Abstract
Living organisms utilize carbohydrates as essential energy storage molecules. Starch is the predominant carbohydrate storage molecule in plants while glycogen is utilized in animals. Starch is a water-insoluble polymer that requires the concerted activity of kinases and phosphatases to solubilize the outer surface of the glucan and mediate starch catabolism. All known plant genomes encode the glucan phosphatase Starch Excess4 (SEX4). SEX4 can dephosphorylate both the starch granule surface and soluble phosphoglucans and is necessary for processive starch metabolism. The physical basis for the function of SEX4 as a glucan phosphatase is currently unclear. Herein, we report the crystal structure of SEX4, containing phosphatase, carbohydrate-binding, and C-terminal domains. The three domains of SEX4 fold into a compact structure with extensive interdomain interactions. The C-terminal domain of SEX4 integrally folds into the core of the phosphatase domain and is essential for its stability. The phosphatase and carbohydrate-binding domains directly interact and position the phosphatase active site toward the carbohydrate-binding site in a single continuous pocket. Mutagenesis of the phosphatase domain residue F167, which forms the base of this pocket and bridges the two domains, selectively affects the ability of SEX4 to function as a glucan phosphatase. Together, these results reveal the unique tertiary architecture of SEX4 that provides the physical basis for its function as a glucan phosphatase.
Keywords: carbohydrate, Lafora disease, laforin, phosphorylation
Plants and animals store carbohydrates as starch and glycogen, respectively. Starch is produced in diurnal cycles and is composed of < 10% w/w amylose and > 80% w/w amylopectin in Arabidopsis thaliana leaves (1). Amylose is a linear molecule composed of glucose moieties linked by α-1,4-glycosidic linkages with very few branches. Amylopectin, which is similar to glycogen, is composed of α-1,4-glycosidic linkages with α-1,6-glycosidic branches, but amylopectin branches are arranged in clusters at regular intervals and the branches form double helices that pack together to form crystalline lamellae (2, 3). The decreased branching and crystalline lamellae of amylopectin are key contributors to the insolubility of starch, while glycogen has more branches and is water-soluble.
Starch is a water-insoluble polymer whose surface is inaccessible to most enzymes. Recent work convincingly demonstrates that reversible starch phosphorylation and dephosphorylation is essential for processive starch metabolism (reviewed in refs. 4–7). An essential signal triggering starch catabolism is phosphorylation on the C6 position of glucose moieties on the surface of starch by glucan water dikinase (GWD/R1) (8, 9). C6 phosphorylation triggers C3 phosphorylation by phosphoglucan water dikinase (PWD) (8, 10, 11). Recent data suggest that C6 phosphorylation fits within the unphosphorylated structure of the amylopectin helix, but C3 phosphorylation imposes significant steric effects and is predicted to induce a conformational change (12–15). This suggests that GWD-directed C6 phosphorylation promotes local hydration of crystalline lamellae and that PWD-directed C3 phosphorylation induces helix strain. This helix strain allows substrate access to β-amylases that release maltose from the surface of the starch molecule (4). However, in order to efficiently digest the starch molecule, glucan phosphatase activity is necessary to prevent accumulation of phosphorylated starch and phosphorylated starch breakdown intermediates (16).
All known Archaeplastida/Kingdom Plantae genomes encode for the glucan phosphatase Starch Excess4 (SEX4), but genomes outside of Kingdom Plantae lack a SEX4 ortholog (17). SEX4 contains an amino-terminal chloroplast Targeting Peptide (cTP), followed by a dual specificity phosphatase (DSP) domain, and carbohydrate-binding module (CBM) (17, 18). Mutations in the SEX4 gene result in substantial accumulations of starch in A. thaliana leaves due to decreased rates of degradation and the accumulation of soluble phospho-oligosaccharides (16, 19–21). In addition to dephosphorylating amylopectin (18), recombinant SEX4 also dephosphorylates crystalline maltodextrins (22), starch granules isolated from A. thaliana leaves (16), and phospho-oligosaccharides (16). While C6 and C3 phosphorylation is carried out by two dikinases, SEX4 is able to dephosphorylate both C6 and C3 positions with similar kinetics (22). Thus, it appears that SEX4 possesses a broader substrate specificity than the kinases, in terms of phosphate position, length of glucan, and solubility of the glucan.
SEX4 is a member of the larger glucan phosphatase family that includes the human protein laforin (17–19). Mutations in the gene encoding laforin result in Lafora disease (LD, OMIM 254780), an autosomal recessive progressive myoclonus epilepsy (23). A hallmark of LD is the accumulation of intracellular water-insoluble glucans, i.e., glucose polymers linked by glycosidic bonds, termed Lafora bodies (LBs) (23–27). Laforin contains a CBM and DSP, like SEX4, but in the opposite orientation. Similar to SEX4, laforin dephosphorylates phospho-glucans in vitro (28), and LBs from LD patients and a LD mouse model have increased phosphate compared to glycogen (29–32). Strikingly, mutations in the genes encoding both laforin and SEX4 result in the accumulation of insoluble glucans (19–21). In addition, laforin partially complements mutations in SEX4, highlighting a functional overlap between divergent glucan phosphatases (18).
The structure, mechanism, and basis for specificity of glucan phosphatases are all unknown. Herein, we report a previously undescribed structure of a glucan phosphatase. The structure reveals that the domains of SEX4 form an integral structural unit, with extensive interdomain interactions. The unique multidomain structure of SEX4 serves to align the phosphatase active site and carbohydrate-binding face. This extended contiguous surface produces an active site incorporating both catalytic and glucan substrate binding functionalities, providing a structural model for the physical basis of SEX4 function as a glucan phosphatase.
Results
Structure of SEX4.
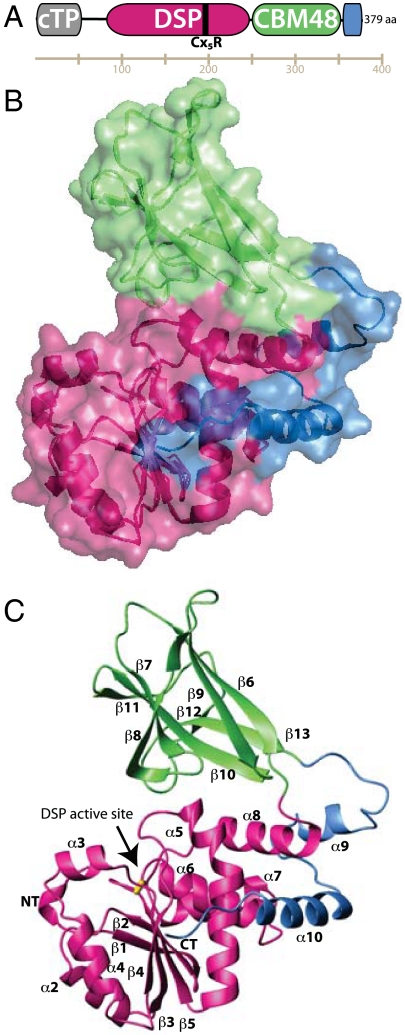
A. thaliana SEX4 (SEX4) is a 379 amino acid protein containing three known domains: an amino-terminal cTP, a DSP domain, and a CBM family member 48 (Fig. 1A). SEX4 (residues 90–379, C198S) was crystallized, selenomethionine single-wavelength anomalous dispersion (SAD) data collected, and the structure determined at 2.4 Å (Table 1). The structure of the SEX4 glucan phosphatase reveals a unique set of extensive interdomain interactions producing a complex tertiary architecture (Fig. 1B). The SEX4 DSP domain consists of a central five-stranded β-sheet flanked by eight α-helices (Fig. 1C and Fig. S1). The SEX4 CBM domain possesses six β-sheets that fold into a characteristic compact β sandwich composed of antiparallel sheets (Fig. 1C). C-terminal to the CBM is an extended region that possesses two α-helices (Fig. 1C). The structure reveals an integrally folded unit composed of an N-terminal DSP domain, CBM, and a previously unrecognized domain at the C terminus.
Fig. 1.
Structure of SEX4. (A) SEX4 domain structure. The active site of SEX4 is denoted with a black line and labeled Cx5R. (B) Structure of SEX4 showing the integrated architecture of DSP (pink), CBM (green), and C-terminal (blue) domains. (C) Ribbon diagram of SEX4 (residues 90–379) with the active site labeled and S198 in gold sticks. Elements of secondary structure are numbered consecutively from N to C termini.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Beamline | APS 22-ID |
| Space group | P212121 |
| Wavelength | 0.9792 |
| Unit-cell parameters | 59.408, 73.934, 162.382 |
| Unique reflections | 28453 |
| Completeness (%) | 99.2 (94.7) |
| Resolution (Å) | 2.4 (2.49–2.40) |
| Rmerge (%) | 10.0 (44.7) |
| Redundancy | 3.2 (2.5) |
| I/σ(I) | 11.4 (2.04) |
| Refinement | |
| Resolution Limits (Å) | 20.0–2.40 |
| # reflections/# to compute Rfree | 24660/2000 |
| R (Rfree) | 19.8 (24.9) |
| # protein residues | 579 |
| # solvent molecules | 197 |
| # phosphate molecules | 2 |
| Ramachandran | |
| Most favored | 93.3 |
| Add. allowed | 6.3 |
| Generously | 0.4 |
| Disallowed | 0.0 |
| RMS deviation | |
| Bond, Å | 0.006 |
| Angle, ° | 0.997 |
The SEX4 DSP Domain.
The DSP domain (residues 90–252) has a characteristic αβα protein tyrosine phosphatase (PTP) fold. DSP domains contain a number of conserved elements (reviewed in ref. 33) (Fig. S1). The SEX4 active site sequence HCTAGMGRA (residues 197–205), also known as the PTP loop (33, 34), lies between β5 and α6 at the base of the active site cleft. The cysteine in this motif functions as a nucleophile during catalysis, attacking the phosphorous atom of the substrate and forming a phosphoenzyme intermediate (34). The D-loop of SEX4 (residues 162–172) is arranged similarly to other DSP domains, with D166 in position to act as the general acid catalyst to enhance hydrolysis of the phosphoenzyme intermediate (33, 34) (Fig. S1). In addition, the variable insert and recognition domain of DSPs, including SEX4, contribute to the depth of the active site (33, 35).
The SEX4 structure is the catalytically inactive C198S mutant and has phosphate tightly bound (Fig. S2A). The SEX4 active site is formed by the variable insert, recognition domain, D-loop, and R-motif (Fig. S2B). A structural search of the Protein Data Bank using DALI identifies relatively modest structural homology with protein phosphatases (36). The closest structural homologue to the SEX4 DSP domain is a PTP from the Archaea thermophile Sulfolobus solfataricus, SsoPTP (PDB 2I6O) with an rmsd of 2.5 Å and 18% identity by sequence (37). The limited structural similarities between the glucan phosphatase SEX4 and PTP family members are due to two features within the DSP domain and the unique C-terminal domain of SEX4.
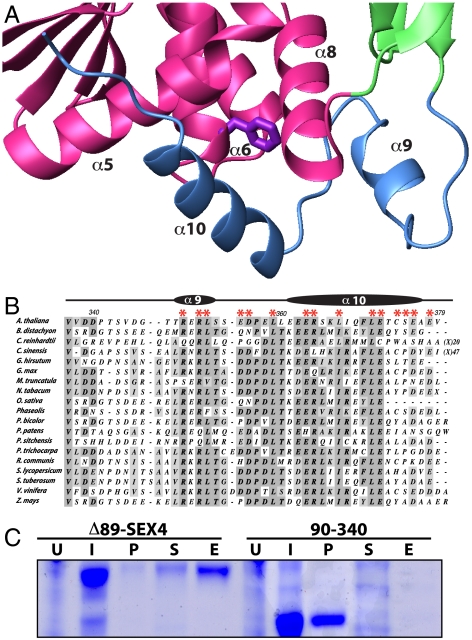
The most striking physical differences between the DSP domain of SEX4 and other DSP domains are found within the variable insert and AYLM motif. The SEX4 variable insert (residues 131–157) is longer than many proteinaceous DSPs, including human Vaccinia virus H1-related phosphatase (VHR) (Fig. S3). Additionally, while the variable insert of DSP domains typically contain little to no secondary structure, the variable insert of SEX4 has two α-helices (α3 and α4) with α3 forming the apex of one side of the active site cleft (Fig. 1C and Fig. S2B). The AYLM motif, located in α6 (residues 211–214, TYMF in SEX4), is one of the few DSP motifs outside the PTP loop that is highly conserved among the DSPs (33). This motif forms part of the extended signature sequence of the DSP family, but it is not conserved in SEX4 (Fig. S1 and S4). In the SEX4 structure, the final residue of this motif, F214, directly interacts with the C-terminal helix (Fig. 2A).
Fig. 2.
Interaction of the C-terminal domain with the DSP domain. (A) The C-terminal domain wraps around the final DSP α8 helix and additionally interacts with both α5 and α6. F214 in α6, the final residue of the AYLM motif, is highlighted in purple. (B) Multiple sequence alignment of SEX4 orthologs reveals that the residues contributing to the DSP interface (highlighted with red asterisks) are highly conserved. (C) The C-terminal helix is essential for soluble expression of SEX4. Δ89-SEX4 lacks the N-terminal 89 residues but has the full-length C terminus, to residue 379. 90–340 has both N- and C-terminal truncations. U = Uninduced cells, I = Cells induced with IPTG, P = Pellet of insoluble protein, S = Soluble protein, E = eluted from IMAC column.
The C Terminus of SEX4 Integrally Folds into the DSP.
The C-terminal domain (residues 338–379) is composed of an extended loop followed by two α-helices (Fig. 1B, 1C and Fig. S1). The C-terminal region of SEX4 makes the most extensive interdomain contacts with the DSP domain, with 1010 Å2 interface accessible surface area (Fig. 2A). The two helical regions in the C-terminal domain intimately associate with the DSP, cradling the final helix of the DSP domain. The region following the CBM (residues 338–358), including α9, interacts with α8 of the DSP domain. The final helix, α10 (residues 361–373), associates with several regions of the DSP, interacting with α5, α6, and α8 (Fig. 2A).
Many dual specificity phosphatases contain little more than a DSP domain and can be readily expressed in Escherichia coli (38–41). The DSP of SEX4 cannot be independently expressed, and we hypothesized that the intimate association of the C-terminal domain with the DSP underlies this phenomenon. We identified and aligned 19 SEX4 orthologs from Kingdom Plantae genomes, including a green alga, a moss, and land plants/trees (Fig. S4). The orthologs are 34–64% identical to At-SEX4 at the amino acid level, with the DSP domain sharing the highest degree of conservation (Table S1). An alignment of SEX4 orthologs demonstrates striking conservation in the DSP-contacting residues in the C terminus (Fig. 2B). Within the C-terminal domain, α9 contains a conserved -RXRL- motif. In addition, L359, located between the two helices, is invariant. Finally, α10 contains a highly conserved -ERXXLXXXL- motif.
To determine the functional significance of this region, we generated a C-terminal deletion construct, SEX4 (90–340). The crystallized construct, containing the C terminus (residues 90–379), is produced upon induction, produces predominantly soluble protein, and is readily purified from the soluble fraction (Fig. 2C). SEX4 lacking the C-terminal domain (residues 90–340) is also produced upon induction but is entirely insoluble with no protein recovered from the soluble fraction (Fig. 2C). Thus, the C-terminal domain of SEX4 is necessary for soluble expression of SEX4.
The DSP and CBM Domains Interact.
Based on CAZy classification (42), the SEX4 CBM domain (residues 253–338) belongs to the CBM48 family, the same family as the AMPKβ CBM. Consistent with this classification, structural analyses of the Protein Data Bank using DALI identifies AMPKβ1 (PDB 1Z0M) as the closest structural homologue (rmsd = 1.3 Å, 30% identity by sequence) (43).
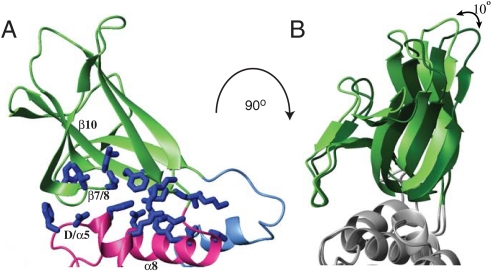
The DSP and CBM domains of SEX4 directly interact with 457 Å2 of interfacial surface area. A broad surface formed by multiple residues creates the extensive interface between the CBM and DSP domain (Fig. 3A). Specifically, the D-loop/α5 region of the DSP possesses a highly conserved FDXFDLR motif (residues 167–173) that packs against the β7/8 loop of the CBM that possesses a corresponding LDIGWG motif (residues 274–279) (Fig. 3A). An alignment of SEX4 orthologs demonstrates striking conservation in these DSP/CBM interacting residues (Fig. S4). Additionally, α8 of the DSP packs against β10 of the CBM. These interactions position the two domains such that they form a continuous surface that runs the length of the enzyme.
Fig. 3.
Interaction of the CBM and DSP domains. (A) The DSP domain and CBM of SEX4 directly interact via several structural elements. The D-loop/α5 region of the DSP (D/α5) possesses packs against the β7/8 loop of the CBM. Additionally, α8 of the DSP packs against β10 of the CBM. Residues at the interdomain interface are highlighted in blue sticks. (B) Comparison of the two molecules in the asymmetric unit reveals that the DSP/CBM interface is maintained but has rotational flexibility. Overlay generated by superimposing the DSP domains of the two molecules.
Intriguingly, the two SEX4 molecules in the asymmetric unit differ slightly in the relative orientation of the DSP and CBM domain via a 10° rotation (Fig. 3B). This rotation is coupled to alteration in the packing of α9 from the C-terminal domain and results in a slight flexing of the hinge region between the CBM and DSP domain.
SEX4 Possesses a Unique Ligand Binding Active Site.
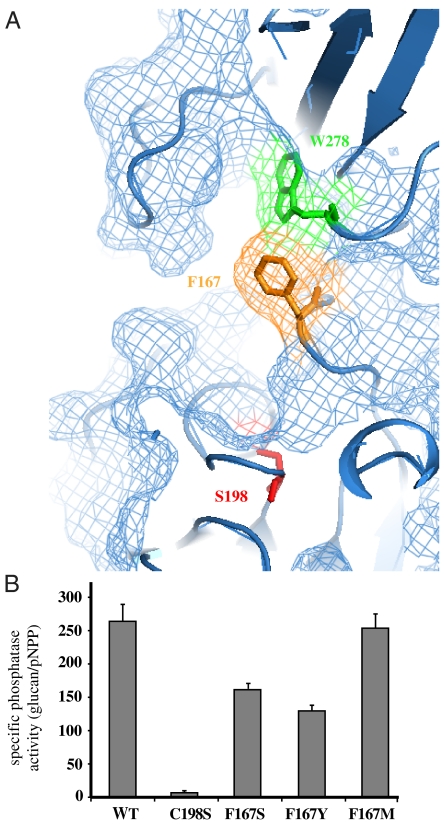
The DSP active site is located between β5 and α6 while the carbohydrate-binding site of SEX4 is located in the deep cleft formed by the extended loop between β6 and β7 and the five-sheet face of the domain (18, 43, 44). The unique multidomain structure of SEX4 serves to align the DSP active site and carbohydrate-binding face of the CBM into a single surface (Fig. 4A and Fig. S5). This extended surface produces a contiguous active site incorporating both catalytic and glucan binding functionalities.
Fig. 4.
SEX4 possesses a contiguous ligand-binding active site. (A) Surface view of the DSP active site to CBM binding site pocket. F167 (yellow), S198 (red), and W278 (green) are all highlighted. (B) SEX4 specific phosphatase activity expressed as the ratio of glucan phosphatase activity/generic p-NPP activity. Glucan phosphatase activity was quantified as pmol min-1 mg-1 of phosphate release. The activity against p-NPP was quantified as μmol min-1 mg-1. Δ89-SEX4 (WT), C198S, and three F167 mutants were analyzed. Error bars indicate means ± the standard deviation.
F167 is an invariant residue located in a highly conserved region within the D-loop (Fig. S5C). F167 is physically oriented between D166 of the DSP domain and W278 of the CBM (Fig. 4A and Fig. S5). D166 acts as the general acid-base catalyst to enhance leaving group expulsion in most DSPs, and W278 is a critical glucan binding residue located at the apex of the interacting regions (Fig. 4A and Fig. S5) (33, 34). F167 forms the base of the bridge that connects the CBM binding site to the DSP active site. Most DSP family members possess a short chain hydrophilic residue, S/T/N/H, at the position corresponding to F167 (Fig. S5C). Only one other DSP, VHZ, contains a phenylalanine residue in the same position.
Based on the structure, we predicted that F167 functions in coupling the substrate binding and catalysis functions of SEX4 by positioning the phosphoglucan directly into the extended SEX4 catalytic cleft. To test this prediction, we mutated F167 to a serine, tyrosine, and methionine. Proteins were expressed, purified, and assayed for phosphatase activity. We assayed both glucan phosphatase activity, utilizing amylopectin (Fig. S5D), and generic phosphatase activity, utilizing the exogenous substrate para-nitrophenylphosphate (p-NPP) (Fig. S5E). We then compared the ratio of these two activities (i.e., glucan activity/generic activity) to determine specific glucan phosphatase activity (Fig. 4B). F167S, a short chain hydrophilic residue most commonly found across the DSP family, showed decreased specific activity as a glucan phosphatase, supporting the hypothesis that F167 specifically functions in SEX4 glucan phosphatase activity. F167Y, which is expected to minimally physically perturb the system, but whose hydroxyl would partially occlude the glucan binding site of the CBM, substantially decreases the glucan phosphatase activity of SEX4 without affecting p-NPP activity. Of note, F167M, the corresponding residue in the glucan phosphatase laforin, did not impact the specific glucan phosphatase activity of SEX4. Cumulatively, these data demonstrate a role for F167 in the glucan phosphatase activity of SEX4.
Discussion
Our data demonstrate the essential tertiary architecture necessary for the glucan phosphatase activity of SEX4. The SEX4 structure reveals multiple features unique to SEX4 compared to other DSPs, including a variable insert that contains two α-helices, adaptation of the AYLM motif for interdomain interaction, a novel C-terminal domain that is structurally integrated into the DSP domain, extensive DSP–CBM interactions that align the DSP active site and CBM binding site into a common pocket, and a channel formed by the CBM-DSP interface. These features serve to structurally differentiate SEX4 from phosphatases that dephosphorylate proteinaceous substrates.
The majority of phosphatases are protein phosphatases (45–48). The PTP superfamily is divided into the classical PTPs that dephosphorylate only phosphotyrosine; the DSPs that dephosphorylate phosphotyrosine, phosphoserine, and phosphothreonine; and a subset of DSPs that dephosphorylate nonproteinaceous substrates (35, 47, 49, 50). Within the PTP superfamily, the depth of the active site contributes a significant portion to substrate specificity (33, 47). The active site depth of proteinaceous DSPs is ≈6 Å, allowing access of both pS/T and pY (33, 51). Alternatively, tyrosine-specific PTPs possess an ≈9 Å cleft, allowing the longer pY access but limiting the shorter pS/T (33, 52). The nonproteinaceous phosphoinositol phosphatases phosphate and tensin homolog (PTEN) and myotubularin-related protein 2 (MTMR2) have an 8 Å and 13 Å deep pocket, respectively, with an elliptical opening nearly twice as wide as the tyrosine-specific PTP1B (53, 54).
The unique active site cleft of SEX4 forms a 21 Å pocket from phosphatase active site to CBM glucan binding region. If SEX4 requires simultaneous engagement of both glucan binding and phosphatase domains by a single substrate, then it would minimally require a glucan composed of approximately five to six glucose moieties (i.e., phospho-maltopentaose or -hexaose) to span this region. Therefore, it would require a linker region of three to four glucans to span the two sites, with an additional glucose bound at the CBM and a phospho-glucose moiety at the active site. Our data demonstrate that F167 is a critical residue for SEX4 glucan phosphatase activity. F167 bridges the two domains and physically links the CBM binding site to the DSP active site. It is tempting to speculate that F167 functions by positioning the linker region so that the phosphoglucan is properly presented to the SEX4 active site. In addition to the 21 Å wide pocket, the DSP–CBM interface also forms a channel on the face of the enzyme that contains both the glucan binding site and DSP active site. This deep interfacial channel terminates at the open binding pocket positioned between DSP active site and the CBM binding site. This open CBM terminus suggests a physical basis by which SEX4 could accommodate both linear phospho-oligosaccharides and also longer nonlinear phosphoglucan substrates, such as those found in the starch granule surface.
In addition to these features, the SEX4 structure reveals that the C-terminal domain of SEX4 integrally binds to the DSP domain, and our results demonstrate that this domain is essential for protein stability. As previously noted, the AYLM motif, located in α6 (residues 211–214), is one of the few DSP motifs outside the PTP loop that is highly conserved among DSPs (33). However, this motif is not conserved in any of the known glucan phosphatases, including SEX4. In SEX4, the final residue of this motif, F214, directly interacts with the C-terminal helix. The interaction between F214 and the C-terminal helix suggests that the AYLM motif is adapted in SEX4 to support DSP domain interaction with the C-terminal domain and may prove a general feature of glucan phosphatases.
While the C-terminal domain is essential for maintaining the fold and stability of SEX4, the CBM is essential for substrate binding. The interdomain orientation of the DSP domain and CBM is essential for glucan phosphatase activity of SEX4. Interestingly, the overall positioning of the SEX4 DSP and CBM domains are quite similar to that of the phosphatase and C2 domains of PTEN, with the equivalent elements in the phosphatase domain, D loop/α5 and α8, employed in the interdomain interaction (54).
Previous data from H/D exchange mass spectrometry of SEX4 indicated that no major interdomain conformational change occurred in SEX4 upon substrate binding (55). Our data are consistent with this conclusion but suggest that some rotation of the DSP/CBM interface is possible. Intriguingly, the two SEX4 molecules in the asymmetric unit differ by a 10° rotation that alters the packing of α9 from the C-terminal domain. This rotation provides a possible manner to regulate substrate entry, which we are currently investigating. SEX4 dephosphorylates both the starch granule surface as well as soluble phosphoglucans (16). In addition, SEX4 can dephosphorylate both the C3 and C6 position of glucose (22). Therefore, SEX4 possesses a broad substrate specificity and must accommodate both different glucans (i.e., soluble and crystalline) as well as differently positioned phosphates (i.e., C3 versus C6). We are currently investigating if this conformational lability accommodates binding to inherently chemical heterogeneous glucans and/or allows processive substrate dephosphorylation. The SEX4 structure begins to address how the enzyme manages these sterically different substrates.
Materials and Methods
Protein Expression and Purification.
Cloning of Arabidopsis thaliana SEX4 was previously described (18). Based on data from secondary structure predictions, disorder predictions, and deuterium exchange mass spectrometry experiments, we generated a construct of A. thaliana SEX4 lacking the first 89 amino acids (Δ89–SEX4) (55). Δ89–SEX4 lacks the cTP (predicted to be residues 1–54) along with residues up to the DSP recognition domain (Fig. S1). Δ89–SEX4, point mutants, and Δ89–SEX4(90–340) were subcloned into pET28 (Novagen) using NdeI and XhoI to encode HIS6, a thrombin cleavage site, and SEX4. BL21-CodonPlus Escherichia coli cells (Stratagene) or T7 Express Crystal (NEB) were transformed with expression vectors for production of native and selenomethionine labeled protein, respectively. Cells were grown at 37 °C in 2 × YT or M9 supplemented with 100 mg/L selenomethionine to an OD600 = 0.6, placed on ice for 20 min, induced with 1 mM isopropyl β-D-thiogalactoside (IPTG), grown at 20 °C for 16 h, and harvested by centrifugation. Cells were lysed in 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 15 mM imidazole, and 2 mM dithiothreitol (DTT), centrifuged, and proteins were purified using a Profinia IMAC column (Bio-Rad) with a Profinia protein purification system (Bio-Rad). Protein was dialyzed in 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 2 mM dithiothreitol (DTT) overnight at 4 °C in the presence of thrombin. Protein was then reverse purified over the Profinia IMAC column. Protein was purified to homogeneity using a HiLoad 16/60 Superdex 200 size exclusion column (GE Healthcare).
Crystal Structure Determination.
Single high quality crystals, with two protein molecules in the asymmetric unit, were obtained via hanging drop vapor diffusion using a Mosquito crystallization robot (TTPLabtech) with a 200 nL drop using a 100∶100 mixture of selenomethionine labeled SEX4 (12 mg/mL): 0.4 M lithium citrate and 24% PEG3350. Crystals were briefly soaked in 21% PEG3350 and 0.5 M ammonium hydrogen phosphate and then flash frozen. SAD data was collected at the selenomethionine peak wavelength, based on a fluorescence energy scan, on the 22-ID beamline of SER–CAT at the Advanced Photon Source, Argonne National Laboratory (Table 1). Data was processed using HKL2000 (56). PHENIX (57), employing HYSS (58), PHASER (59), and RESOLVE (60), was used to locate all twelve expected selenomethionine sites, obtain phase information, perform density modification, and generate an initial structural model. The structure was then fully built and refined via iterative model building and refinement using Coot (61) and Refmac5 (62), respectively (Table 1).
Structural and Sequence Analysis.
Interdomain interfaces were analyzed using PROTORP (63). Molecular graphics were prepared using the programs Pymol and MOLMOL (64). The sequences of A. thaliana SEX4 orthologs were obtained by performing tBLASTn searches using the GenBank “dbEST” database or BLASTp and PSI-BLAST searches using GenBank “eukaryote genome” and “nonredundant” (nr) databases, PlantGDB, Department of Energy Joint Genome Institute Resource, and Phytozome. Amino-acid sequences of SEX4 orthologs were aligned by ClustalW and refined manually using MacVector.
Phosphatase Assays.
Phosphatase assays utilizing p-NPP have been previously described and were performed with the following modifications (18, 28): Hydrolysis of p-NPP was performed in 50 μl reactions containing 1X phosphatase buffer (0.1 M sodium acetate, 0.05 M bis-Tris, 0.05 M Tris-HCl pH 6.0, 2 mM dithiothreitol), 50 mM p-NPP, and 100–500 ng of enzyme at 37 °C for 15–20 min. The reaction was terminated by the addition of 200 μl of 0.25 M NaOH and absorbance was measured at 410 nm. Malachite green assays were performed as previously described with the following modifications (65): 1X phosphatase buffer (0.1 M sodium acetate, 0.05 M bis-Tris, 0.05 M Tris-HCl pH 8.0, 2 mM dithiothreitol), 100–500 ng of SEX4, and 45 μg of amylopectin (Sigma) in a final volume of 20 μl. The reaction was stopped by the addition of 20 μl of 0.1 M N-ethylmaleimide and 80 μl of malachite green reagent. Absorbance was measured after 40 minutes at 620 nm. Specific glucan phosphatase activity is expressed as phosphate release from amylopectin divided by the specific activity against p-NPP.
Supplementary Material
Acknowledgments.
We thank Dr. Qingjun Wang for technical assistance, Dr. Carol Beach in the Molecular Basis of Human Disease COBRE Proteomics Core, the Molecular Basis of Human Disease COBRE administrative and protein analytical cores, and Drs. Carolyn Worby, Oliver Kötting, Doug Andres, Mike Begley, and Choel W. Kim for technical assistance and discussions. This work was supported by National Institutes of Health (NIH) grants R00NS061803, P20RR020171, and R01NS070899; University of Kentucky College of Medicine startup funds to M.S.G.; NIH grant P20RR0202171 to C.W.V.K.; and the Korea Research Foundation grant KRF-2008-331-E00031 to Y.J.K.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3NME).
See Commentary on page 15312.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009386107/-/DCSupplemental.
References
- 1.Zeeman SC, Smith SM, Smith AM. The diurnal metabolism of leaf starch. Biochemical J. 2007;401:13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- 2.Buleon A, Colonna P, Planchot V, Ball S. Starch granules: Structure and biosynthesis. Int J Biol Macromol. 1998;23(2):85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- 3.Gallant DJ, Bouchet B, Baldwin PM. Microscopy of starch: Evidence of a new level of granule organization. Carbohyd Polym. 1997;32(3–4):177–191. [Google Scholar]
- 4.Blennow A, Engelsen SB. Helix-breaking news: Fighting crystalline starch energy deposits in the cell. Trends Plant Sci. 2010;15(4):236–240. doi: 10.1016/j.tplants.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Fettke J, et al. Eukaryotic starch degradation: Integration of plastidial and cytosolic pathways. J Exp Bot. 2009;60(10):2907–2922. doi: 10.1093/jxb/erp054. [DOI] [PubMed] [Google Scholar]
- 6.Kotting O, Kossmann J, Zeeman SC, Lloyd JR. Regulation of starch metabolism: The age of enlightenment? Curr Opin Plant Biol. 2010;13(3):321–329. doi: 10.1016/j.pbi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Zeeman SC, Kossmann J, Smith AM. Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- 8.Ritte G, et al. Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006;580(20):4872–4876. doi: 10.1016/j.febslet.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 9.Ritte G, et al. The starch-related R1 protein is an alpha -glucan, water dikinase. Proc Natl Acad Sci USA. 2002;99(10):7166–7171. doi: 10.1073/pnas.062053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baunsgaard L, et al. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J . 2005;41(4):595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- 11.Kotting O, et al. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 2005;137(1):242–252. doi: 10.1104/pp.104.055954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen PI, et al. Structure and hydration of the amylopectin trisaccharide building blocks—Synthesis NMR and molecular dynamics. Biopolymers. 2008;89(12):1179–1193. doi: 10.1002/bip.21075. [DOI] [PubMed] [Google Scholar]
- 13.Hansen PI, et al. Starch phosphorylation–maltosidic restrains upon 3'- and 6'-phosphorylation investigated by chemical synthesis, molecular dynamics and NMR spectroscopy. Biopolymers. 2009;91(3):179–193. doi: 10.1002/bip.21111. [DOI] [PubMed] [Google Scholar]
- 14.Kozlov SS, Blennow A, Krivandin AV, Yuryev VP. Structural and thermodynamic properties of starches extracted from GBSS and GWD suppressed potato lines. Int J Biol Macromol. 2007;40(5):449–460. doi: 10.1016/j.ijbiomac.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Sanderson JS, Daniels RD, Donald AM, Blennow A, Engelsen SrB. Exploratory SAXS and HPAEC-PAD studies of starches from diverse plant genotypes. Carbohyd Polym. 2006;64(3):433–443. [Google Scholar]
- 16.Kotting O, et al. STARCH-EXCESS4 is a Laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell. 2009;21(1):334–346. doi: 10.1105/tpc.108.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentry MS, Pace RM. Conservation of the glucan phosphatase laforin is linked to rates of molecular evolution and the glycogen metabolism of the organism. BMC Evol Biol. 2009;9(1):138. doi: 10.1186/1471-2148-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentry MS, et al. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178(3):477–488. doi: 10.1083/jcb.200704094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niittyla T, et al. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem. 2006;281(17):11815–11818. doi: 10.1074/jbc.M600519200. [DOI] [PubMed] [Google Scholar]
- 20.Sokolov LN, Dominguez-Solis JR, Allary AL, Buchanan BB, Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA. 2006;103(25):9732–9737. doi: 10.1073/pnas.0603329103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeeman SC, Northrop F, Smith AM, Rees T. A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J . 1998;15(3):357–365. doi: 10.1046/j.1365-313x.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 22.Hejazi M, Fettke J, Kotting O, Zeeman SC, Steup M. The Laforin-like dual-specificity phosphatase SEX4 from Arabidopsis hydrolyzes both C6- and C3-phosphate esters introduced by starch-related dikinases and thereby affects phase transition of alpha-glucans. Plant Physiol. 2010;152(2):711–722. doi: 10.1104/pp.109.149914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafora GR. Uber des Vorkommen amyloider KJrperchen im innern der Ganglienzellen. Virchows Arch A. 1911;205:295–303. [Google Scholar]
- 24.Harriman DG, Millar JH, Stevenson AC. Progressive familial myoclonic epilepsy in three families: Its clinical features and pathological basis. Brain. 1955;78(3):325–349. doi: 10.1093/brain/78.3.325. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz GA, Yanoff M. Lafora’s Disease. Distinct clinico-pathologic form of Unverricht’s Syndrome. Archives of neurology. 1965;12:172–188. doi: 10.1001/archneur.1965.00460260062008. [DOI] [PubMed] [Google Scholar]
- 26.Yokoi S, Austin J, Witmer F. Isolation and characterization of Lafora bodies in two cases of myoclonus epilepsy. J Neuropath Exp Neur. 1967;26(1):125–127. [PubMed] [Google Scholar]
- 27.Yokoi S, Austin J, Witmer F, Sakai M. Studies in myoclonus epilepsy (Lafora body form). I. Isolation and preliminary characterization of Lafora bodies in two cases. Arch Neurol. 1968;19(1):15–33. doi: 10.1001/archneur.1968.00480010033002. [DOI] [PubMed] [Google Scholar]
- 28.Worby CA, Gentry MS, Dixon JE. Laforin: A dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281(41):30412–30418. doi: 10.1074/jbc.M606117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai M, Austin J, Witmer F, Trueb L. Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology. 1970;20(2):160–176. doi: 10.1212/wnl.20.2.160. [DOI] [PubMed] [Google Scholar]
- 30.Schnabel R, Seitelberger F. Histophysical and histochemical investigations of myoclonus bodies. Pathol Eur. 1968;3(2):218–226. [PubMed] [Google Scholar]
- 31.Tagliabracci VS, et al. Abnormal metabolism of glycogen phosphate as a cause for lafora disease. J Biol Chem. 2008 doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tagliabracci VS, et al. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci USA. 2007;104(49):19262–19266. doi: 10.1073/pnas.0707952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alonso A, Rojas A, Godzik A, Mustelin T. The dual-specific protein tyrosine phosphatase family. Berlin: Springer; 2003. pp. 333–358. [Google Scholar]
- 34.Denu JM, Stuckey JA, Saper MA, Dixon JE. Form and function in protein dephosphorylation. Cell. 1996;87(3):365–368. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 35.Gentry MS, Dixon JE, Worby CA. Lafora disease: Insights into neurodegeneration from plant metabolism. Trends Biochem Sci. 2009;34(12):628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20(11):478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 37.Chu HM, Wang AH. Enzyme-substrate interactions revealed by the crystal structures of the archaeal Sulfolobus PTP-fold phosphatase and its phosphopeptide complexes. Proteins. 2007;66(4):996–1003. doi: 10.1002/prot.21262. [DOI] [PubMed] [Google Scholar]
- 38.Agarwal R, Burley SK, Swaminathan S. Structure of human dual specificity protein phosphatase 23, VHZ, enzyme-substrate/product complex. J Biol Chem. 2008;283(14):8946–8953. doi: 10.1074/jbc.M708945200. [DOI] [PubMed] [Google Scholar]
- 39.Koksal AC, Nardozzi JD, Cingolani G. Dimeric quaternary structure of the prototypical dual specificity phosphatase VH1. J Biol Chem. 2009;284(15):10129–10137. doi: 10.1074/jbc.M808362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun JP, et al. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation, and tumor invasion. Biochemistry. 2005;44(36):12009–12021. doi: 10.1021/bi0509191. [DOI] [PubMed] [Google Scholar]
- 41.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272(5266):1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 42.Boraston AB, Bolam DN, Gilbert HJ, Daview GJ. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polekhina G, et al. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 2005;13(10):1453–1462. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 44.Polekhina G, et al. AMPK beta subunit targets metabolic stress sensing to glycogen. Curr Biol. 2003;13(10):867–871. doi: 10.1016/s0960-9822(03)00292-6. [DOI] [PubMed] [Google Scholar]
- 45.Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: In cauda venenum (the sting is in the tail) Trends Biochem Sci. 2008;33(3):113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 46.Shi Y. Serine/threonine phosphatases: Mechanism through structure. Cell. 2009;139(3):468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Tonks NK. Protein tyrosine phosphatases: From genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 48.Rigden DJ. The histidine phosphatase superfamily: Structure and function. Biochem J . 2008;409(2):333–348. doi: 10.1042/BJ20071097. [DOI] [PubMed] [Google Scholar]
- 49.Alonso A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117(6):699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 50.Robinson FL, Dixon JE. Myotubularin phosphatases: Policing 3-phosphoinositides. Trends Cell Biol. 2006;16(8):403–412. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Guan KL, Broyles SS, Dixon JE. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature. 1991;350(6316):359–362. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- 52.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263(5152):1397–1404. [PubMed] [Google Scholar]
- 53.Begley MJ, et al. Crystal structure of a phosphoinositide phosphatase, MTMR2: Insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol Cell. 2003;12(6):1391–1402. doi: 10.1016/s1097-2765(03)00486-6. [DOI] [PubMed] [Google Scholar]
- 54.Lee J-O, et al. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99(3):323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 55.Hsu S, et al. Structural insights of glucan phosphatase dynamics using amide hydrogen/deuterium exchange mass spectrometry. Biochemistry. 2009;48(41):9891–9902. doi: 10.1021/bi9008853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 57.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr D. 2003;59(Pt 11):1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 59.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D. 2008;64(Pt 1):61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murshudov GN. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 63.Reynolds C, Damerell D, Jones S. ProtorP: A protein-protein interaction analysis server. Bioinformatics. 2009;25(3):413–414. doi: 10.1093/bioinformatics/btn584. [DOI] [PubMed] [Google Scholar]
- 64.Koradi R, Billeter M, Wuthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 65.Harder KW, et al. Characterization and kinetic analysis of the intracellular domain of human protein tyrosine phosphatase beta (HPTP beta) using synthetic phosphopeptides. Biochemical J. 1994;298:395–401. doi: 10.1042/bj2980395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.