and p110δ has been described previously (23). To facilitate cloning, the SfiI site in WT p85 was destroyed by point mutation. The p85 mutant constructs were generated by using the QuikChange site-directed mutagenesis kit (Stratagene) and the following primers: G376R (+): 5′GATTATACTCTTACACTAAGGAAAGGGAGAAATAACAAATTAATCAAAATATTTC 3′; G376R (−): 5′ GAAATATTTTGATTAATTTGTTATTTCTCCCTTTCCTTAGTGTAAGAGTATAATC 3′; E439del (+): 5′ CAAATACCAACAGGATCAAGTTGTCAAAGATAATATTGAAGCTGTAGGG 3′; E439del (−): 5′ CCCTACAGCTTCAATATTATCTTTGACAACTTGATCCTGTTGGTATTTG 3′; KS459delN (+): 5′ GAATATAACACTCAGTTTCAAGAAAATCGAGAATATGATAGATTATATG 3′; KS459delN (−): 5′ CATATAATCTATCATATTCTCGATTTTCTTGAAACTGAGTGTTATATTC 3′; DKRMNS560del (+): 5′ GGCAGCTGAGTATCGAGAAATCATTAAACCAGACCTTATCCAGCTGAG 3′; DKRMNS560del (−): 5′ CTCAGCTGGATAAGGTCTGGTTTAATGATTTCTCGATACTCAGCTGCC 3′; D560Y (+): 5′ GAAGCAGGCAGCTGAGTATCGAGAAATTTACAAACGTATGAACAGCATTAAACC 3′; D560Y (−): 5′ GGTTTAATGCTGTTCATACGTTTGTAAATTTCTCGATACTCAGCTGCCTGCTTC 3′; N564K (+): 5′ GTATCGAGAAATTGACAAACGTATGAAGAGCATTAAACCAGACCTTATCCAGCTG 3′; N564K (−): 5′ CAGCTGGATAAGGTCTGGTTTAATGCTCTTCATACGTTTGTCAATTTCTCGATAC 3′; R574fs (+): 5′ GCATTAAACCAGACCTTATCCAGCTGAAAGACGAGAGACCAATACTTG 3′; R574fs (−): 5′ CAAGTATTGGTCTCTCGTCTTTCAGCTGGATAAGGTCTGGTTTAATGC 3′; T576del (+): 5′ CCAGACCTTATCCAGCTGAGAAAGAGAGACCAATACTTGATGTGGTTG 3′; T576del (−): 5′ CAACCACATCAAGTATTGGTCTCTCTTTCTCAGCTGGATAAGGTCTGG 3′; W583del (+): 5′ GAAAGACGAGAGACCAATACTTGATGTTGACTCAAAAAGGTGTTCGG 3′; W583del (−): 5′ CCGAACACCTTTTTGAGTCAACATCAAGTATTGGTCTCTCGTCTTTC 3′; K379E (+): 5′ GGAAAGGGGGAAATAACGAATTAATCAAAATATTTCATC 3′; K379E (−): 5′ GATGAAATATTTTGATTAATTCGTTATTTCCCCCTTCC 3′.
The mutated genes were subsequently cloned as SfiI DNA fragments into the avian retrovirus vector RCAS(A).Sfi (39). To examine the binding of p85 mutants with p110 isoforms, the mutated p85 genes were FLAG-tagged and cloned into the pCAGGs vector by standard PCR and cloning. All clones were confirmed by sequencing.
Cell Culture and Transfection.
Fertilized chicken eggs (white Leghorn) were obtained from Charles River Breeding Laboratories. Primary CEF were prepared and cultured as described previously (40, 41). For transfection, cells were plated at 80% confluence in F-10 containing 5.8% iron-supplemented FCS (Omega Scientific) and 1% L-glutamine–penicillin–streptomycin solution (Sigma-Aldrich). On the following day, CEF were transfected with the RCAS vectors using the dimethyl sulfoxide/Polybrene method (42). After two passages in the presence of serum, the cells were harvested for further analysis.
HEK 293-T cells were cultured in DMEM (Gibco, Invitrogen) supplemented with 10% FCS and 1% L-glutamine–penicillin–streptomycin solution. Transfections were carried using lipofectamine-PLUS (Invitrogen) according to the manufacturer's protocol. HEK293T cells in MP6 plates at 70% confluency were washed once with Opti-MEM medium and incubated in 0.8 mL of Opti-MEM. A total of 1 μg of plasmid DNA was mixed with 0.1 mL of Opti-MEM and 2 μL of Lipofectamine PLUS for 15 min at room temperature. Opti-MEM (0.1 mL) with 6 μL of Lipofectamine was added to the DNA-PLUS mixture and incubated for 15 min at room temperature. The mixture of DNA, PLUS, and Lipofectamine was added to the cells and incubated overnight. The second day, the medium was changed to DMEM containing 10% FCS and 1% L-glutamine–penicillin–streptomycin solution. Forty hours after transfection, the cells were collected, lysed, and analyzed for specific proteins.
Focus Assay.
Focus assays with infectious retroviral vectors were performed as previously described (40, 41). CEF were transfected with the appropriate RCAS constructs using the dimethyl sulfoxide/Polybrene method and overlayed with nutrient agar every other day for 2 to 3 wk until focus formation was observed. The plates were stained with crystal violet, and foci of transformed cells were counted. To examine inhibition of focus formation by different compounds, 10 μM LY294002, 100 nM NVP-BEZ-235, 2 nM rapamycin, 5 μM ZK-93, 250 nM TGX221, 5 μM IC87114, or 5 μM AS604850 were added to the nutrient agar in every overlay (29).
Cell Proliferation.
After transfection CEF were split into proliferation assay media containing F-10 supplemented with 2% FCS and 1% chicken serum. At the second split cells were seeded into a 96-well plate at 4,000 cells per well. On days 1 to 5 after seeding, CEF were incubated with 10 μg/mL Resazurin-Na in proliferation assay media for 4 h at 37 °C (43). Fluorescence was determined at an excitation wavelength of 560 nm and an emission of 590 nm.
Western Blot and Immunoprecipitation.
Western blotting was performed as previously described (44), with minor modifications. Cells were lysed in modified Nonidet P-40 lysis buffer (20 mM Tris-Cl, 150 mM NaCl, 1 mM MgCl2, 1% Nonidet P-40, and 10% glycerol with 1 mM PMSF, 1 mM DTT, 50 mM NaF, 1 mM Na3VO4, 50 mM β-glycerophosphate, and a protease inhibitor mixture from Roche). After centrifugation for 10 min at 18,000 × g at 4 °C, the protein concentration of the supernatant was determined. For the examination of inhibitor signaling, the cells were treated with 250 nM TGX-221 or 5 μM IC87114 for 2 h in the serum-containing condition ahead of collecting. For immunoprecipitation, cell lysates containing 40 μg of protein were incubated with anti-FLAG M2 Agarose (Sigma-Aldrich) overnight at 4 °C. Agarose beads were washed four times with lysis buffer and heated to 95 °C before separation on an SDS/PAGE gel. After transfer to Immobilon P membranes (Millipore) these membranes were blocked with 5% BSA in Tris-buffered saline with 0.1% Tween-20 (TBS-T) for 2 h at room temperature and then incubated with a dilution of 1:2,000 of anti-FLAG or 1:1,000 of anti-p110α or anti-p110β primary antibody overnight. Membranes were washed three times in TBS-T and incubated with peroxidase-coupled goat anti-mouse or goat anti-rabbit (Thermo Scientific) antibody for 1 h in 5% BSA/TBS-T at room temperature. The reactive bands were visualized by SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific).
For Western blotting, cell lysates containing 10 μg of total protein were separated on SDS/PAGE gels and transferred to Immobilon P membranes (Millipore). Membranes were incubated with 1:1,000 dilutions of primary antibodies directed against p85, pAkt (T308), Akt, p4E-BP, 4E-BP, and β-actin. Western blots were developed as described above. Anti-FLAG antibody (Flag-M2 F3165) was purchased from Sigma-Aldrich. Anti-p110α antibody (#4255), anti-p110 β antibody (#3011S), anti-p85 antibody (#4292), anti-Akt (#2967), anti-phospho-Akt (Thr-308) (#9275S), anti-4E-BP1 (#9452), and anti-phospho-4E-BP1 (Ser-65) (#9451S) antibodies were obtained from Cell Signaling Technology. Anti-p110δ antibody (sc-7176) was purchased from Santa Cruz Biotechnology.
Isoform-Specific Inhibitors.
The isoform-specific inhibitors for p110β, p110γ, and p110δ have been described in a previous publication (45). The specific inhibitor A66 for p110α was synthesized and characterized as described previously (compound 6) (46).
Supplementary Material
Acknowledgments
We thank Lynn Ueno for expert technical support. ZK-93 was a kind gift from Kevan Shokat (University of California, San Francisco, CA). We thank Yen Hoang Le Nguyen for stimulating and insightful discussions. This work was supported by grants from the National Cancer Institute. This is Manuscript 20600 of The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009652107/-/DCSupplemental.
References
- 1.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 2.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikenoue T, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 4.Isakoff SJ, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 5.Zhao JJ, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 7.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shekar SC, et al. Mechanism of constitutive phosphoinositide 3-kinase activation by oncogenic mutants of the p85 regulatory subunit. J Biol Chem. 2005;280:27850–27855. doi: 10.1074/jbc.M506005200. [DOI] [PubMed] [Google Scholar]
- 10.Jücker M, et al. Expression of a mutated form of the p85alpha regulatory subunit of phosphatidylinositol 3-kinase in a Hodgkin's lymphoma-derived cell line (CO) Leukemia. 2002;16:894–901. doi: 10.1038/sj.leu.2402484. [DOI] [PubMed] [Google Scholar]
- 11.Philp AJ, et al. The phosphatidylinositol 3′-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- 12.Mizoguchi M, Nutt CL, Mohapatra G, Louis DN. Genetic alterations of phosphoinositide 3-kinase subunit genes in human glioblastomas. Brain Pathol. 2004;14:372–377. doi: 10.1111/j.1750-3639.2004.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jimenez C, et al. Identification and characterization of a new oncogene derived from the regulatory subunit of phosphoinositide 3-kinase. EMBO J. 1998;17:743–753. doi: 10.1093/emboj/17.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaiswal BS, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CH, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 19.Wu H, et al. Regulation of Class IA PI 3-kinases: C2 domain-iSH2 domain contacts inhibit p85/p110alpha and are disrupted in oncogenic p85 mutants. Proc Natl Acad Sci USA. 2009;106:20258–20263. doi: 10.1073/pnas.0902369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miled N, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007;317:239–242. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 21.Hughes SH, Vogt PK, Stubblefield E, Bishop JM, Varmus HE. Integration of avian sarcoma virus DNA in chicken cells. Virology. 1981;108:208–221. doi: 10.1016/0042-6822(81)90539-0. [DOI] [PubMed] [Google Scholar]
- 22.Oh J, Julias JG, Ferris AL, Hughes SH. Construction and characterization of a replication-competent retroviral shuttle vector plasmid. J Virol. 2002;76:1762–1768. doi: 10.1128/JVI.76.4.1762-1768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc Natl Acad Sci USA. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA. 2008;105:2652–2657. doi: 10.1073/pnas.0712169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang CH, Mandelker D, Gabelli SB, Amzel LM. Insights into the oncogenic effects of PIK3CA mutations from the structure of p110alpha/p85alpha. Cell Cycle. 2008;7:1151–1156. doi: 10.4161/cc.7.9.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Z, Aronoff-Spencer E, Wu H, Gerfen GJ, Backer JM. The iSH2 domain of PI 3-kinase is a rigid tether for p110 and not a conformational switch. Arch Biochem Biophys. 2004;432:244–251. doi: 10.1016/j.abb.2004.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elis W, Lessmann E, Oelgeschlager M, Huber M. Mutations in the inter-SH2 domain of the regulatory subunit of phosphoinositide 3-kinase: Effects on catalytic subunit binding and holoenzyme function. Biol Chem. 2006;387:1567–1573. doi: 10.1515/BC.2006.195. [DOI] [PubMed] [Google Scholar]
- 28.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 29.Knight ZA, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maira SM, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 31.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 32.Amzel LM, et al. Structural comparisons of class I phosphoinositide 3-kinases. Nat Rev Cancer. 2008;8:665–669. doi: 10.1038/nrc2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sen KI, Wu H, Backer JM, Gerfen GJ. The structure of p85ni in class IA PI 3-kinase exhibits inter-domain disorder. Biochemistry. 2010;49:2159–2166. doi: 10.1021/bi902171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandelker D, et al. A frequent kinase domain mutation that changes the interaction between PI3Kalpha and the membrane. Proc Natl Acad Sci USA. 2009;106:16996–17001. doi: 10.1073/pnas.0908444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, Wjasow C, Backer JM. Regulation of the p85/p110alpha phosphatidylinositol 3′-kinase. Distinct roles for the n-terminal and c-terminal SH2 domains. J Biol Chem. 1998;273:30199–30203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- 36.Yu J, et al. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rordorf-Nikolic T, Van Horn DJ, Chen D, White MF, Backer JM. Regulation of phosphatidylinositol 3′-kinase by tyrosyl phosphoproteins. Full activation requires occupancy of both SH2 domains in the 85-kDa regulatory subunit. J Biol Chem. 1995;270:3662–3666. doi: 10.1074/jbc.270.8.3662. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez-Garcia A, et al. A new role for the p85-phosphatidylinositol 3-kinase regulatory subunit linking FRAP to p70 S6 kinase activation. J Biol Chem. 2002;277:1500–1508. doi: 10.1074/jbc.M103808200. [DOI] [PubMed] [Google Scholar]
- 39.Aoki M, Batista O, Bellacosa A, Tsichlis P, Vogt PK. The akt kinase: molecular determinants of oncogenicity. Proc Natl Acad Sci USA. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bos TJ, et al. Efficient transformation of chicken embryo fibroblasts by c-Jun requires structural modification in coding and noncoding sequences. Genes Dev. 1990;4:1677–1687. doi: 10.1101/gad.4.10.1677. [DOI] [PubMed] [Google Scholar]
- 41.Duff RG, Vogt PK. Characteristics of two new avian tumor virus subgroups. Virology. 1969;39:18–30. doi: 10.1016/0042-6822(69)90344-4. [DOI] [PubMed] [Google Scholar]
- 42.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwack K, Lynch RG. A new non-radioactive method for IL-2 bioassay. Mol Cells. 2000;10:575–578. doi: 10.1007/s10059-000-0575-6. [DOI] [PubMed] [Google Scholar]
- 44.Aoki M, et al. The catalytic subunit of phosphoinositide 3-kinase: Requirements for oncogenicity. J Biol Chem. 2000;275:6267–6275. doi: 10.1074/jbc.275.9.6267. [DOI] [PubMed] [Google Scholar]
- 45.Denley A, Kang S, Karst U, Vogt PK. Oncogenic signaling of class I PI3K isoforms. Oncogene. 2008;27:2561–2574. doi: 10.1038/sj.onc.1210918. [DOI] [PubMed] [Google Scholar]
- 46.Fairhurst RA, Imbach P. Preparation of N1-bithiazolyl pyrrolidinedicarboxamides and related compounds as phosphatidylinositol 3-kinase inhibitors. WIPO WO2009080705. 2009 (July 2, 2009) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.