Abstract
Objective
Maternal food restriction during pregnancy results in growth restricted newborns and reduced glomerular number, contributing to programmed offspring hypertension. We investigated whether reduced nephrogenesis may be programmed by dysregulation of factors controlling ureteric bud branching and mesenchyme to epithelial transformation.
Study Design
10 to 20 days gestation, Sprague Dawley pregnant rats (n=6/group) received ad libitum food; FR rats were 50% food restricted. At embryonic day 20, mRNA and protein expression of WT1, Pax2, FGF2, GDNF, cRET, WNT4, WNT11, BMP4, BMP7, and FGF7 were determined by real-time PCR and Western blotting.
Results
Maternal FR resulted in up-regulated mRNA expression for WT1, FGF2, and BMP7 whereas Pax2, GDNF, FGF7, BMP4, WNT4, and WNT11 mRNAs were down-regulated. Protein expression was concordant for WT1, GDNF, Pax2, FGF7, BMP4 and WNT4.
Conclusion
Maternal FR altered gene expression of fetal renal transcription and growth factors, and likely contributes to development of offspring hypertension.
Keywords: Branching morphogenesis, Fetal programming, Hypertension, Maternal food restriction, Metanephric development
Introduction
Human and animal studies have demonstrated that the in utero environment affects fetal development and adult phenotype.1–5 This concept of fetal origins of adult disease was first described by Barker, who observed an increased incidence of adult cardiovascular disease in association with low birth weight.6 The mechanism proposes that variations in fetal nutrition during pregnancy lead to developmental changes, which are thought to elicit a permanent response in tissue structure or function in the adult, possibly leading to an increased risk of disease later in life.7 Several epidemiological studies have confirmed this association for heart disease8;9 and extended the theory to other conditions such as diabetes,10 obesity,11 cancer,12 and hypertension.13 The mechanistic pathways underlying this association only now are being investigated using rat gestational under-nutrition as a model for in utero derived plasticity. In the rat, global under-nutrition (50% maternal food restriction [FR] during gestation) results in offspring that develop metabolic syndrome (i.e., hypertension, obesity, hyperleptinemia, hyperinsulinism, and hyperphagia).2;4;5;11
Hypertension is intrinsically linked with kidney function. Although development of hypertension is clearly multifactorial, a recent study demonstrates a strong correlation between low nephron number and hypertension with hypertensive patients having a mean reduction of 46% in the number of intact glomeruli.14 Animal studies confirm the association of low birth weight, reduced glomerular number and adult hypertension. Nephron reduction of as little as 11% in sheep15 and 13% in the rat16 is associated with changes in postnatal renal function and elevations in arterial blood pressure.7 Intrauterine growth restriction, including nutrient and protein restriction, results in kidneys with reduced nephron number per kidney. The mechanism of how maternal FR results in reduced nephron number is unclear. Nephron development could be impacted by FR at several developmental points which include signaling between the mesenchyme and the developing ureteric bud (UB), UB branching, the metanephric mesenchymal to epithelial transformation, and possibly glomerulogenesis.17–19 Following initial ureteric bud branching, metanephric mesenchyme aggregates and condenses around the tips of the UB branches in response to signaling.20,21 Mesenchymal to epithelial transformation begins to occur from rat embryo day 14 (e14) with UB branching and mesenchymal trans-differentiation occurring actively at e20. The resulting epithelium forms primitive nephrons, which are invaded by capillaries at e15–16 and undergo further differentiation to form glomeruli.22 An individual nephron develops in five to seven days, and the entire kidney develops over 18 days, with early functional nephrons in place by e17.23 This coordinated progression of nephron development, if disrupted by maternal FR, could lead to decreased nephron numbers.24;25
We examined several key transcription and growth factors controlling fetal nephrogenesis following maternal FR at e20, a time point in which maternal FR (10 days total) should have a detectable effect. At this time point, mRNA and protein expression of transcription and growth factors WT1, Pax2, FGF2 (mesenchyme condensation and transdifferentiation factors), GDNF, cRET (UB signaling), and WNT4, WNT11, BMP4, BMP7, FGF7 (ureteric branching and mesenchymal to epithelial transformation factors) were determined in fetal kidneys by real-time PCR and Western blotting, respectively. We detected significant changes in gene expression of several of these factors, suggesting maternal FR has a large impact on the regulation of the developing fetal kidney.
Materials and Methods
Maternal rat diets
Studies were approved by the Animal Care and Use Committee of the Los Angeles BioMedical Research Institute at Harbor–University of California, Los Angeles and were in accordance with the American Association for Accreditation of Laboratory Care and National Institutes of Health guidelines. A model of rat dams that were 50% food-restricted (FR) during pregnancy was used. First-time pregnant Sprague Dawley rats (Charles River Laboratories, Hollister, CA) were housed in a facility with constant temperature and humidity and a controlled 12:12-hour light/dark cycle. At 10 days of gestation, rats were provided either an ad libitum diet of standard laboratory chow (Lab Diet 5001, Brentwood, MO; protein, 23%; fat, 4.5%; metabolizable energy, 3030 kcal/kg) or a 50% FR diet that was determined by quantification of normal intake in the ad libitum fed rats. The respective diets were given from 10 days of pregnancy until gestational day 20 (e20).
Fetal kidney removal
At e20, dams (6 control (C) and 6 FR) were anesthetized with isoflurane, and fetuses were quickly removed and placed on ice cold dishes. Litter sizes ranged from 12 to 16. No fetuses from dams with litter sizes of fewer than 12 or more than 16 were used. Fetuses were dissected, intact kidneys removed, and kidneys flash frozen in liquid nitrogen. Due to the limiting size of the e20 kidneys, all kidneys were pooled from unsexed fetuses derived from each dam for a total of 6 control and 6 FR pools.
Real-time reverse transcriptase polymerase chain reaction (PCR)
RNA extraction
Total RNA was extracted from 75 mgs of pooled kidneys (approximately 4–6 kidneys/sample; C n=6 samples, FR n=6 samples) with the use of the RNAqueous-4PCR column kit (Ambion, Austin, TX). Residual DNA was digested with DNase I enzyme and inactived with DNase removal reagent (Turbo DNase kit, Amibion). Purified total RNA was quantitated by spectrophotmetry at 260 nm and quality assayed by running RNA samples on a 1% formaldehyde denaturing agarose gel.
Primers
Optimized and validated primer sets were supplied by realtimeprimers.com. The primer sets were: WT1 (gene accession no., NM_031534: Forward, 5′-AGGACTGCGAGAGAAGGTTT-3′; Reverse, 5′-ACGTGGAGTTTGGTCATGTT3′); Pax2 (XM_239083: Forward, 5′-GAGACTCCCAGAGTGGTGTG-3′; Reverse, 5′-CATTCCCCTGTTCTGATTTG-3′); WNT4 (NM_053402: Forward, 5′-GCCACGCACTAAAGGAGAAG-3′; Reverse, 5′-GGCCTTAGACGTCTTGTTGC-3′); WNT11 (XM_238122: Forward, 5′-TACCTGCTTGACCTGGAGAG-3′; Reverse, 5′-TAGGAGCATCGGAAAACTTG-3′); FGF2 (NM_019305: Forward, 5′-CCAGTTGGTATGTGGCACTG-3′; Reverse, 5′-CAGGGAAGGGTTTGACAAGA-3′); FGF7 (NM_022182: Forward, 5′-GCTCTACAGACCGTGCTTCC-3′; Reverse, CCCCTCCTTCCATGTAGTCA-3′); cRET (NM_012643: Forward, 5′-CAAGTAGCCTCAGGGCTGTC-3′; Reverse, 5′-CAACAGGGGGACTGTGTCTT-3′); GDNF (NM_019139: Forward, 5′-CCCGAAGATTATCCTGACCA-3′; Reverse, 5′-TAGCCCAAACCCAAGTCAGT-3′); BMP7 (XM_342591: Forward, 5′-AGACCAAGCACCTCTCCTGA-3′; Reverse, 5′-CCAACGGATTCTTTCTTGGA-3′); BMP4 (NM_012827: Forward, 5′-GACTTCGAGGCGACACTTCTG-3′; Reverse, 5′-AGCCGGTAAAGATCCCTCATG-3′); and 18S (M11188: Forward, 5′-ATTCCGATAACGAACGAGACT-3′; Reverse, 5′-AGCTTATGACCCGCACTTACT-3′). 18S was used as the normalization gene because it was found not to vary between control and FR samples in kidney mRNA samples (data not shown).
Real-time PCR
Complementary DNA (cDNA) was generated from 5 μg of total RNA using Superscript III reverse transcriptase kit (Invitrogen, Carlsbad, CA). The RNA was incubated in 20 μL of a reverse transcription reaction mixture (1 × reverse transcription buffer, 6.25 mM MgCl2, 10 mM DTT, 0.5 mM deoxyribonucleoside triphosphates, 50 ng random hexamers, 40 U RNaseOUT [RNase inhibitor], and 10 U Superscript III reverse transcriptase) at 50°C for 50 minutes. PCR was performed in 96-well optical reaction plates (Applied Biosystems, Foster City, CA) on cDNA equivalent to 500 ng RNA (50 ng for 18S reactions) in a volume of 25 μL that contained 1X reaction buffer (Eurogentec North American Inc, San Diego, CA), and forward and reverse appropriate primers (300 nM). PCR was performed for respective mRNAs and the 18S ribosomal RNA (normalization), with the ABI-Prism 7000 Sequence System (Applied Biosystems) at the following conditions: 10 minutes at 95°C for 1 cycle, 15 seconds at 95°C, and 1 minute at 60°C for 40 cycles. Three independent experiments for each RT-PCR assay were performed, and all samples were run in triplicate with the means determined. Each primer set was verified for similar efficiencies (90–110% range) of polymerization by using 10 fold dilutions of pooled e20 kidney cDNA and calculating the slope and efficiency based on linear regression. The efficiency data were: PAX2, 90.0%, WT1, 91.0%, FGF2, 98.9%, FGF7, 104.0%, BMP4, 105.7%, BMP7, 102.6%; WNT4, 90.9%; WNT11, 99.7%; cRET, 99.7%, GDNF, 104.9%; and 18S, 96.8%. Control PCR samples replaced cDNA with water and gave a threshold level (CT) value of 40, which indicated no detectable PCR product. ABI Sequence Detection System 1.6 software (Applied Biosystems) was used to select a threshold level of fluorescence that was in the linear phase of the PCR product accumulation. Results from the reverse transcription–PCR assay was determined as the difference between the CT for a specific mRNA gene and the CT for a reference mRNA, normalized to 18S threshold expression, and was expressed as fold change with the formula 2-ΔΔCT.26 Changes in gene expression were then expressed as fold changes with control set + 1.0 for each mRNA. SEM was determined based on the CT averages and proportionally transformed into fold changes.
Western blot
The primary and secondary antibodies were: Pax2 (Zymed 71–6000) Primary 1:400, Secondary 1:10000; FGF2 (Upstate 05-118): Primary 1:500, Secondary 1:10000. FGF7 (Santa Cruz SC-7882): Primary 1:300, Secondary 1:25,000; cRET (Santa Cruz SC-167): Primary 1:500, Secondary 1:10000; GDNF (Santa Cruz SC-328): Primary 1:500, Secondary 1:10000; WT1 (Santa Cruz SC-192): Primary 1:500, Secondary 1:10000; BMP7 (Santa Cruz SC-6899): Primary 1:300, Secondary 1:10000; BMP4 (Santa Cruz SC-6896): Primary 1:200, Secondary 1:5000; WNT4 (Santa Cruz SC-13962): Primary 1:300, Secondary 1:10000; WNT11 (Santa Cruz SC-23862): Primary 1:300, Secondary 1:10000. B-actin (Sigma A5441): Primary 1:10000, Secondary 1:10000. Secondary horse radish peroxidase conjugated antibodies were anti-goat (Santa Cruz SC-2020), anti-rabbit (Bio-Rad 170–6515), and anti-mouse (Bio-Rad 170–6515). All commercial antibodies were optimized for binding specificity and bands depicted have the expected molecular weights.
Protein extraction
Protein was extracted in radioimmuno precipitation assay (RIPA) buffer that contained protease inhibitors (HALT cocktail, Pierce) by homogenization of frozen kidneys on ice. Supernatants were clarified by 20 minute micro centrifugation at 12,000g. Supernatant protein concentration was determined by bicinchoninic acid (BCA) solution (PIERCE, Rockford, IL). Supernatants were frozen at −80°C until use.
Western Blot
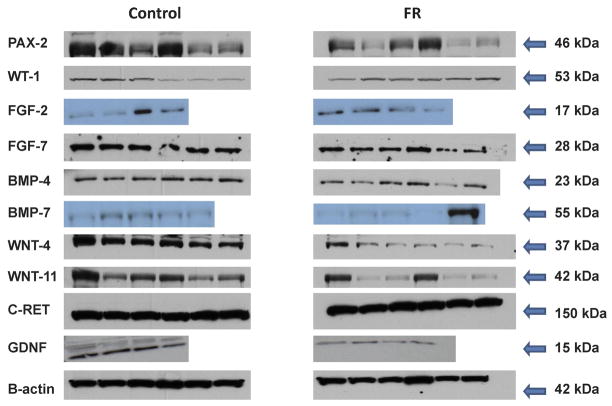
Protein expression was analyzed as previously done by our group and each blot was performed at least twice.27 4–6 samples were run for control and FR as detailed in Figure 3. Equal amounts of protein (30 μg) were mixed with Criterion sodium dodecyl sulfate sample buffer (Bio-Rad, Hercules, CA), boiled for 3 minutes, and separated on a Criterion 12% Bis-Tris denaturing gel. The separated proteins were transferred electrophoretically to a nitrocellulose membrane (Bio-Rad Laboratories) for 1.5 hours at 120 volts, 4°C. Nonspecific antibody binding was blocked by incubation overnight at 4°C with 5% nonfat dry milk in Tris-buffered saline solution containing 0.1% Triton X-100 (TBST; Bio-Rad). The membrane was incubated with the appropriate primary antibody in 5% milk in TBST for one hour, washed 3 times for 10 minutes each with TBST+0.1% Triton X-100 at room temperature. Blots were incubated for 1 hour with anti-goat, anti-rabbit, or anti-mouse secondary antibodies depending on the primary antibody, followed by washing as before and a final TBS only wash. SuperSignal West Pico Chemiluminescent Substrate (Pierce) was used to detect the targeted protein. The band density on the X-ray film was optically scanned and quantitated using Image J software, version 1.37 (NIH, Bethesda, MD). Blots were stripped with Restore stripping buffer (Pierce), reprobed and normalized to the reference protein (β-actin) and presented as fold change relative to control level (Fold = 1.0).
Figure 3.
Representative Western blots of fetal kidney maternal FR protein expression. Blots represented control samples (left hand panels) and FR samples (right hand panels). 4–6 pooled samples per blot.
Statistical analysis
Differences between control and FR groups were compared by unpaired t-test. P values <0.05 were considered significant. Means were transformed into fold changes and SEM were expressed as a percentage proportional to the original mean and SEM.
Results
Expression of ureteric bud branching signaling growth factors
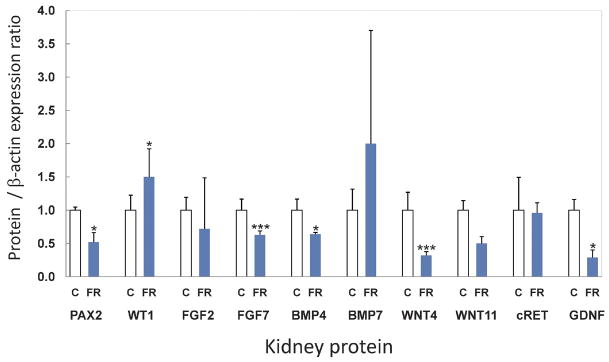
GDNF, a key mediator of UB branching is released from the mesenchyme and binds to its heterodimeric receptor complex, cRET-GDFRα1, which in turn activates branching.28 Maternal FR was found to reduce both the mRNA (Figure 1) and protein (Figures 2 and 3) expression of GDNF (mRNA 0.5 fold, p=0.03; protein 0.3 fold, p=0.03) whereas its receptor on the UB, cRET, was not significantly modulated (mRNA 0.6 fold, p=0.17; protein 1.0 fold, p=0.18).
Figure 1.
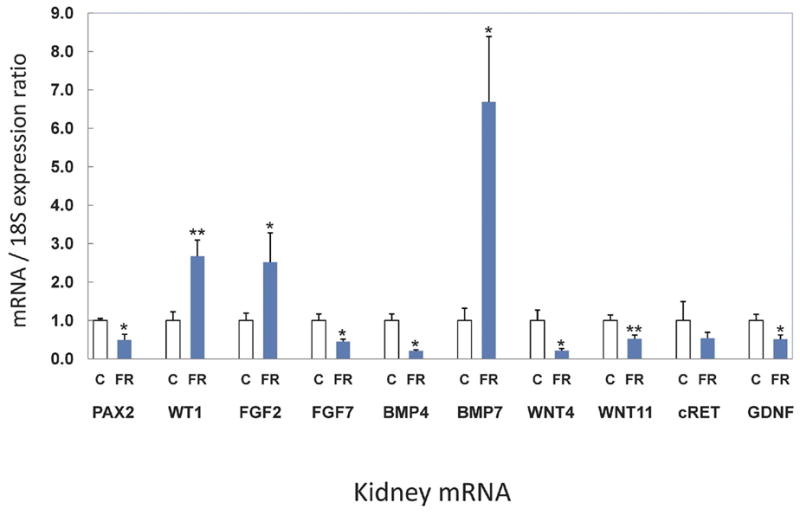
Fetal kidney maternal FR mRNA expression. Fold change in mRNA expression compared with controls. Control fold = 1.0 *p<0.05, **p<0.01. Error bars represent SEM of the fold change.
Figure 2.
Fetal kidney maternal FR protein expression. Fold change in protein expression compared with controls. Control fold = 1.0 *p<0.05, **p<0.01, ***p<0.001. Error bars represent SEM of the fold change.
Expression of factors controlling UB branching and mesenchymal transdifferentiation
WT1 is expressed in mesenchyme while Pax2 is found in mesenchyme and ureteric bud.29;30 Both are involved in regulation of branching and mesenchymal to epithelial transformation. Pax2, a transcriptional activator that is regulated by WT1, demonstrated significantly decreased mRNA expression by half (0.5 fold, p=0.02) (Figure 1) with concomitant reduction in protein expression (0.5 fold, p=0.03). In contrast, the transcriptional repressor WT1 was increased by 2.7 fold (p=0.01) for mRNA (Figure 1) and protein expression also changed significantly (Figures 2 and 3) (1.5 fold, p=0.02,) in FR as compared to C e20 kidneys. WNT4 and WNT11, transcription factors that are regulated by Pax2, were decreased in mRNA; 0.2 fold, p=0.04 and 0.5 fold, p=0.01, respectively (Figure 1). Protein expression also was significantly lower for WNT4 (0.3 fold, p=0.001) and trended down but was not significant for WNT11 (0.5 fold, p=0.06). FGF2 is critical for maintenance of the mesenchyme and had increased mRNA expression (2.5 fold, p=0.05) although protein expression remained unchanged (0.7 fold, p=0.25;). Growth factors BMP4 and FGF7 were reduced in mRNA expression by 0.2 fold (p=0.05) and 0.5 fold (p=0.02), respectively (Figure 1). Protein expression for BMP4 and FGF7 also were reduced significantly (0.6 fold, p=0.02, 0.6 fold, p=0.001 respectively). (Figures 2 and 3). A negatively acting growth factor controlling branching, BMP7, was significantly increased for mRNA at 6.7 fold (p=0.02) whereas the protein level trended higher at 2.0 fold but was not statistically significant (p=0.32) (Figures 1, 2 and 3).
Comment
Previously, we and others have determined that restricted nutrition in both humans and animal studies reduces the number of offspring nephrons.31–36 These findings suggest that maternal FR impacts nephrogenesis during fetal development, which may lead to the higher incidence of hypertension seen in the maternal FR rat model.35,37
Our study evaluated transcriptional and growth factor expression at e20, a time during which ureteric bud branching and mesenchymal to epithelial transformation are ensuing. Maternal FR inhibition of UB branching was evidenced by a reduction in GDNF gene expression. The importance of GDNF signal transduction to renal development is demonstrated in knock-out mice, as well as in mice lacking its receptor, c-Ret, or its co-receptor, GDNFR-α1; these animals have no kidneys as a result of a failure of UB outgrowth, while mice heterozygote for GDNF have small kidneys with 30% fewer nephrons.38 This, together with the ability of ectopically expressed or applied GDNF to induce the formation of supernumerary UBs from the Wolffian Duct in explants of e10–11 urogenital regions39 or in vivo40 confirms the critical role of GDNF in promoting UB outgrowth.
FGF7, a TGFβ family member, is strongly expressed in the developing rat kidney from e17 until birth,41 and also is important in branching morphogenesis.42 The RNA and protein expression of FGF7 were reduced by maternal FR and also may contribute to a reduction in ureteric bud branching and nephron number.
E20 kidney WT1 mRNA and protein are upregulated significantly while Pax2 mRNA and protein are down-regulated, suggesting that the Pax2 cascade is suppressed by maternal FR. WT1 inhibits transcription of Pax2,43;44 and may be, in part, responsible for its downregulation. Others have reported a decrease in Pax2 mRNA expression by 1.4 fold in the rat growth restricted fetus,45 and some Pax2 alleles in humans are associated with renal hypoplasia in adults,46 which support our findings. WT1 inhibits and Pax2 upregulates transcription of WNT4,47 and the observed dysregulation of these factors is associated with reduction in WNT4 RNA and protein, in this model.. The Wnt superfamily, are secreted glycoproteins that have important regulatory functions during vertebrate embryonic development.48 WNT4, an autocrine epithelializing signal, is expressed in several compartments in the developing nephron and is necessary for nephron formation.48;49 Wnt signaling is considered sufficient to trigger tubule development experimentally in the separated, uninduced kidney mesenchyme.48 WNT4 binds to the frizzled family of transmembrane receptors, activates the canonical WNT signaling pathway50,51 and activates mTOR, an important nutrient sensing pathway.52 The importance of this transcription factor is evidenced by WNT4 null mice, which fail to epithelialize the mesenchyme.53 Therefore, the observed significant decrease in WNT4 mRNA and protein expression in the offspring of food restricted dams may play a key role resulting in the reduced number of nephrons.
BMP4 is localized to the metanephric mesenchyme and functions to inhibit branching morphogenesis possibly acting through a GDNF reduction mechanism at a specific point in kidney nephrogenesis to contribute to lowered nephron number.54 BMP4 also prevents apoptosis and promotes growth of the mesenchyme depending on the specific localization.55,56 Mice heterozygote for BMP4 with reduced protein levels do not show abnormal ureteric bud branching.55 In contrast, upregulation of BMP4 mRNA has been identified in offspring of glucocorticoid treated spiny mouse dams in association with reduced nephron number.57 RNA and protein levels were significantly downregulated in the FR model, and the significance of this reduction in BMP4 is uncertain; it may result in increased apoptosis and inhibit mesenchymal growth although further studies in this area are needed.
We identified several factors in maternal FR offspring with dysregulated mRNA levels for which associated protein expression was not significantly different. The lack of change in protein expression or discrepant mRNA and protein level findings for these factors may be due to compensatory mechanisms present at e20 versus earlier points in fetal kidney development. It is possible that protein translation, post-translational modification, and degradation, may influence the level of a protein present within the kidney. Additionally, gene expression and protein concentrations may vary within the different compartments of the developing kidney and during the different stages of development. Changes in nuclear proteins may not be fully reflected in analysis of whole tissue homogenates. Further studies localizing these factors during renal development may help elucidate the significance of these changes. Finally, a confounding factor in this study is the lack of stratification of the e20 kidneys by sex. There is evidence that male offspring are more affected by maternal FR than females.36 Separate assessment of embryonic kidneys by gender and with larger cohorts may uncover additional significant aberrations in growth and signaling factor mRNA levels and proteins during renal development.
In conclusion, in this study we have identified maternal FR-induced dysregulation of GDNF, WT1, Pax2, WNT4, BMP4 and FGF7, factors impacting ureteric bud branching and mesenchymal to epithelial transformation in the developing fetal kidney at embryonic day 20. Our previous work has identified dysregulated TGF-β receptor 1 in the kidney earlier in nephrogenesis at embryonic day 16.58 These data suggest that dysregulation of critical renal genes at different stages of development is a key mechanism of the nephropenia induced by maternal FR. Further studies elucidating this process will provide additional insights into our understanding of reduced nephron number associated with fetal growth restriction. This is necessary to develop appropriately timed interventions to correct this maladaptive programming.
We would like to thank Guang Han, Bindu Cherian, Linda Day, and Stacy Behare for their technical assistance. This study was supported in part by grants from the March of Dimes Birth Defects Foundation #6-FY04-72 (MGR) and NIH 5P20MD000545 (TRM).
Acknowledgments
Financial Support:: This study was supported in part by grants from the March of Dimes Birth Defects Foundation #6-FY04-72 (MGR) and NIH 5P20MD00545 (TRM).
Footnotes
Accepted for an oral presentation at the Society of Maternal-Fetal Medicine 28th Annual Meeting, Dallas, Texas, January 28-February 2, 2008
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Shankar K, Harrell AM, Liu X, Gilchrist JM, Ronis MJ, Badger TM. Maternal Obesity at Conception Programs Obesity in the Offspring. Am J Physiol Regul Integr Comp Physiol. 2007 doi: 10.1152/ajpregu.00316.2007. [DOI] [PubMed] [Google Scholar]
- 2.de R, Sr, Painter RC, Holleman F, Bossuyt PM, Roseboom TJ. The metabolic syndrome in adults prenatally exposed to the Dutch famine. Am J Clin Nutr. 2007;86:1219–24. doi: 10.1093/ajcn/86.4.1219. [DOI] [PubMed] [Google Scholar]
- 3.Thompson JN. Fetal nutrition and adult hypertension, diabetes, obesity, and coronary artery disease. Neonatal Netw. 2007;26:235–40. doi: 10.1891/0730-0832.26.4.235. [DOI] [PubMed] [Google Scholar]
- 4.Gluckman PD, Lillycrop KA, Vickers MH, Pleasants AB, Phillips ES, Beedle AS, et al. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. Proc Natl Acad Sci USA. 2007;104:12796–800. doi: 10.1073/pnas.0705667104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross MG, Desai M. Gestational programming: population survival effects of drought and famine during pregnancy. Am J Physiol Regul Integr Comp Physiol. 2005;288:R25–R33. doi: 10.1152/ajpregu.00418.2004. [DOI] [PubMed] [Google Scholar]
- 6.Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261:412–17. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 7.Langley-Evans SC. Developmental programming of health and disease. Proc Nutr Soc. 2006;65:97–105. doi: 10.1079/pns2005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 9.Barker DJ, Lackland DT. Prenatal influences on stroke mortality in England and Wales. Stroke. 2003;34:1598–602. doi: 10.1161/01.STR.0000077257.27430.7E. [DOI] [PubMed] [Google Scholar]
- 10.Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 11.Taylor PD, Poston L. Developmental programming of obesity in mammals. Exp Physiol. 2007;92:287–98. doi: 10.1113/expphysiol.2005.032854. [DOI] [PubMed] [Google Scholar]
- 12.Coleman MP, Babb P, Sloggett A, Quinn M, De SB. Socioeconomic inequalities in cancer survival in England and Wales. Cancer. 2001;91:208–16. doi: 10.1002/1097-0142(20010101)91:1+<208::aid-cncr6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 13.Bagby SP. Maternal nutrition, low nephron number, and hypertension in later life: pathways of nutritional programming. J Nutr. 2007;137:1066–72. doi: 10.1093/jn/137.4.1066. [DOI] [PubMed] [Google Scholar]
- 14.Mackenzie HS, Lawler EV, Brenner BM. Congenital oligonephropathy: The fetal flaw in essential hypertension? Kidney Int Suppl. 1996;55:S30–S34. [PubMed] [Google Scholar]
- 15.Gilbert JS, Lang AL, Grant AR, Nijland MJ. Maternal nutrient restriction in sheep: hypertension and decreased nephron number in offspring at 9 months of age. J Physiol. 2005;565:137–147. doi: 10.1113/jphysiol.2005.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langley-Evans SC, Welham SJ, Jackson AA. Fetal exposure to a maternal low protein diet impairs nephrogenesis and promotes hypertension in the rat. Life Sci. 1999;64:965–974. doi: 10.1016/s0024-3205(99)00022-3. [DOI] [PubMed] [Google Scholar]
- 17.Costantini F. Renal branching morphogenesis: concepts, questions, and recent advances. Differentiation. 2006;74:402–21. doi: 10.1111/j.1432-0436.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- 18.Moritz KM, Dodic M, Wintour EM. Kidney development and the fetal programming of adult disease. Bioessays. 2003;25:212–20. doi: 10.1002/bies.10240. [DOI] [PubMed] [Google Scholar]
- 19.Davies JA, Fisher CE. Genes and proteins in renal development. Exp Nephrol. 2002;10:102–13. doi: 10.1159/000049905. [DOI] [PubMed] [Google Scholar]
- 20.Davies JA. Mesenchyme to epithelium transition during development of the mammalian kidney tubule. Acta Anat(Basel) 1996;156:187–201. doi: 10.1159/000147846. [DOI] [PubMed] [Google Scholar]
- 21.Sariola H. Nephron induction revisited: from caps to condensates. Curr Opin Nephrol Hypertens. 2002;11:17–21. doi: 10.1097/00041552-200201000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Abrahamson DR. Glomerulogenesis in the developing kidney. Semin Nephrol. 1991;11:375–89. [PubMed] [Google Scholar]
- 23.Bard J. The metanephros. In: Vize PD, Woolf AS, Bard JBL, editors. The Kidney. Academic Press, Eselvier Science; San Diego: 2003. pp. 139–148. [Google Scholar]
- 24.Hostetter TH, Olson JL, Rennke HG, Venkatachalam MA, Brenner BM. Hyperfiltration in remnant nephrons: a potentially adverse response to renal ablation. J Am Soc Nephrol. 2001;12:1315–25. [Google Scholar]
- 25.Oliver JD, III, Simons JL, Troy JL, Provoost AP, Brenner BM, Deen WM. Proteinuria and impaired glomerular permselectivity in uninephrectomized fawn-hooded rats. Am J Physiol. 1994;267:F917–F925. doi: 10.1152/ajprenal.1994.267.6.F917. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–08. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Choi GY, Tosh DN, Garg A, Mansano R, Ross MG, Desai M. Gender-specific programmed hepatic lipid dysregulation in intrauterine growth-restricted offspring. Am J Obstet Gynecol. 2007;196:477. doi: 10.1016/j.ajog.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Costantini F, Shakya R. GDNF/Ret signaling and the development of the kidney. Bioessays. 2006;28:117–27. doi: 10.1002/bies.20357. [DOI] [PubMed] [Google Scholar]
- 29.Brodbeck S, Englert C. Genetic determination of nephrogenesis: the Pax/Eya/Six gene network. Pediatr Nephrol. 2004;19:249–55. doi: 10.1007/s00467-003-1374-z. [DOI] [PubMed] [Google Scholar]
- 30.Dressler GR. Pax-2, kidney development, and oncogenesis. Med Pediatr Oncol. 1996;27:440–4. doi: 10.1002/(SICI)1096-911X(199611)27:5<440::AID-MPO9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 31.Schreuder MF, Nauta J. Prenatal programming of nephron number and blood pressure. Kidney Int. 2007;72:265–68. doi: 10.1038/sj.ki.5002307. [DOI] [PubMed] [Google Scholar]
- 32.Welham SJ, Riley PR, Wade A, Hubank M, Woolf AS. Maternal diet programs embryonic kidney gene expression. Physiol Genomics. 2005;22:48–56. doi: 10.1152/physiolgenomics.00167.2004. [DOI] [PubMed] [Google Scholar]
- 33.Hoppe CC, Evans RG, Bertram JF, Moritz KM. Effects of dietary protein restriction on nephron number in the mouse. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1768–R1774. doi: 10.1152/ajpregu.00442.2006. [DOI] [PubMed] [Google Scholar]
- 34.Woods LL, Weeks DA, Rasch R. Programming of adult blood pressure by maternal protein restriction: role of nephrogenesis. Kidney Int. 2004;65:1339–48. doi: 10.1111/j.1523-1755.2004.00511.x. [DOI] [PubMed] [Google Scholar]
- 35.Henry T, Torday J, Magee T, Abdel-Hakeem A, Desai M, Ross MG, Rehan V, Nast C. Maternal food restriction inhibits nephrogenesis by disrupting mesonephric mesenchyme ureteric bud signaling. Early Hum Dev. 2007;83(Suppl 1):S176. [Google Scholar]
- 36.Desai M, Gayle D, Babu J, Ross MG. Permanent reduction in heart and kidney organ growth in offspring of undernourished rat dams. Am J Obstet Gynecol. 2005;193:1224–32. doi: 10.1016/j.ajog.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 37.Almeida JR, Mandarim-de-Lacerda CA. Maternal gestational protein-calorie restriction decreases the number of glomeruli and causes glomerular hypertrophy in adult hypertensive rats. Am J Obstet Gynecol. 2005;192:945–51. doi: 10.1016/j.ajog.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Cullen-McEwen LA, Drago J, Bertram JF. Nephron endowment in glial cell line-derived neurotrophic factor (GDNF) heterozygous mice. Kidney Int. 2001;60:31–36. doi: 10.1046/j.1523-1755.2001.00767.x. [DOI] [PubMed] [Google Scholar]
- 39.Vega QC, Worby CA, Lechner MS, Dixon JE, Dressler GR. Glial cell line-derived neurotrophic factor activates the receptor tyrosine kinase RET and promotes kidney morphogenesis. Proc Natl Acad Sci USA. 1996;93:10657–61. doi: 10.1073/pnas.93.20.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shakya R, Watanabe T, Costantini F. The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev Cell. 2005;8:65–74. doi: 10.1016/j.devcel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Cancilla B, Ford-Perriss MD, Bertram JF. Expression and localization of fibroblast growth factors and fibroblast growth factor receptors in the developing rat kidney. Kidney Int. 1999;56:2025–39. doi: 10.1046/j.1523-1755.1999.00781.x. [DOI] [PubMed] [Google Scholar]
- 42.Bush KT, Sakurai H, Steer DL, et al. TGF-beta superfamily members modulate growth, branching, shaping, and patterning of the ureteric bud. Dev Biol. 2004;266:285–98. doi: 10.1016/j.ydbio.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 43.Discenza MT, He S, Lee TH, Chu LL, Bolon B, Goodyer P, et al. WT1 is a modifier of the Pax2 mutant phenotype: cooperation and interaction between WT1 and Pax2. Oncogene. 2003;22:8145–55. doi: 10.1038/sj.onc.1206997. [DOI] [PubMed] [Google Scholar]
- 44.Ryan G, Steele-Perkins V, Morris JF, Rauscher FJ, III, Dressler GR. Repression of Pax-2 by WT1 during normal kidney development. Development. 1995;121:867–75. doi: 10.1242/dev.121.3.867. [DOI] [PubMed] [Google Scholar]
- 45.Buffat C, Boubred F, Mondon F, Chelbi ST, Feuerstein JM, Lelievre-Pegorier M, et al. Kidney gene expression analysis in a rat model of intrauterine growth restriction reveals massive alterations of coagulation genes. Endocrinol. 2007;148:5549–57. doi: 10.1210/en.2007-0765. [DOI] [PubMed] [Google Scholar]
- 46.Dziarmaga A, Quinlan J, Goodyer P. Renal hypoplasia: lessons from Pax2. Pediatr Nephrol. 2006;21:26–31. doi: 10.1007/s00467-005-2039-x. [DOI] [PubMed] [Google Scholar]
- 47.Torban E, Dziarmaga A, Iglesias D, Chu LL, Vassilieva T, Little M, et al. PAX2 activates WNT4 expression during mammalian kidney development. J Biol Chem. 2006;281:12705–12. doi: 10.1074/jbc.M513181200. [DOI] [PubMed] [Google Scholar]
- 48.Vainio SJ. Nephrogenesis regulated by Wnt signaling. J Nephrol. 2003;16:279–85. [PubMed] [Google Scholar]
- 49.Sariola H, Sainio K. Cell lineages in the embryonic kidney: their inductive interactions and signalling molecules. Biochem Cell Biol. 1998;76:1009–16. [PubMed] [Google Scholar]
- 50.Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–39. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- 51.Iglesias DM, Hueber PA, Chu L, Campbell R, Patenaude AM, Dziarmaga AJ, et al. Canonical WNT signaling during kidney development. Am J Physiol Renal Physiol. 2007;293:F494–F500. doi: 10.1152/ajprenal.00416.2006. [DOI] [PubMed] [Google Scholar]
- 52.Nijland MJ, Schlabritz-Loutsevitch NE, Hubbard GB, Nathanielsz PW, Cox LA. Non-human primate fetal kidney transcriptome analysis indicates mammalian target of rapamycin (mTOR) is a central nutrient-responsive pathway. J Physiol. 2007;579:643–56. doi: 10.1113/jphysiol.2006.122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–83. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- 54.Michos O, Goncalves A, Lopez-Rios J, et al. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development. 2007;134:2397–2405. doi: 10.1242/dev.02861. [DOI] [PubMed] [Google Scholar]
- 55.Cain JE, Bertram JF. Ureteric branching morphogenesis in BMP4 heterozygous mutant mice. J Anat. 2006;209:745–55. doi: 10.1111/j.1469-7580.2006.00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oxburgh L, Dudley AT, Godin RE, et al. BMP4 substitutes for loss of BMP7 during kidney development. Dev Biol. 2005;286:637–46. doi: 10.1016/j.ydbio.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 57.Dickinson H, Walker DW, Wintour EM, Moritz K. Maternal dexamethasone treatment at midgestation reduces nephron number and alters renal gene expression in the fetal spiny mouse. Am J Physiol Regul Integr Comp Physiol. 2007;292:R453–61. doi: 10.1152/ajpregu.00481.2006. [DOI] [PubMed] [Google Scholar]
- 58.Nast CC, Desai M, Ross MG, Rehan VK, Magee TR, Torday JS. Maternal undernutrition dysregulates fetal rat kidney growth factor signaling in association with decreased nephrogenesis and hypertension. J Am Soc Nephrol. 2007;18:358A. [Google Scholar]