Abstract
Chromosomal instability and the subsequent genetic mutations are considered to be critical factors in the development of the majority of solid tumors, but the mechanisms by which a stable diploid cell loses the ability to maintain genomic integrity are not well characterized. We have approached this critical issue through the use of high-throughput screens in untransformed diploid epithelial cells. In a screen of a cDNA library, we identified 13 kinases whose overexpression leads to increased ploidy. In a series of shRNA screens, we identified 16 kinases whose loss leads to increased ploidy. In both cDNA and shRNA screens, the majority of hits have not been linked previously to genomic stability. We further show that sustained loss of the shRNA screening hits leads to multipolar spindles and heterogeneous chromosome content, two characteristics of chromosomal instability. Loss of several of the kinases leads to loss of contact inhibition and to anchorage-independent growth, vital traits acquired during tumor development. We anticipate that this work will serve as a template for the comprehensive identification of pathways whose dysregulation can drive tumorigenesis through impaired karyotypic maintenance.
Keywords: aneuploidy, kinases, RNAi screen, tumorigenesis
Genomic instability promotes heterogeneity within a population of developing tumor cells, allowing for more rapid adaptation and acquisition of characteristics favorable for tumor development. Chromosomal instability is one type of genomic instability involving impaired maintenance of chromosome structure and number. Aneuploidy, defined here as an abnormal number of chromosomes, is a manifestation of chromosomal instability and was identified over 100 ago by von Hansemann (1). Aneuploidy is now known to characterize the majority of solid tumors, and chromosomal instability likely represents a common mechanism by which nascent tumor cells acquire beneficial mutations (2).
If the acquisition of chromosomal instability and the resulting aneuploidy represent important steps in the development of many solid tumors, understanding how a chromosomally stable diploid cell becomes aneuploid is a critical issue. Evidence from in vitro and in vivo studies suggests that one route leading from diploidy to aneuploidy passes through tetraploidy (3–5). Little is known, however, about the events that result in tetraploidy.
Here, we use cDNA and shRNA screens of human kinases in immortal but untransformed human diploid epithelial cells to look for proteins involved in the generation of tetraploidy and subsequent chromosomal instability. By approaching the question of chromosomal instability from the direction of a diploid cell, we hope to expand on screens that have been carried out in aneuploid tumor cells (6–11). This will allow us to follow in a controlled manner the progression from diploid to tetraploid to aneuploid. From these screens, we have identified a number of kinases whose overexpression or loss leads to increased ploidy. Further characterization reveals that certain of these kinases may function in pathways that are perturbed during the development of tumors driven by chromosomal instability.
Results
Screening Strategy for the Identification of Factors That Regulate Chromosomal Stability.
Our screening strategy was based on perturbing diploid cells to generate an increase in ploidy. We considered several factors in selecting a cell line for screens. First, the cell line had to be near diploid, immortalized but untransformed, and genetically stable. Second, the cell line had to continue proliferating as a tetraploid rather than arresting [i.e., lack a tetraploidy checkpoint (12, 13)] so that a population of cells with greater than 4N DNA content could be observed on a DNA content histogram. Third, the cell line had to be amenable to infection and processing for flow cytometry in high throughput.
We identified several bronchial epithelial cell lines that satisfied these criteria: NL-20, HBE135, and Beas2B. As shown in Fig. 1A for NL-20 cells, these cell lines continue cycling when tetraploids are generated with the spindle poison colcemid. All three cell lines have been immortalized with viral oncoproteins targeting the p53 and retinoblastoma tumor suppressor signaling pathways; karyotypic analysis indicated that the cell lines maintain diploid or near-diploid chromosome content.
Fig. 1.
Demonstration of chromosomal instability screen using a kinase cDNA library. (A) Mitotic escape in NL-20 cells results in viable tetraploids. NL-20 cells were treated overnight with colcemid or DMSO. Cells were fixed and stained for flow cytometry. (B) Kinase overexpression screen identifies kinases that regulate chromosomal stability. NL-20 cells were transduced with kinase cDNA constructs and grown with or without puromycin selection for 4 d. Cells were fixed and stained for flow cytometry. The percentage of cells with increased ploidy was calculated. The table represents those kinases that showed a reproducible increased ploidy phenotype in both NL-20 and HBE135 cells. (C) FISH analysis verifies increased ploidy. NL-20 cells were infected with the indicated kinase cDNAs and fixed for FISH analysis after 4 d. Fixed nuclei were stained with a chromosome 8-specific probe and counterstained with propidium iodide. The table shows the percentage of cells with greater than two copies of chromosome 8 for the indicated kinases.
To test our screening approach and begin to identify factors that lead to increased ploidy, we performed pilot screens in NL-20 and HBE135 cells using a cDNA library containing ≈250 human kinases. We hypothesized that overexpression of a kinase that is involved in maintaining chromosome content would cause an increase in the population of cells with greater than 4N DNA content or would generate a population of cells with exactly 8N DNA content (Fig. 1A). The table in Fig. 1B shows those kinases that increased ploidy in both cell lines across multiple experiments. Importantly, for two of the kinases known to have a role in the regulation of chromosomal stability in cancer cells, Plk1 and Nek2, the expression levels in tumors [2- to 5-fold increase above normal (14, 15)] are comparable to the relatively low level of overexpression that has been observed with Moloney murine leukemia-based expression vectors (16).
We next verified that the phenotype we were observing was indeed attributable to increased ploidy by using chromosome-specific interphase FISH. We introduced several of the screening hits into NL-20 cells and determined the number of copies of chromosome 8. As shown in Fig. 1C, all the kinases tested showed an increase in the number of copies of chromosome 8, revealing that we were, in fact, observing increased ploidy attributable to kinase expression.
shRNA Screen Identifies Kinases That Regulate Chromosomal Stability.
With the screening strategy in place, we carried out a larger scale screen using a kinase shRNA library targeting 460 human kinases, with an average of 4.5 shRNAs targeting each kinase (17, 18). Screens were performed by transducing shRNA constructs into target cells and fixing the cells for flow cytometric analysis of DNA content after 4 d. Data were analyzed by measuring the population of cells with greater than 4N DNA content and calculating z scores relative to either plate median or negative control samples.
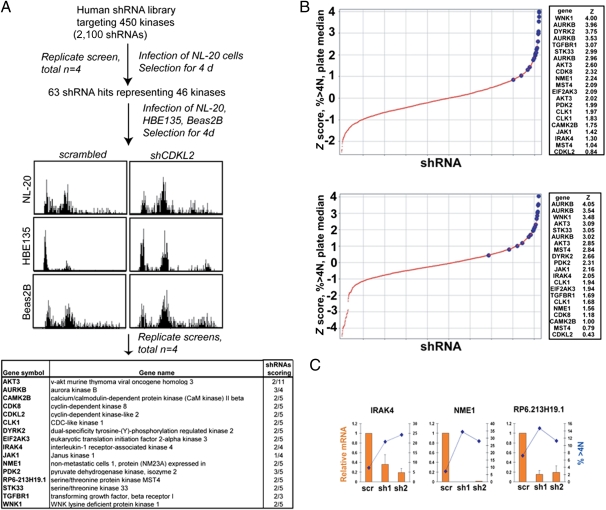
In a primary screen of 2,100 kinase shRNAs in NL-20 cells, 63 shRNAs targeting 46 kinases were classified as hits (a detailed description of hit determination in primary and secondary screens is included in Materials and Methods). Our screening approach was validated through the identification of several kinases whose knockdown is known or would be expected to promote aneuploidy, including AURKB, AURKC, and NEK6. Secondary screens were performed using NL-20, HBE135, and Beas2B cells to find those shRNAs that gave similar phenotypes across multiple cell lines. Our final list of kinase shRNAs was composed of kinases that reproducibly scored in multiple cell lines or strongly scored in one cell line (Fig. 2A). Notably, shRNAs targeting these kinases had reproducibly shown the highest z scores in the primary screens (Fig. 2B and Fig. S1).
Fig. 2.
shRNA screen identifies kinases that regulate chromosomal stability. (A) Flow chart describing the screening of a kinase shRNA library for kinases whose knockdown causes an increase in ploidy. NL-20 cells were infected in quadruplicate in 96-well plates, and ploidy was assessed by flow cytometry 4 d postinfection. Primary hits were rescreened against three different cell lines to find the strongest hits shown in the table. (Right) Column indicates the number of unique shRNAs that showed a ploidy phenotype. (B) Z score distribution (>4N) for two of the replicate screens. Z scores relative to the plate median of the percentage of cells with greater than 4N DNA content were calculated for each data point in each of four replicate screens. The large blue markers represent shRNAs that scored in follow-up screens in additional cell lines. (Right) Z scores for these shRNAs are shown. (C) Correlation of increased ploidy with gene knockdown. HBE135 cells were transduced with the indicated shRNAs. After 4 d, mRNA was prepared and cells were fixed for flow cytometric measurement of DNA content. The level of mRNA was calculated by measuring the ratio of the indicated gene to GAPDH for the indicated shRNAs vs. the same ratio in control samples. For flow cytometry, the percentage of cells with greater than 4N DNA content is shown. scr, scrambled; sh, short hairpin.
A primary concern in shRNA screens is the production of phenotypes through off-target effects. As shown in the table in Fig. 2A, for 15 of the 16 kinases, shRNAs targeting multiple sequences of the target mRNA were found to give a phenotype, arguing against generalized off-target effects. To test this in more detail, we correlated gene knockdown with phenotypic strength for two distinct shRNAs in each of three genes. As shown in Fig. 2C, in each case, the shRNA showing the highest reduction in mRNA level also gave rise to the stronger phenotype (percentage of cells with greater than 4N DNA content). These data argue that increased ploidy is the result of on-target effects of the shRNAs targeting each kinase.
Comparison with shRNA Screens Carried Out in Aneuploid Tumor Cell Lines.
An important component of our screening strategy was our use of stable near-diploid cells. Given the multiple changes that occur during tumor development, one might expect that the pathways regulating chromosomal stability in tumor cells are quite different from those in normal cells. To test this hypothesis, we compared the data from our screens with those obtained in two screens in the aneuploid tumor cell lines HeLa and U2OS (6, 10). Because these screens were carried out at the genome level to look for general cell cycle regulators, we extracted data for kinases whose loss led to an increase in the population of cells with greater than 4N DNA content. A comparison of the phenotypic consequences of knocking down the kinases found in our shRNA screens in aneuploid cells reveals that only for one kinase (AURKB) did knockdown lead to increased ploidy in all three cell lines (Table S1). It is important to note that expression of each of these genes was verified in HeLa cells (6). Conversely, none of the genes found in the other screens had any ploidy phenotype upon knockdown in our screens. Notably, as observed in the other screening papers, there is very little overlap between HeLa and U2OS cells, suggesting that aneuploid cells of different origins have evolved different strategies to control chromosome content.
Validation of shRNA Hits with FISH.
As described above for the cDNA screening hits, we used FISH to count the number of copies of chromosome 12 as a proxy for ploidy in cells expressing kinase shRNAs. The strongest shRNA against each kinase that scored in our secondary screen was introduced into both NL-20 and HBE135 cells by viral transduction. Following selection for 4 d, the cells were fixed and processed for FISH analysis. As shown in Fig. 3A and B, the majority of shRNAs in both NL-20 and HBE135 cells induced an increase in the number of cells with greater than two copies of chromosome 12.
Fig. 3.
Measurement of increased ploidy by FISH. (A) Example of shRNAs that increase ploidy as measured by FISH. NL-20 cells were infected with the indicated kinase shRNAs and fixed for FISH analysis after 4 d. A centromeric probe specific for chromosome 12 (red) was hybridized to nuclei, which were then counterstained with DAPI (blue). sh, short hairpin. (B) Quantitation of FISH analysis for chromosome 12. FISH analysis was carried out with the indicated shRNAs in NL-20 and HBE135 cells. The number of nuclei with greater than two copies of chromosome 12 for each shRNA was measured using CellProfiler software. For each shRNA, at least 50 nuclei were counted.
For several shRNAs, we directly compared the two phenotypic readouts (flow cytometry and FISH) within the same set of infections. Fig. S2 demonstrates a good correlation between the percentage of cells with a chromosome content of greater than 4N and the percentage of cells with more than two copies of either chromosome 12 or chromosome 8. These data both validate the use of flow cytometry in our primary and secondary screens as a strategy for assessing increased ploidy and demonstrate that the knockdown of the kinases we have identified does indeed induce changes in ploidy.
Knockdown of Several Kinases Leads to Tumor Cell Phenotypes.
We hypothesized that the tetraploidy we observed on knockdown of our kinase hits would serve as a precursor to aneuploidy and chromosomal instability. Because chromosomal instability gives rise to heterogeneity in a cell population, we reasoned that examining the distribution of phenotypic and genotypic markers across a population of cells after sustained knockdown of the kinases would indicate the presence of chromosomal instability.
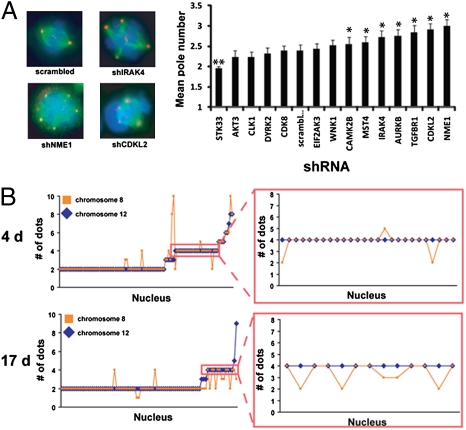
The first phenotype we investigated was mitotic spindle pole multiplicity. Abnormal spindles (i.e., a number of poles not equal to 2) are commonly found in aneuploid tumors and are indicative of the errors in the cell cycle that give rise to chromosomal instability. At 21 d postinfection, HBE135 cells were fixed and stained to identify the mitotic spindle (tubulin), centrosomes (pericentrin), and chromosomes (DAPI). We quantified the imaging data by calculating the average number of poles observed in multiple mitotic figures (Fig. 4A). For seven of the kinases tested, knockdown resulted in a significant increase in the mean number of poles above the scrambled control. We also noted that the modal number of poles was three for abnormal mitotic figures (Table S2). Table S2 also notes additional mitotic abnormalities that we observed on kinase knockdown. These data are consistent with the presence of chromosomal instability.
Fig. 4.
Knockdown of several kinases results in multipolar mitoses and aneuploidy. (A) Knockdown of kinases results in multipolar mitoses. Kinase shRNAs were transduced into HBE135 cells, and the cells were selected for 21 d. Cells were fixed for immunofluorescence and stained with antibodies against tubulin (green) and pericentrin (red). DNA was stained with DAPI (blue). (Left) Examples of multipolar mitosis phenotypes are shown. (Right) Graph shows the mean number of poles for each sample (n = 25 for each). *P < 0.05 by Student's t test. (B) Sustained knockdown of NME1 results in a progression from tetraploidy to aneuploidy. HBE135 cells were transduced with the NME1 shRNA and selected for 4 or 17 d. Cells were fixed for FISH analysis and stained with centromeric probes for chromosomes 8 and 12. The number of copies of these two chromosomes in each nucleus is shown. (Right) Number of copies of chromosome 8 in nuclei with four copies of chromosome 12.
We next investigated the ploidy of cells expressing one of our more penetrant shRNA hits, NME1, which allowed us not only to investigate heterogeneity but to demonstrate the evolution of aneuploidy from tetraploidy directly. We performed chromosome counting FISH analysis on HBE135 cells transduced with the NME1 shRNA 4 and 17 d postinfection. As shown in Fig. 4B, at 4 d, the distribution of chromosome number was suggestive of a bimodal population of cells with either two or four copies of both chromosomes 8 and 12. At 17 d, however, the bimodal distribution is less apparent. The differences between the cell populations at 4 and 17 d can most clearly be observed by comparing the distribution of chromosome number in cells with four copies of chromosome 12 (Fig. 4B Right). At 4 d, the majority (88%) of nuclei were tetraploid for both chromosomes 8 and 12. At 17 d, however, only 61% of nuclei were tetraploid for both chromosomes. It should also be noted that these data likely greatly underrepresent the extent of aneuploidy, because only two chromosomes are being counted. Even with this underestimation of the degree of aneuploidy, these data are consistent with a progression from tetraploidy to aneuploidy and further highlight the heterogeneity of the cell population after kinase knockdown.
To test whether the chromosomal instability we observed by knockdown of the kinases of interest was correlated with the acquisition of tumorigenic characteristics, we assayed cells for the loss of contact inhibition and anchorage dependence, two hallmarks of tumor cells. Control or kinase shRNAs were introduced into HBE135 cells, and the cells were allowed to grow under selection for ≈14 d. At this point, the cells were plated on tissue culture plastic or within a soft agar matrix and allowed to grow for an additional 14 or 21 d, respectively. As shown in Fig. S3, knockdown of several kinases significantly increased the number of foci formed on plastic relative to the control condition. Further, knockdown of two kinases, IRAK4 and NME1, with shRNAs targeting two regions of each mRNA, allowed for a significant increase in soft agar colony formation (Fig. 5A). For IRAK4, in which a noticeable difference in mRNA knockdown efficiency was observed between the two shRNAs (Fig. 2C), the number of colonies was directly correlated with the degree of knockdown efficiency and the induction of increased ploidy.
Fig. 5.
Knockdown of kinases results in growth in soft agar. (A) Knockdown of IRAK4 and NME1 promotes growth in soft agar. HBE135 cells were transduced with scrambled shRNAs or two shRNAs targeting IRAK4 or NME1. sh, short hairpin. Cells were selected for 21 d, seeded in soft agar, and incubated for an additional 21 d. Colonies were stained with crystal violet. (B) Soft agar growth requires sustained knockdown of NME1. HBE135 cells were transduced with NME1 shRNA and plated in a soft agar matrix at 4 or 17 d postinfection. The colony number is the average of three (4 d) or two (17 d) wells ± SEM. (C) Soft agar colonies are aneuploid. A single colony was picked from soft agar in cells containing the NME1 shRNA. The colony was expanded for several passages, and the cells were fixed for FISH analysis as described in Fig. 4B.
Because only two of the kinases tested promoted soft agar colony formation on knockdown, one might argue that it was not chromosomal instability that was driving this phenotype. Two pieces of evidence support the idea that growth in soft agar is the result of chromosomal instability. First, soft agar colony formation in cells expressing the NME1 shRNA occurred only 17 d postinfection and not 4 d postinfection (Fig. 5B). This is consistent with our hypothesis that passage through several cell cycles is required for the accumulation of the appropriate advantageous chromosomal changes. Second, the genetic content of the soft agar colonies is clearly aneuploid. For cells with the NME1 shRNA, we picked soft agar colonies from the matrix after 3 wk of incubation and expanded the clones out on tissue culture plastic; the cells were then prepared for chromosome counting FISH analysis as described for Fig. 4B. As shown in Fig. 5C for the cells from one soft agar colony, the nuclei are much more heterogeneous than are cells either 4 or 17 d postinfection (Fig. 4B). There is no indication of a bimodal population of diploids and tetraploids; instead, the population seems to consist of cells either diploid for both chromosomes or aneuploid. Although we cannot eliminate the possibility of a different underlying cause for growth in soft agar, these results strongly support the hypothesis that knockdown of our kinase hits leads to tetraploidy and chromosomal instability, which, in certain contexts, can promote focus formation and anchorage-independent growth.
Discussion
Through the use of combined cDNA and shRNA screens in untransformed diploid cells, we identified several kinases that regulate chromosomal stability. In a pilot screen of 250 kinase cDNAs, we identified 13 kinases whose overexpression led to increased ploidy. Of these 13 kinases, NEK2 and PLK1 are known to function in the maintenance of chromosomal stability, validating our screening strategy (19–21). In a screen of a kinase shRNA library, we obtained a list of 16 largely uncharacterized kinases whose knockdown in diploid cells caused an increase in ploidy, among which was AURKB, whose dysregulation is known to lead to increased ploidy through failed cell division (22). Thus, we were able to validate the utility of both screens through the identification of known regulators of chromosomal stability. Importantly, however, the majority of the genes that we identified have no known role in the maintenance of genomic integrity. A scan of the literature reveals that in addition to AURKB and NME1 (which we examined in detail in the companion paper), 9 of the remaining 14 kinase hits from the loss-of-function screen have been shown to be deregulated in some manner in a variety of tumors (23–31), lending strong support to the physiological significance of our findings to tumor biology.
We initiated this work with the assumption that tetraploidy is a precursor of chromosomal instability, an assumption that was supported by our characterization of the consequences of sustained kinase knockdown. The mitosis phenotypes observed on knockdown of our shRNA screening hits strongly support the existence of chromosomal instability. We observed several instances of multipolar spindles with four poles, which would be indicative of a tetraploid cell undergoing mitosis. More commonly among abnormal spindles was the presence of three spindle poles, suggestive of an aneuploid cell undergoing mitosis. This is consistent with the occurrence of multiple cell divisions following the initial tetraploid mitosis during the several weeks after introduction of the kinase shRNA. In addition to spindle abnormalities, various segregation errors were observed, including chromosome capture defects and lagging chromosomes during anaphase. These segregation errors are consistent with the consequences of supernumerary centrosomes in aneuploid cells and are presumed to be the direct cause of chromosomal instability (32). Thus, knockdown of our kinase hits leads to tetraploidy, and for several of the kinase hits, subsequent mitoses lead to aneuploidy and chromosomal instability.
On the basis of our experiments, we concluded that the tetraploid cells present several days after kinase knockdown moved toward an aneuploid state in stable knockdown cells and in cells isolated from soft agar. However, we cannot exclude the possibility that chromosome missegregation in the diploid population resulting from kinase knockdown led to aneuploid cells with a proliferative advantage over the rest of the population. In support of our conclusions, previous work in which diploid and tetraploid populations were separated showed that the tetraploid but not the diploid population promoted tumorigenesis (33).
Chromosomal instability generates heterogeneity within a population of cells, providing the opportunity for rapid adaptation to different environments. Our focus formation and soft agar experiments were designed to test whether this heterogeneity driven by chromosomal instability was sufficient to promote the development of tumor-like characteristics. The focus formation experiments revealed that four of the kinase shRNAs led to a significant increase in focus formation over the control. Further, of these shRNAs, only those targeting IRAK4 and NME1 led to soft agar colony formation with two unique shRNAs. The focus formation and soft agar results are consistent with survival and proliferation in soft agar being a much stronger selective pressure than loss of contact inhibition.
Overall, these results indicated that chromosomal instability itself is not sufficient for either loss of contact inhibition or anchorage-independent growth; there are clearly additional contributing factors. One factor is the inability of cells to tolerate continued knockdown of certain kinases. In culturing cells stably expressing the kinase shRNAs, we found that cells containing certain shRNAs (most clearly, those against AURKB and RP6-213H19.1) either could not be cultured for long-term experiments or required significantly more time in culture before enough cells were obtained for further experiments. In this case, the negative effect of the shRNA on proliferation masks its potential positive effect on tumorigenicity. In cases like this, transient down-regulation of a kinase or pathway essential for proliferation may be sufficient to generate a level of chromosomal instability that favors tumorigenesis. An example of such a phenomenon comes from work in transgenic mice overexpressing the spindle checkpoint protein MAD2. In transgenic mice in which tumors had already formed as a result of the chromosomal instability resulting from MAD2 overexpression, shutting off the MAD2 transgene had no effect (34).
A second contributing factor is the degree of induced chromosomal instability. One of the strongest hits in our shRNA screens was EIF2AK3. In follow-up experiments, knockdown of EIF2AK3 was found to promote an increase (below the significance threshold) in multipolar mitoses and a significant increase in focus formation. However, in screening for soft agar colony formation, we found that cells in which EIF2AK3 was stably knocked down formed fewer colonies than the control. Similar observations were made with cells stably expressing a shRNA against CAMK2B. In these cases, one might imagine that the stress of proliferating with a large degree of chromosomal instability is compounded by the stress of adapting to anchorage-independent growth. In support of this observation, work on mice with decreased expression of the centromeric motor protein CENP-E found that the positive effect of aneuploidy on tumorigenicity is inversely proportional to the level of aneuploidy (35). These observations suggest that the relationship between chromosomal instability and tumorigenesis is much more than a matter of the degree of instability.
Taken together, this work has identified several factors that may play a role in early stages of tumorigenesis driven by chromosomal instability. Experiments are underway to place these factors within the cellular signaling context using genetic and cell biological experiments. We anticipate that by understanding the mechanisms by which chromosomal instability is acquired, we can better follow the progression from normal diploid cells to aneuploid tumor cells.
Materials and Methods
Cell Lines and Antibodies.
NL-20, HBE135 E6/E7, Beas2B, and 293T cells were obtained from the American Type Culture Collection and cultured according to the supplier's instructions. Beas2B cells were subcloned to create tetraploid and diploid variants (according to flow cytometry and karyotyping), and the diploid variant was used in secondary screens. The karyotype of cell lines used in screens was verified at the Harvard/Dana Farber Cancer Center cytogenetics core facility. For NL-20, 100% of cells examined were near diploid, and for HBE135 and Beas2B, 90% were near diploid. Primary antibodies used were anti-α-tubulin (mouse monoclonal TU01; Zymed Laboratories) and anti-pericentrin (rabbit polyclonal; Covance). Secondary antibodies for immunofluorescence were obtained from Invitrogen and used at a ratio of 1:400.
cDNA Library, shRNA Library, Virus Production, and Infections.
The kinase cDNA library has been described elsewhere (36), and kinases were expressed from a retroviral vector based on the Moloney murine leukemia virus. The kinase shRNA library used for screens is a subset of the lentiviral shRNA library in the LKO.1 vector from the RNAi Consortium. DNA preparation, transfections, and virus preparation methods have been published elsewhere (37). For primary screens in NL-20 cells, 3,000 cells were plated in a 96-well plate. The next day, lentivirus was added to each well along with 8 μg/mL polybrene (Sigma). Plates were centrifuged at 1,100 × g for 30 min and incubated overnight at 37 °C. After ≈16 h, the virus was removed and fresh media with or without 0.5 μg/mL puromycin (Sigma) were added. Cells were incubated for a total of 4 d, with an additional media change on day 3 postinfection. The efficiency of the infection was assessed by adding the viability dye Resazurin (Sigma) on day 4 before cell fixation.
Cell Fixation and Flow Cytometry Analysis.
Following viability measurement, cells were dissociated from the dish and washed once with PBS; 175 μL of ice-cold 70% ethanol (vol/vol) then was added to the cell pellet. Cells were prepared for flow cytometry analysis by washing once with PBS and staining with 20 μg/mL propidium iodide (Sigma)/20 μg/mL RNase A (Sigma) in 0.1% Triton X-100 for 30 min. Flow cytometry analysis on 1,000 events was carried out using an Easycyte Flow Cytometer (Millipore). Following data acquisition, the percentage of cells containing 2N, 2N–4N, 4N, >4N, and 8N DNA was quantified for data analysis and hit determination, the details of which appear in SI Text.
Supplementary Material
Acknowledgments
We thank J. Sawyer for preparation of the kinase shRNA virus; A. L. Conery, J. Doench, A. Rolfes, and W. Endege for critical reading of the manuscript; and members of the E.H. laboratory for helpful discussions. A.R.C. was supported by Postdoctoral Fellowship PF-07-030-01-CCG from the American Cancer Society.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010627107/-/DCSupplemental.
References
- 1.von Hansemann D. Ueber asymmetrische Zelltheilung in epithel Krebsen und deren biologische Bedeutung. Virchows Arch A Pathol Anat Histopathol. 1890;119:299. [Google Scholar]
- 2.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Galipeau PC, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett's esophagus. Proc Natl Acad Sci USA. 1996;93:7081–7084. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 5.Caldwell CM, Green RA, Kaplan KB. APC mutations lead to cytokinetic failures in vitro and tetraploid genotypes in Min mice. J Cell Biol. 2007;178:1109–1120. doi: 10.1083/jcb.200703186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kittler R, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- 7.Swanton C, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Draviam VM, et al. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat Cell Biol. 2007;9:556–564. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- 9.Björklund M, et al. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- 10.Mukherji M, et al. Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci USA. 2006;103:14819–14824. doi: 10.1073/pnas.0604320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kittler R, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 12.Margolis RL, Lohez OD, Andreassen PR. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J Cell Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- 13.Wong C, Stearns T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6:6–17. doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayward DG, et al. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004;64:7370–7376. doi: 10.1158/0008-5472.CAN-04-0960. [DOI] [PubMed] [Google Scholar]
- 15.He ZL, Zheng H, Lin H, Miao XY, Zhong DW. Overexpression of polo-like kinase1 predicts a poor prognosis in hepatocellular carcinoma patients. World J Gastroenterol. 2009;15:4177–4182. doi: 10.3748/wjg.15.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi K, et al. Engineered long terminal repeats of retroviral vectors enhance transgene expression in hepatocytes in vitro and in vivo. Mol Ther. 2003;8:796–803. doi: 10.1016/j.ymthe.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 18.Grueneberg DA, et al. Kinase requirements in human cells: I. Comparing kinase requirements across various cell types. Proc Natl Acad Sci USA. 2008;105:16472–16477. doi: 10.1073/pnas.0808019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayward DG, Fry AM. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006;237:155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 22.Meraldi P, Honda R, Nigg EA. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr Opin Genet Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellacosa A, Kumar CC, Di Cristofano A, Testa JR. Activation of AKT kinases in cancer: Implications for therapeutic targeting. Adv Cancer Res. 2005;94:29–86. doi: 10.1016/S0065-230X(05)94002-5. [DOI] [PubMed] [Google Scholar]
- 25.Miller CT, et al. Amplification and overexpression of the dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 (DYRK2) gene in esophageal and lung adenocarcinomas. Cancer Res. 2003;63:4136–4143. [PubMed] [Google Scholar]
- 26.Kadota M, et al. Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. Cancer Res. 2009;69:7357–7365. doi: 10.1158/0008-5472.CAN-09-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porcu M, Gielen O, Cools J, De Keersmaecker K. JAK1 mutation analysis in T-cell acute lymphoblastic leukemia cell lines. Haematologica. 2009;94:435–437. doi: 10.3324/haematol.13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung V, et al. The Ste20 kinase MST4 plays a role in prostate cancer progression. Cancer Res. 2003;63:3356–3363. [PubMed] [Google Scholar]
- 29.Lyng H, et al. Gene expressions and copy numbers associated with metastatic phenotypes of uterine cervical cancer. BMC Genomics. 2006;7:268–272. doi: 10.1186/1471-2164-7-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Q, et al. Tgfbr1 haploinsufficiency is a potent modifier of colorectal cancer development. Cancer Res. 2009;69:678–686. doi: 10.1158/0008-5472.CAN-08-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang CL, Cha SK, Wang HR, Xie J, Cobb MH. WNKs: Protein kinases with a unique kinase domain. Exp Mol Med. 2007;39:565–573. doi: 10.1038/emm.2007.62. [DOI] [PubMed] [Google Scholar]
- 32.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 34.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Park J, et al. Building a human kinase gene repository: Bioinformatics, molecular cloning, and functional validation. Proc Natl Acad Sci USA. 2005;102:8114–8119. doi: 10.1073/pnas.0503141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearlberg J, et al. Screens using RNAi and cDNA expression as surrogates for genetics in mammalian tissue culture cells. Cold Spring Harb Symp Quant Biol. 2005;70:449–459. doi: 10.1101/sqb.2005.70.047. [DOI] [PubMed] [Google Scholar]
- 38.Carpenter AE, et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100–R111. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.