Abstract
Prostaglandin endoperoxide H synthases (PGHSs)-1 and -2 (also called cyclooxygenases (COXs)-1 and -2) catalyze the committed step in prostaglandin biosynthesis. Both isoforms are targets of nonsteroidal anti-inflammatory drugs (NSAIDs). PGHSs are homodimers that exhibit half-of-sites COX activity; moreover, some NSAIDs cause enzyme inhibition by binding only one monomer. To learn more about the cross-talk that must be occurring between the monomers comprising each PGHS-1 dimer, we analyzed structures of PGHS-1 crystallized under five different conditions including in the absence of any tightly binding ligand and in the presence of non-specific NSAIDs and of a COX-2 inhibitor. When crystallized with sub-stoichiometric amounts of an NSAID, both monomers are often fully occupied with inhibitor; thus, the enzyme prefers to crystallize in a fully occupied form. In comparing the five structures, we only observe changes in the positions of residues 123-129 and residues 510-515. In cases where one monomer is fully occupied with an NSAID and the partner monomer is incompletely occupied, an alternate conformation of the loop involving residues 123-129 is seen in the partially occupied monomer. We propose, based on this observation and previous cross-linking studies, that cross-talk between monomers involves this mobile 123-129 loop, which is located at the dimer interface. In ovine PGHS-1 crystallized in the absence of an NSAID, there is an alternative route for substrate entry into the COX site different than the well-known route through the membrane binding domain.
Keywords: COX-1, Prostaglandin Endoperoxide H Synthase, NSAIDs, Aspirin, Nimesulide, Flurbiprofen, Diclofenac, Heterodimer, Crystallography, Merohedral twinning
Prostaglandin endoperoxide H synthases (PGHSs)+-1 and -2, also known as cyclooxygenases (COXs)-1 and -2, are ER resident proteins that are bound to a single face of the ER membrane bilayer. PGHSs-1 and -2 are responsible for catalyzing the conversion of arachidonic acid (AA) to prostaglandin H2 (PGH2) in the committed step of prostaglandin (PG) biosynthesis (1-5). PGH2 is then converted to biologically active PGs including PGD2, PGE2, PGF2α or PGI2 (prostacyclin) or thromboxane A2 (TxA2) (6). Both PGHS isoforms are targets of non-selective nonsteroidal anti-inflammatory drugs (nsNSAIDs), such as aspirin and ibuprofen, while PGHS-2 can also be blocked selectively by diarylheterocyclic “COX-2 specific” inhibitors called coxibs (7).
Both PGHS-1 and -2 are homodimers composed of ~72 kDa subunits that are tightly bonded to one another via an interface spanning 2500 Å2 (8). Each PGHS monomer contains an epidermal growth factor-like domain of unknown function, a membrane binding domain (MBD), and a large catalytic core (9). The bifunctional catalytic subunit houses both COX and peroxidase (POX) enzymatic activities (10). During catalysis, the proper orientation of the cis-methylene interrupted double bond system in AA facilitates delocalization of an electron following abstraction of the 13-proS hydrogen atom from AA. The electron is abstracted by the catalytic Tyr385 radical located at the roof of the hydrophobic COX channel (11). Formation of an endoperoxide bridge and cyclization of the carbon skeleton generates PGG2. This product is subsequently reduced to PGH2 by the POX activity located at a physically distinct POX active site.
We have provided evidence that both PGHS isoforms exhibit half-of-sites COX activity (12, 13). When a ligand--either a substrate or non-substrate fatty acid, an nsNSAID or a coxib--binds to one monomer of an unoccupied dimer, that monomer becomes the allosteric subunit, and the partner monomer becomes the catalytic monomer. The catalytic efficiency of the catalytic monomer, is determined by its interaction with its allosteric partner and this, in turn, is different for each different ligand that binds the allosteric monomer. Binding of certain ligands increases catalytic efficiency (13), some ligands have no detectable effect (14) and others decrease catalytic efficiency (12, 13, 15) Moreover, the same ligand can have different effects with PGHS-1 vs. PGHS-2 (12, 13) (15) With respect to enzyme inhibition, it appears that time-dependent inhibition by nsNSAIDs and COX-2 specific inhibitors involves binding to only the allosteric monomer (15, 16), whereas inhibition by reversible, competitive nsNSAIDs such as ibuprofen requires that the inhibitor bind both the allosteric and catalytic monomers (14). The importance of understanding the cross talk between the allosteric and catalytic monomers was highlighted by recent work showing that COX-2 inhibitors can bind tightly to one monomer of PGHS-1 and attenuate the actions of aspirin on PGHS-1 in vitro and in vivo (15).
PGHS crystal structures have provided limited information about the nature of the differences between monomers of the biological dimer. Recently, we determined that ovPGHS-1 crystals frequently display merohedral twinning (15, 17-19) and that solving a structure in space group P65, which contains the biological dimer in the asymmetric unit, can unmask asymmetry between the partner monomers. Here, we prepared crystals of ovine (ov) PGHS-1 in the presence and absence of several different nsNSAIDs and COX-2 inhibitors to delineate what interactions are important in cross talk between the allosteric and catalytic subunits of the enzymes. The most prominent differences were seen at the dimer interface in some complexes and in a loop region in the COX active site “side pocket”.
Methods
Materials
Flag-affinity resin, hemin, and acetylsalicylic acid (aspirin) were purchased from Sigma Chemical Company. The NSAIDs flurbiprofen, nimesulide, and diclofenac were purchased from Cayman Chemical company. Complete EDTA free protease inhibitor was from Roche Applied Biosciences. The non-ionic detergents C10E6 and n-octyl-β-D-glucopyranoside (β-OG) were from Anatrace. Ni-NTA was from Qiagen. BCA protein reagent was purchased from Pierce Biochemical. All other chemicals and reagents were purchased from Sigma Chemical Company
ovPGHS-1 Expression and Purification
Generation of recombinant baculovirus, expression of recombinant octa-histidine- tagged ovPGHS-1, and purification of ovPGHS-1 were essentially as described previously (13, 20). Protein expression from FastBac or pFastBacDual expression vectors was performed according to the instructions of the manufacturer for the Invitrogen Bac-to-Bac expression system. Sf21 cells at a density of 1.2 – 1.5 × 106 cells/ml were infected with recombinant virus at a multiplicity of infection of ~0.1. After expression for 3-4 days, the cells were harvested, washed with lysis buffer (20 mM TrisHCl pH 8.0, 100 mM KCl) and stored at -80 °C
Cell pellets from 3-5 L cultures were resuspended in lysis buffer containing Complete EDTA-free Protease Inhibitor and disrupted by sonication. PGHS protein was solubilized from the membranes using 0.8% C10E6 at 4 °C for at least 1 hr with gentle agitation. The solubilized lysate was then centrifuged at 100,000 × g at 4 °C for 2 hours. The supernatant was carefully removed and incubated overnight for at 4 °C with Ni-NTA resin pre-equilibrated buffer (20 mM TrisHCl pH 8.0, 100 mM KCl, 5 mM Imidazole, 0.1% C10E6, and 5% glycerol). The slurry was poured into a column that was then washed with 10 column volumes of wash buffer I (20 mM TrisHCl pH 8.0, 1 M KCl, 20 mM imidazole, 20 mM galactose 0.1% C10E6, and 5% glycerol) followed by five column volumes of wash buffer II (20 mM TrisHCl, 40 mM KCl, 50 mM imidazole, 0.1% C10E6, and 5% glycerol). Octa-histidine-tagged ovPGHS-1 was eluted with buffer containing 20 mM TrisHCl pH 8.0, 40 mM KCl, 250 mM imidazole, 0.1% C10E6, and 5% glycerol. The eluted protein was pooled and buffer exchanged with 50mM TrisHCl pH 8.55, 150mM KCl, and 0.1% C10E6 using a Millipore Ultrafree-15 spin concentrator (100-kDa nominal molecular weight cut-off). The octa-histidine tag was removed using TEV protease at a 1:20 protease to protein ratio and overnight incubation at 4 °C. Ni-NTA chromatography was performed, and the flow-through fraction was collected and adjusted to 300 mM KCl and 10 mM imidazole before a final detergent exchange was performed using Amicon Ultra (Milipore) centrifugal filter device with crystallization buffer containing 20 mM HEPES, pH 7.3, 40 mM NaCl, and 0.4% n-octyl-β-D-glucopyranoside. The protein concentrations were determined using BCA protein assays (Pierce).
pFastBac ovPGHS-1 Mutations or pFastBac Dual for Heterodimer Expression
The Arg120Gln mutation of ovPGHS-1 was performed using a modified pFastBac vector (Invitrogen). A QuikChange site-directed mutagenesis kit (Stratagene) was used to create mutations, which were confirmed using DNA sequencing performed by the University of Michigan DNA Sequencing Core. The construction of expression vectors encoding the ovPGHS-1 heterodimer was performed in two steps. In the first step, the desired DNA fragments with hexa-histidine tags were obtained from PCR and sequentially digested with XhoI-KpnI and then subcloned into the Pp10 site of pFastBacDual, which had also been digested with XhoI-KpnI. In the second step, the other DNA fragment with a FLAG epitope tag (derived from PCR and treated with EcoRI-HINDIII) was subcloned into the PpH site of pFastBacDual, which had been cleaved with the same restriction enzymes. The correct orientation of the insert was confirmed by restriction digestion (12, 13).
Crystallization of ovPGHS-1 complexes
ovPGHS-1 was washed extensively by buffer exchange with crystallization buffer. Protein was prepared for crystallization (typically 100 μM dimer) by the addition of two moles of Fe (III) protoprophyrin IX per mole of protein dimer. Diclofenac (or nimesulide) was added to the protein at a one mole per mole of dimer ratio (typically 50 μM) and incubated for 10 min before setting up crystallization experiments. ovPGHS-1 was acetylated by incubation with 500 mM aspirin for 1 hour at room temperature prior to washing only for the aspirin/diclofenac structure. Using the sitting drop vapor diffusion method, 3 μl of protein was mixed with 3 μl of buffer (0.64 M sodium citrate, 0.3-0.9 M LiCl, 0.4% (w/v) β-OG, and 1 mM NaN3) and was equilibrated within a reservoir containing 0.68-0.88 M sodium citrate, 0.3-0.6 M LiCl, and 1 mM NaN3. Crystals appeared within 4-21 days.
Data collection and processing
Diffraction data were collected from single crystals in a -135° C nitrogen stream at beam line 21 ID-F, Life Sciences-CAT (Argonne National Laboratory, Argonne, IL) except for native ovPGHS-1/flurbiprofen, which was collected at DND-CAT (Argonne National Laboratory, Argonne, IL). Data processing and scaling were performed using HKL2000 (21) except for native ovPGHS-1/flurbiprofen, which was integrated using MOSFLM (22) and merged using SCALA (23). Statistics for the data collection, processing and crystallographic refinement are shown in Tables 1 and 2.
Table 1.
Data scaling of all ovPGHS-1 structures used in this study and from the previously published structure of celecoxib/ovPGHS-1 in space groups P6522 and P65.a
| Unoccupied | Flurbiprofen (Native/R120Q) | Nimesulide | ASA-Diclofenac | FBP 1 per dimer | Celecoxib | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Resolution Range |
P6522 Linear R-fac |
P65 Linear R-fac |
Resolution Range |
P6522 Linear R-fac |
P65 Linear R-fac |
Resolution Range |
P6522 Linear R-fac |
P65 Linear R-fac |
Resolution Range |
P6522 Linear R-fac |
P65 Linear R-fac |
Resolution Range |
P6522 Linear R-fac |
P65 Linear R-fac |
Resolution Range |
P6522 Linear R-fac |
P65 Linear R-fac |
| 50.00-6.57 | 0.043 | 0.043 | 50.00-5.92 | 0.051 | 0.047 | 30.00-6.00 | 0.041 | 0.040 | 50.00-5.49 | 0.040 | 0.039 | 31.63-9.17 | - | 0.029 | 50.00-5.92 | 0.038 | 0.038 |
| 6.57-5.21 | 0.076 | 0.075 | 5.92-4.70 | 0.072 | 0.066 | 6.00-4.77 | 0.069 | 0.067 | 5.49-4.36 | 0.066 | 0.065 | 9.17-6.38 | - | 0.052 | 5.92-4.70 | 0.060 | 0.062 |
| 5.21-4.56 | 0.079 | 0.078 | 4.70-4.11 | 0.068 | 0.060 | 4.77-4.17 | 0.072 | 0.070 | 4.36-3.81 | 0.065 | 0.064 | 6.38-5.29 | - | 0.126 | 4.70-4.11 | 0.058 | 0.059 |
| 4.56-4.14 | 0.087 | 0.086 | 4.11-3.73 | 0.078 | 0.069 | 4.17-3.79 | 0.086 | 0.084 | 3.81-3.46 | 0.080 | 0.078 | 5.29-4.59 | - | 0.076 | 4.11-3.73 | 0.065 | 0.064 |
| 4.14-3.84 | 0.107 | 0.106 | 3.73-3.46 | 0.098 | 0.089 | 3.79-3.51 | 0.138 | 0.134 | 3.46-3.21 | 0.120 | 0.116 | 4.59-4.10 | - | 0.080 | 3.73-3.46 | 0.082 | 0.080 |
| 3.84-3.62 | 0.140 | 0.138 | 3.46-3.26 | 0.150 | 0.139 | 3.51-3.31 | 0.167 | 0.163 | 3.21-3.02 | 0.187 | 0.182 | 4.10-3.74 | - | 0.106 | 3.46-3.26 | 0.130 | 0.126 |
| 3.62-3.43 | 0.194 | 0.190 | 3.26-3.10 | 0.214 | 0.202 | 3.31-3.14 | 0.247 | 0.240 | 3.02-2.87 | 0.298 | 0.290 | 3.74-3.47 | - | 0.146 | 3.26-3.10 | 0.196 | 0.190 |
| 3.43-3.29 | 0.308 | 0.302 | 3.10-2.96 | 0.322 | 0.308 | 3.14-3.01 | 0.356 | 0.346 | 2.87-2.75 | 0.410 | 0.395 | 3.47-3.24 | - | 0.232 | 3.10-2.96 | 0.291 | 0.283 |
| 3.29-3.16 | 0.424 | 0.416 | 2.96-2.85 | 0.462 | 0.442 | 3.01-2.89 | 0.478 | 0.458 | 2.75-2.64 | 0.478 | 0.455 | 3.24-3.06 | - | 0.376 | 2.96-2.85 | 0.433 | 0.418 |
| 3.16-3.05 | 0.545 | 0.530 | 2.85-2.75 | 0.596 | 0.561 | 2.89-2.79 | 0.600 | 0.565 | 2.64-2.55 | 0.607 | 0.556 | 3.06-2.90 | - | 0.614 | 2.85-2.75 | 0.575 | 0.547 |
| | |||||||||||||||||
| All reflections | 0.101 | 0.099 | 0.090 | 0.083 | 0.090 | 0.088 | 0.084 | 0.082 | Not Done | 0.105 | 0.080 | 0.079 | |||||
| | |||||||||||||||||
| Multivariate Z score | 2.516 | 1.431 | 4.872 | 4.086 | 3.786 | 2.892 | 8.864 | 7.112 | 3.614 | 7.532 | 6.153 | ||||||
Data are Rsym values in each resolution shell and the overall Rsym for the entire resolution range. Multivariate Z scores are from the phenix.xtriage analysis of the scaled data. All data were processed and scaled using HKL2000 except the flurbiprofen (FBP)/ovPGHS-1 structure which was processed using MOSFLM.
Table 2.
Data collection and refinement statistics.
| ovPGHS-1 Structure | Unoccupied | Flurbiprofen (Native/R120Q) | Nimesulide | ASA/Diclofenac | Flurbiprofen |
|---|---|---|---|---|---|
| Space group | P65 | P65 | P65 | P65 | P65 |
| Twin operator | -h-k, k, -l | -h-k, k, -l | -h-k, k, -l | -h-k, k, -l | h+k, -k, -l |
| Refined twin fraction | 0.488 | 0.467 | 0.496 | 0.499 | 0.498 |
| a = b, c (Å) | 181.907, 102.579 | 182.483, 103.095 | 181.709, 103.454 | 182.327, 103.106 | 181.49, 103.661 |
| α = β, γ | 90, 120 | 90, 120 | 90, 120 | 90, 120 | 90, 120 |
| Resolution (Å) | 50.0 – 3.05 | 50.0-2.75 | 30.0 – 2.75 | 30.0 – 2.60 | 31.63 - 2.90 |
| No. of unique reflections | 35,349 | 48,759 | 46,907 | 57,157 | 41,602 |
| I/sigma (outer shell) | 24.00 (4.00) | 24.33(2.23) | 17.04 (2.03) | 23.74 (2.04) | 14.7 (3.6) |
| Completeness (outer shell) | 99.6 (97.6) | 98.99 (87.36) | 95.51(70.0) | 97.8 (79.3) | 99.75 (97.86) |
| Average redundancy | 11.0 (8.6) | 7.2(4.1) | 6.8 (3.3) | 7.1 (3.8) | 9.7 (9.4) |
| Rsym, all data (outer shell) | 9.9 (53.0) | 7.5(55.5) | 8.8 (56.5) | 7.7 (49.7) | 10.5 (61.4) |
| No. of atoms in refinement | 9486 | 9498 | 9308 | 9423 | 9434 |
| Rfactor | 20.39 | 17.76 | 18.49 | 18.38 | 17.78 |
| Rfree | 23.45 | 20.18 | 21.46 | 19.20 | 20.32 |
| RMSD in bond distance (Å) | 0.006 | 0.007 | 0.006 | 0.006 | 0.006 |
| RMSD in bond angle (°) | 1.044 | 1.135 | 0.972 | 1.025 | 0.909 |
| Average B factor, protein (Å2) | 45.106 | 34.471 | 42.449 | 24.738 | 40.43 |
| Ligand in monomer A | 58.30 | 44.80 | 50.40 | 33.00 | 58.90 |
| Ligand in monomer B | N/A | 46.50 | 36.10 | 54.70 | |
| Residues in preferred regions (%) | 96.01 | 95.64 | 95.27 | 97.18 | 97.19 |
| Residues in allowed regions (%) | 3.45 | 4.27 | 4.73 | 2.55 | 2.72 |
| Outliers | 0.54 | 0.09 | 0 | 0.27 | 0.09 |
| Clash score | 9.79 | 8.2 | 10.25 | 7.86 | 8.56 |
| Molprobity score | 2.18 | 2.27 | 2.27 | 2.05 | 2.12 |
| Residues with bad bonds (%) | 0 | 0.09 | 0 | 0 | 0 |
| Residues with bad angles (%) | 0.18 | 0.27 | 0.09 | 0 | 0 |
| Cβ deviations >0.25Å | 0 | 0 | 0 | 0 | 0 |
| Rotamer outliers (%) | 3.37 | 4.86 | 3.47 | 4.16 | 4.71 |
Crystallographic refinement
As described previously (15), data were processed in space groups P65 and P6522. The Rsym was marginally, but consistently, lower in space group P65 under identical scaling conditions for all datasets tested (Table 1). The molecular replacement solution was obtained using Phaser (24) with starting coordinates taken from the 2.0Å ovPGHS-1 alpha-methyl-4-biphenylacetic acid structure (1Q4G) (25). One round of rigid body refinement was performed followed by iterative cycles of refinement with TLS (generated by input of the initial model to the TLSMD server (26)) using REFMAC 5.5.0088 or 5.5.0102 (27) and model building using COOT (28). In both space groups, refinement stalled with an Rwork of 20-21% and Rfree of 29-30% despite building the model to completion. Analysis of the data for twinning suggested the presence of merohedral twinning or pseudosymmetry. Following data refinement in space group P65 using the automatically determined twin operators -- (h+k,-k,-l) for the flurbiprofen structure or (h,-h-k,-l) for all other structures -- Rfree immediately dropped between 5-7%. Importantly, the Rfree test set was maintained during refinement and REFMAC accounts for the symmetry related reflections. Further model building resulted in progressive improvement of refinement statistics. After the early stages of refinement loose or medium NCS restraints were applied. Data converged with Rwork values between 17.76-20.56 and Rfree values between 19.20-22.53. The twin fractions for the final refined structures determined by REFMAC ranged from 0.467-0.499 for all the crystals. Structure validation was performed using PROCHECK (29) and Molprobity (30). All figures were generated using MacPyMOL (31).
Determination of ligand occupancies in monomers
In order to estimate the occupancy in the active sites of both monomers, when the model of the dimer was complete we maintained occupancy of the ligand/inhibitor in one monomer at 1.0 and varied the occupancies in the partner monomer (between 0.25–1.0) or set ligands/inhibitors in both monomers to occupancies of 0.5. Following refinement, we examined and compared the resulting electron density maps, Rwork/Rfree values, and B-factors. We repeated this process for ligands/inhibitors in the second monomer. The final occupancies for ligands in both active sites were chosen based on the lowest R factors and B values.
Determination of the solvent pathway to the active site
Tunnels from the active site to the exterior of the protein were calculated using Caver (32). The path to the protein's exterior was determined from a starting point in the active site (center between residues Val349, Ser530, and Tyr385) using a 0.8Å grid resolution.
Results
Refinement of twinned data
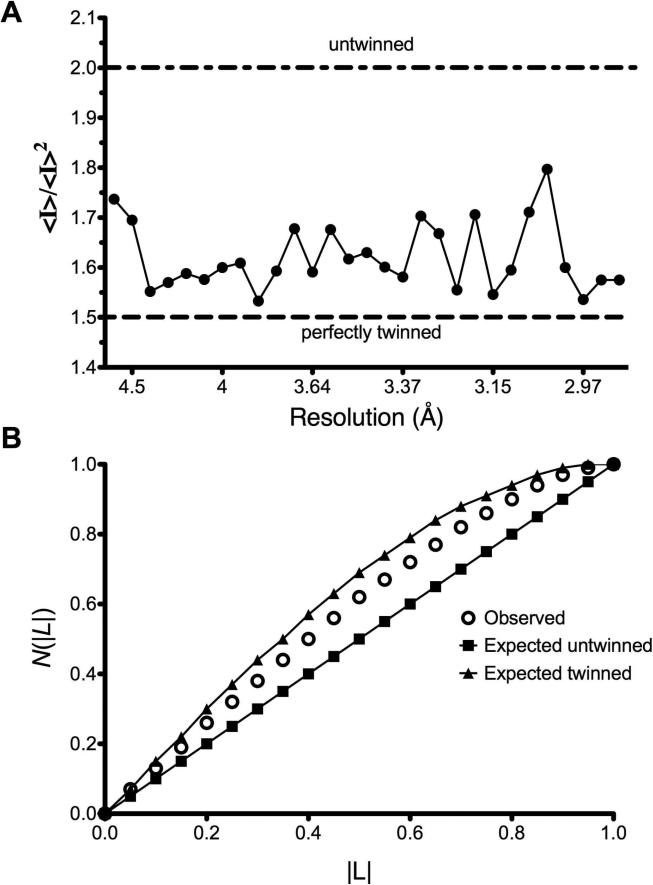
The crystals obtained for this study were generated with a molar ratio of one ligand per dimer of protein, with exception of aspirin (ASA) in the ASA/diclofenac structure and flurbiprofen in the R120Q/native ovPGHS-1 mutant heterodimer structure. All structures were tested for twinning using phenix.xtriage (33) and SCALA (23) software. Even with careful data processing, results from phenix.xtriage suggested that scaling in P6522 was likely to be overmerged pseudosymmetric or twinned. Figure 1 shows a representative output of the twinning tests. The tests for perfect twinning and the cumulative distribution function for |L| (34) on the observed data do not behave as expected for either untwinned or perfectly twinned data. The multivariate Z scores from the phenix,xtriage tests in both space groups were higher than expected; however, twinning was suspected in space group P65 (Table 1). Therefore, all structures were initially refined without twinning in both space groups P6522 and P65. As model building proceeded in either space group, Rfree stalled between 29-30% in all structures while the Rfactor continued to fall. This indicated that the correct solution was not P6522 but could be P65 twinned as suggested by the results from phenix.xtriage. Subsequent refinement using the twin operation in space group P65 improved statistics between 6-8% for all structures and reduced residual B factors between ~20-60 Å2 confirming the presence of twinning. In addition, some improvement of electron density was observed in some structures. Refinement of the data as merohedrally twinned allowed us to unmask the pseudosymmtery present in space group P6522, containing one monomer in the asymmetric unit, and observe differences between partner monomers present in space group P65, which contains the biological dimer in the asymmetric unit.
Figure 1. Analysis of x-ray diffraction data for twinning.
Data was integrated, scaled and merged in space groups P65 and P6522 as described in the Methods section. Representative results are shown from the test for (A) perfect twinning and (B) the cumulative probability distribution, N(|L|), for the parameter L. Where L is the difference between two twin related reflections divided by their sum (34). All data analyzed showed similar results, and during refinement, Rwork and Rfree statistics improved significantly.
Discriminating between monomers of ovPGHS-1 dimers
As described below we observe differences between the structures of the active sites of the two monomers (designated A and B) of the dimer in the asymmetric unit. Regions termed P-loops involving residues 279-284 are located on the surface of PGHS monomers and are involved in crystal contacts between ovPGHS-1 dimers. The structures of the P-loops in monomers A vs. monomers B differ significantly in four of the five structures presented here (Figure 2, Table 3); however, only modest differences exist between the P loops of monomer A vs. B in the unoccupied ovPGHS-1 structure. These results establish that the monomer A subunits and monomer B subunits in the various structures can be discriminated from one another based on their surface interactions and suggest that differences seen in the active sites of monomers A and B are significant.
Figure 2. Crystal structure of residues 279-284 comprising the “P-loop” of acetylated ovPGHS-1 complexed with diclofenac.
Residues 279-284 in monomer A (green) are superimposed onto monomer B (magenta) of the same structure.
Table 3.
Distances (Å) between the Cα atoms in the P-loops of monomers A and B of various ovPGHS-1 crystal structures.
| Residue | Unoccupied | Flurbiprofen (Native/R120Q) | Nimesulide | ASA/Diclofenac | Flurbiprofen (Native) | Celecoxib |
|---|---|---|---|---|---|---|
| 275 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.2 |
| 276 | 0.7 | 0.7 | 1.0 | 0.9 | 1.2 | 0.5 |
| 277 | 0.4 | 1.2 | 0.6 | 1.3 | 1.9 | 1.8 |
| 278 | 0.4 | 1.0 | 0.8 | 1.0 | 1.8 | 1.5 |
| 279 | 0.4 | 1.2 | 1.1 | 1.3 | 1.9 | 2.0 |
| 280 | 0.4 | 2.4 | 1.5 | 3.2 | 3.4 | 2.6 |
| 281 | 0.4 | 1.6 | 2.0 | 2.4 | 2.7 | 3.0 |
| 282 | 0.3 | 1.4 | 1.4 | 1.3 | 2.8 | 1.3 |
| 283 | 0.4 | 1.6 | 0.7 | 1.3 | 2.1 | 0.5 |
| 284 | 0.2 | 0.4 | 0.7 | 0.5 | 0.6 | 0.3 |
Structure of unoccupied ovPGHS-1
During crystallization trials of ovPGHS-1 in the absence of a ligand, we obtained data sets that diffracted to a limit of 3.05 Å. Following molecular replacement and iterative cycles of model building and map inspection, no electron density was visible in the active site of monomer B (Figure 3). (Table 4 provides a summary of the ligands bound and the structural changes observed in the ovPGHS-1 crystal structures reported here.) However, a small amount of density too large to be a water molecule was visible in the active site of monomer A that could be accounted for by the placement of a glycerol molecule coming from the solvent (Figure 3A). A notable difference between monomer B with no bound ligand and monomer A with glycerol bound was a shift in the positions of residues 510-515 from Conformation X, the conformation most commonly observed in the crystal structures (Table 4). In the new conformation designated Conformation Y in Table 4, the peptide backbone is closer to the COX active site in monomer A.. The side chain of His513 in monomer A is disordered while in the same region of monomer B, His90 is disordered (Figure 3B). There was no evidence for alternate conformations at the dimer interface (e.g. in the position of the loops involving residues 123-129).
Figure 3. Crystal structure of unoccupied ovPGHS-1.
(A) A stereo view showing the Fo-Fc omit electron density contoured at 2.8σ (blue mesh) around the active site and 2Fo-Fc electron density contoured at 1.5σ (green mesh) around the indicated residues in monomer B. A glycerol molecule (pink sticks), from the buffer, is modeled into the omit electron density map shown in the active site of monomer B. (B) A stereo view showing the position of residues in the side pocket region (residues 512-516) in monomer B as yellow and blue sticks. The accompanying 2Fo-Fc electron density map is shown contoured at 1.5σ. The presumed location of the disordered His513 side chain, omitted from the final PDB, is shown.
Table 4.
Comparison of ligands bound in the monomers of the biological dimer of ovPGHS-1.
| Monomer A |
Monomer B |
|||||||
|---|---|---|---|---|---|---|---|---|
| ovPGHS-1 Structure | Ligand | Relative Occupancy | Conformation Of Dimer Interface Residues 123-129 | Side Pocket Residues 510-515 | Ligand | Relative Occupancy | Conformation of Dimer Interface Residues 123-129 | Side Pocket Residues 510-515 |
| 1. Unoccupied | Glycerol | 1.0 | upa | Xb | None | 0 | up | Yc |
| 2. Celecoxib (15)f | Celecoxib | 1.0 | up | X | Celecoxib | 0.5 | up/downd | X |
| 3. Flurbiprofen (R120Q/Native)g | Flurbiprofen | 1.0 | up | X | Water or Flubiprofen | 1.0 0.25 |
up/down | X |
| 4. Nimesulideh | Nimesulide | 1.0 | up | X | Nimesulide | 1.0 | up | X |
| 5. Diclofenac/ASA co-complexi | Diclofenac | 1.0 | up | Combination of X and Ze | ASA Diclofenac | 0.8 0.2 |
up | X |
“up” means a single conformation is present at dimer interface (i.e. the “up” conformation only).
“up/down” means two conformations are present at the dimer interface (i.e. an “up” and a “down” conformation).
Z is an alternative conformation slightly different from X and Y.
Activity tested with one mole of celecoxib per mole of ovPGHS-1 dimer resulted in no loss of cyclooxygenase activity (15).
Activity tested with an excess of flurbiprofen resulted in complete inhibition of cyclooxygenase activity.
Activity tested with an excess of nimesulide resulted in no loss in cyclooxygenase activity (15).
Activity tested with acetylated ovPGHS-1 (maximum acetylation of one mole of [1-14C-acetyl] per mole of ovPGHS-1 dimer) resulted in complete inhibition of cyclooxygenase activity (15).
Structures of Flurbiprofen/ovPGHS-1 and a Flurbiprofen/R120Q/Native ovPGHS-1 Heterodimer Complexes
Some inhibitors (e.g. celecoxib in PGHS-1) have a clear preference for binding to one monomer versus both monomers (Table 4). We generated the R120Q/native heterodimer to promote crystallization with single site occupancy. We co-crystallized the ovPGHS-1 heterodimer having an Arg120Gln point mutation in one monomer of the biological dimer in a complex with flurbiprofen, a time-dependent nsNSAID. In order to verify the integrity of the heterodimer we tested the activity of the enzyme using a standard oxygen electrode assay and found the enzyme had ~70% of the activity of the native enzyme using 20 or 100 μM AA as substrate; we also found that flurbiprofen caused a time-dependent inhibition of the COX activity (data not shown). Finally, we found that essentially complete instantaneous enzyme inhibition occurred with 100 μM flurbiprofen when the COX activity was measured with 20 μM AA as substrate. We observed that flurbiprofen was bound to monomer A of the heterodimer while the partner monomer contained density inconsistent with flurbiprofen (Figure 4A and B). We also crystallized native ovPGHS-1 with flurbiprofen using a one per dimer molar ratio of flurbiprofen to protein. Despite limiting the inhibitor concentration, flurbiprofen was observed at full occupancy in both monomers of native ovPGHS-1.
Figure 4. Crystal structures of flurbiprofen/ovPGHS-1 and flurbiprofen/R120Q/Native ovPGHS-1 complexes.
(A) Fo-Fc omit electron density contoured at 2.7σ in the active site of monomer A (native monomer) with flurbiprofen (green sticks) modeled into the crystal structure. Lower portion of panel A shows the Fo-Fc omit electron density (red) and 2Fo-Fc electron density (green) contoured at 3.0σ and 1.5σ, respectively, at position 120 in the active site of monomer A model with arginine; (B) The Fo-Fc omit electron density contoured at 2.7σ in monomer B that has the Arg120Gln substitution at the base of the cyclooxygenase channel. A series of water molecules (not shown) were placed into the electron density of the final model. Lower portion of panel B shows Fo-Fc omit electron density (red) and 2Fo-Fc electron density (green) contoured at 3.0σ and 1.5σ, respectively, modeled with glutamine in place of arginine at residue 120 in the active site of monomer B; (C) Alternate conformation of a loop (residues 123-127) in the dimer interface with 2Fo-Fc electron density contoured at 1.3σ; and (D) a close up of the dimer interface showing alternate conformations of the loop. The coloring is as follows: epidermal growth factor-like domain (magenta), membrane binding domain (green), and remaining catalytic domain (blue). The alternate conformations of residues in the active site are shown in blue and orange sticks, representing the “up” and “down” configurations, respectively. Flurbiprofen is shown as yellow spheres in monomer A.
In both monomers of native ovPGHS-1 with bound flurbiprofen (9) and in monomer A of the R120Q/native heterodimer, the carboxylate of flurbiprofen was within hydrogen bonding distance of the guanidinium group of Arg120. In the heterodimer structure, we could distinguish between the native monomer with flurbiprofen bound and the mutant monomer because placing an arginine at position 120 in monomer B resulted in negative Fo-Fc electron density near the guanidinium group and an increase in the Rfree statistic. This information confirmed that the heterodimer had been crystallized. We tested flurbiprofen in the active site of monomer B and found the Rfree to increase when occupancy by flurbiprofen was raised from 0.25 to 1.00. This indicated that this site was not fully occupied with flurbiprofen. We tested other possible ligands including glycerol and citrate anion and found varying improvements in the Rfree; however, electron density was not accurately modeled by any one ligand. Thus, the COX site of monomer B is incompletely occupied by flurbiprofen and possibly occupied by a combination of flurbiprofen and/or glycerol and/or water. In the final model, the flurbiprofen was not modeled into the electron density (Table 4).
Examination of other parts of the model of the heterodimer suggested the appearance of density consistent with an alternate conformation of residues 123-129 in a loop within the dimer interface of monomer B (Figure 4C and 4D). This density was reminiscent of that observed in a crystal of ovPGHS-1 in complex with celecoxib (15). An alternate conformation was modeled into the density and no negative Fo-Fc electron density appeared in the region. However, 2Fo-Fc density remained weak for some residues in the loop region.
Structure of Nimesulide/ovPGHS-1 complex
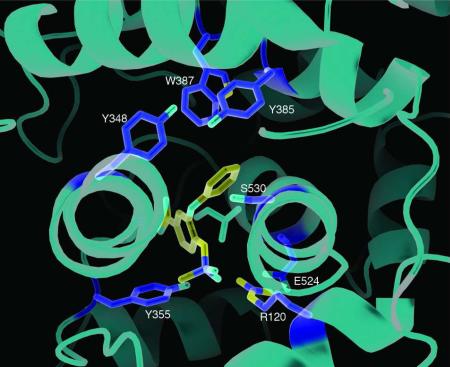
The PGHS-2 selective inhibitor nimesulide was modeled into the electron density within the active site with the methanesulfonamide group next to the mouth of the active site. The larger scattering due to the presence of the sulfur atom was modeled adjacent to Arg120 while the methyl group was placed next to Tyr355. The nitrogen from the nitrophenyl group was placed into the back of the COX active site near the opening of the side pocket, while the cyclohexane ring extended into the active site near Ser530. No alternate conformations could be modeled into the resulting density (Figure 5A). As observed in the celecoxib/ovPGHS-1 crystal structure, Ile523 adopts an extended rotamer conformation to accommodate the bulky nitrosyl group of nimesulide. Figure 5B shows a steric clash with Ile523 (dot representation) in the position of the model used for the molecular replacement solution (yellow sticks).
Figure 5. Crystal structure of nimesulide/ovPGHS-1 complex.
(A) Stereo image showing the Fo-Fc omit electron density of nimesulide in the active site in one of the monomers contoured at 2.7σ. (B) Stereoview of nimesulide bound in the active site of ovPGHS-1 as viewed with the opening from the MBD into the ovPGHS-1 cyclooxygenase active site oriented along the plane of the page. Comparison of nimesulide/ovPGHS-1 complex (green) superimposed onto the starting model (ovPGHS-1 in complex with α-methyl-4-biphenylacetic acid; PDB ID 1Q4G) (superimposed yellow ribbon and yellow side chains) showing Ile523, homologous to Val523 in PGHS-2, in an extended rotamer conformation allowing access to the otherwise inaccessible hydrophobic side pocket. Nimesulide is rendered as sticks and spheres while dots highlight the steric clash of Ile523 with the starting model. Dashed lines represent hydrogen bonds between the sulfonamide moiety of nimesulide and Arg120.
We performed occupancy calculations with nimesulide in one or both monomers and found that the nimesulide electron density was equivalent in both monomers, despite using a one per dimer molar ratio of ligand to protein. Although some density in the loop region between the dimer interface was observed, we were unable to build in an alternate conformation of residues 123-127. Occupancy of both monomers is probably about 100% because the B factors for the inhibitor determined in each COX site were only slightly different.
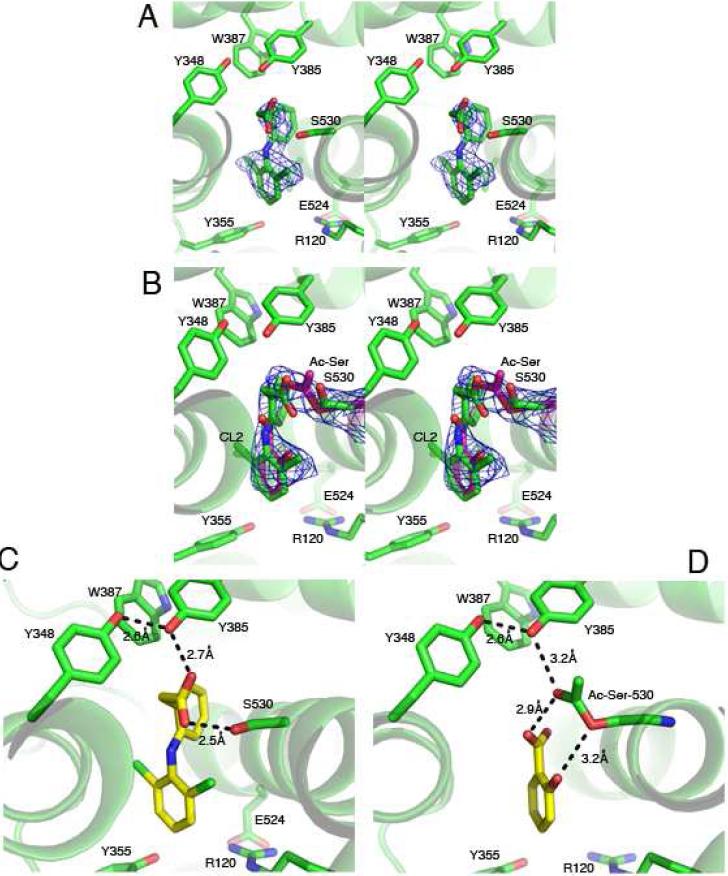
Structure of Diclofenac/Aspirin-acetylated ovPGHS-1 Complex
A complex of aspirin-acetylated ovPGHS-1 with diclofenac, a time-dependent inhibitor, was crystallized to 2.6 Å resolution (Table 4 and Figure 6). These crystals were prepared with diclofenac in the protein drops following an extensive wash after pretreatment of the protein with aspirin. Diclofenac placed in monomer A was unambiguous (Figure 6A). However, modeling of diclofenac in monomer B resulted in significant negative electron density at the position of one chlorine atom (CL2) in diclofenac. Other conformations of diclofenac were also tested but did not fit the electron density. Because salicylic acid, a product of the acetylation reaction, had been observed in the crystal structure of bromoacetylated PGHS-1, we speculated that following acetylation of our enzyme the byproduct of the reaction may have remained. When salicylate was placed in monomer B, no negative electron density was present, but some residual positive Fo-Fc electron density near the two carbons on the benzene ring was present at the apex of the channel (where C11 and C12 of diclofenac had initially been placed). This suggested that both salicylate and diclofenac are present in different molecules of monomer B. A series of refinements were performed with varying occupancies of salicylate and diclofenac in monomer B. After careful inspection of the electron density and comparison of the resulting Rfactor/Rfree values, we concluded that the salicylate:diclofenac ratio was 0.7-0.9. Changing the ratio of salicylate:diclofenac in favor of diclofenac resulted in negative electron density at the position of the CL2 chlorine atom of diclofenac, confirming that diclofenac alone could not occupy monomer B (Figure 6B).
Figure 6. Crystal structure of diclofenac/aspirin-acetylated ovPGHS-1 complex.
(A) Stereo images of the Fo-Fc omit electron densities contoured at 2.7σ in monomer A bound with diclofenac and (B) in the acetylated monomer B bound to salicylate or to diclofenac. (C) Putative hydrogen bonding interactions involving the carboxylate of diclofenac with Tyr385 and Ser530 in monomer A and (D) the carboxylate of salicylate to Tyr385 and acetyl-Ser530 in monomer B.
In contrast to what had been previously observed in the 3.40 Å structure of bromoacetylated aspirin ovPGHS-1 (35), we found that the carboxylate group of the salicylate bound to monomer B was not ion paired with Arg120. The phenolic oxygen and carboxylate group of salicylate and the serine oxygen and the carbonyl oxygen of the acetylated serine, respectively, were within H-bonding distances. The lack of an interaction with Arg120 and the presence of hydrogen bonding with Ser530 is a pattern similar to that of diclofenac in monomer A (Figure 6C and 6D) and was previously reported for diclofenac with PGHS-2 (36). The binding of salicylate must be quite stable because washing the protein following the aspirin treatment did not remove the salicylate.
Comparison of monomer A (diclofenac occupied) with monomer B (salicylate/diclofenac occupied) suggested that there are two alternate conformations of residues 510-516 in monomer A. One is the same as that observed in most of our crystal structures (Conformation X; Table 4), and the other is similar to that observed in the unoccupied PGHS-1 structure (Conformation Z; Fig. 2B; Table 4). Only Conformation X is observed in monomer B. Additionally, as was observed in the unoccupied ovPGHS-1 structure, His513 is disordered in monomer A while His90 is disordered in monomer B.
Comparisons of ovPGHS-1 Structures
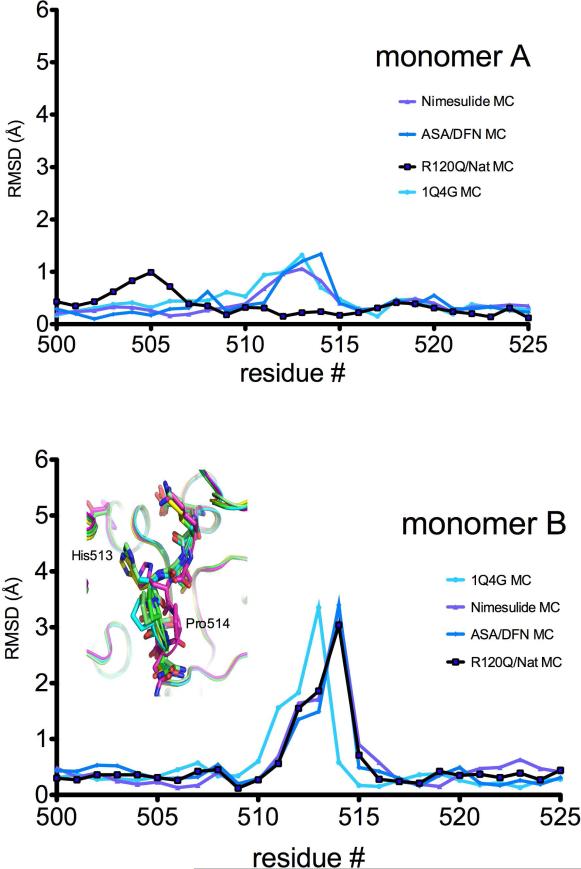
The individual monomers of the ovPGHS dimers were compared and, with the exceptions described above, little deviation was seen between the partner monomers in any of the structures examined. The main chain and side chain atoms for both chains were compared between the various models and between the partner monomers. Comparison between the main chain atoms of residues 510-516 in monomer B of the unoccupied ovPGHS-1 structure showed the most significant deviation (Figure 7 bottom panel). Chain A of the unoccupied structure, which appears to contain a glycerol molecule showed a less significant shift (Figure 7 top panel).
Figure 7. Comparison of the side pocket β-turn loop residues in structures of ligand/ovPGHS-1 complexes.
Comparison of the main chain (MC) atoms in residues 500-525 of various structures. Inset in lower panel shows the position of the main chains (ribbons) in the unoccupied monomer (magenta) superimposed with other structures. Nimesulide, nimesulide/ovPGHS-1 complex; ASA/DFN, diclofenac/aspirin-acetylated ovPGHS-1 complex; R120Q/Nat, flurbiprofen/R120/native ovPGHS-1 complex; 1Q4G, α-methyl-4-biphenylacetic acid/ovPGHS-1.
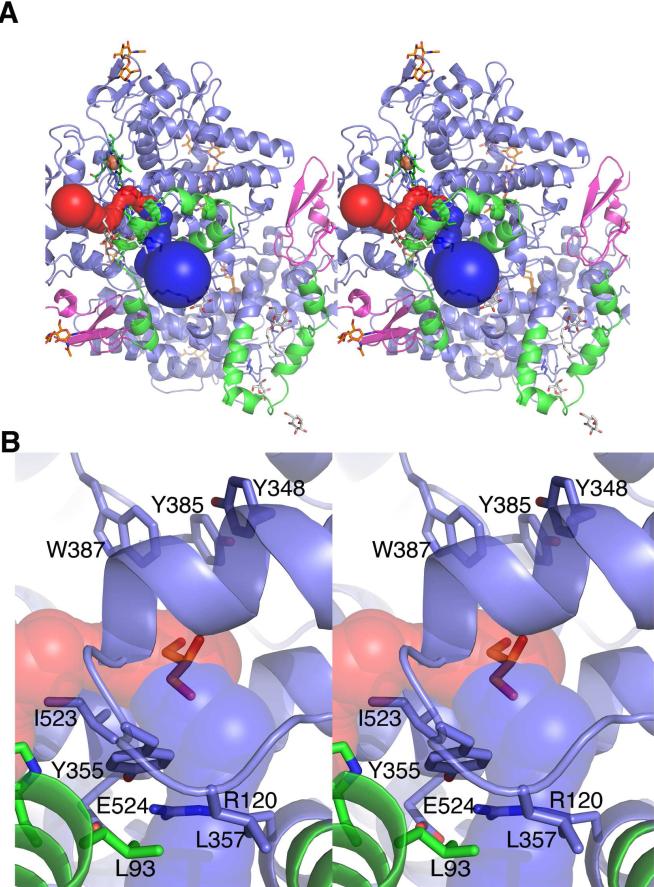
In order to compare the active sites between the various PGHS-1 structures we used the Caver software, which calculates the path to the exterior of a protein and the size of the tunnel. The top three scoring pathways from the mid point of three residues in the active site (Ser530, Tyr385, Val349) leading to the outside were calculated. In all models, there was a primary tunnel with a path through the membrane binding domain (green ribbons), and this is shown as continuous blue spheres (Figure 8). A secondary tunnel in two of the structures (monomer B of the unoccupied and aspirin-diclofenac structures) led to the exterior of the protein along the residues of the COX active site side pocket (shown as continuous red spheres) (Figure 8). In both of the structures that have the secondary tunnel running along the side pocket, there is also an alternate conformation of residues 510-515.
Figure 8. Potential routes for the entry and exit of ligands to and from the cyclooxygenase active site of ovPGHS-1.
All calculations were made using Caver Software as described in the Methods section. Top panel (A) and enlarged view (B) show the exit paths from the active site: the MBD (green), EGF-like domain (magenta), and the catalytic domain (blue). The position of glycerol, superimposed from monomer A, is displayed as red sticks at the center of the active site to provide orientation. The glycosylation sites (amino acids 68, 144 and 410) are indicated by the orange sticks while detergents and detergent head groups are colored grey. The heme in the peroxidase site is indicated by the iron (orange sphere) coordinated by the protoporphyrin ring in green sticks. Only the exit paths from monomer B are shown. The first pathway follows through the MBD indicated by the blue tunnel. Tunnel calculation reveals a second potential exit point from the cyclooxygenase channel (red). The second exit point along the surface of the membrane lies perpendicular to the first entrance through the MBD.
Discussion
PGHSs are homodimers that behave as heterodimers during catalysis and inhibition. Thus, cross-talk between monomers comprising a dimer must occur. The purpose of this study was to delineate differences in the protein brought about by partial or complete binding of the protein by various NSAIDs or coxibs. ovPGHS-1 is particularly amenable to crystallographic analysis, and we examined crystals of four different ligand complexes of native ovPGHS-1 as well as a complex of flurbiprofen with an R120Q/native ovPGHS-1 heterodimer. All structures solved in this study were analyzed for the presence of crystal twinning. Consistently, all data refined with the twin operator included showed dramatically improved refinement statistics. The structures presented here provide models to describe the existence of cross talk in PGHS-1 homodimers; however, due to the presence of crystal twinning and the limitations of the data quality, the possibility of model bias cannot be ruled out and the results needs to be verified experimentally. Recently, Vecchio et al. (37) have crystallized muPGHS-2 in complex with fatty acids in alternate conformations of the homodimer. Consistent with the work presented here only minor changes were observed between the partner monomers.
There were only two differences among the five crystal structures (Table 4). First, shifts in the protein backbone at positions 510-515 were seen in one unoccupied monomer of the ligand free ovPGHS-1 complex and in a monomer fully occupied by diclofenac in the diclofenac/aspirin-treated ovPGHS-1 complex; it should be emphasized that diclofenac is unique in that its binding does not engage Arg120. Second, two different orientations of the loop comprised of residues 123-127 were observed in the incompletely occupied R120Q monomer of the R120Q/native ovPGHS-1 heterodimer. It is notable that two orientations of this loop had previously been observed in another partially occupied monomer of a celebrex/native ovPGHS-1 complex. In contrast, whenever both subunits were fully occupied by the same ligand, the 123-127 loops of both monomers are in the same configuration—referred to here as the “up” conformation (Table 4). The “up” configuration of this loop is designated as one in which the side chains of residues Leu123 and Ile124 face toward the partner monomer and the Ser127 hydroxyl group is slightly distant from Glu541 of the partner monomer. Only this “up” configuration has been observed in PGHS crystal structures reported prior to 2010 when all crystallization was performed in the presence of a large molar excess of ligand.
The 123-127 loop is present at the dimer interface, and in both PGHS-1 and PGHS-2 the 123-127 loop interacts with a loop containing residues 541-543 in the partner monomer. Cross-linking studies of PGHS-2 have shown that the orientations of these loops change relative to one another upon ligand binding. This implies that cross talk between the monomers involves these loops. Our crystallography data suggest that it is only the 123-127 loop that is mobile. Because of the many similarities between the PGHS isoforms, we presume that cross talk in PGHS-1, although sometimes leading to a different effect on activity, involves the same general pathway as in PGHS-2.
Whenever a monomer is fully occupied by a ligand whose binding involves Arg120, the 123-127 loop is only observed in the “up” configuration. Therefore, we presume that when a 123-127 loop is seen to be in the “down” configuration in a partially occupied, adjoining monomer (Table 4) that the “down” orientation is the orientation of the unoccupied monomer. In the “down” conformation the side chains of residues Leu123 and Ile124 face away from the partner monomer while the Ser127 hydroxyl group lies next to Glu541 of the partner monomer. Finally, we speculate that the “down” configuration can only be captured crystallographically, when the partner monomer is fully occupied by ligand.
We cannot directly link the “down” configuration to COX catalytic efficiency. The “down” configuration of the 123-127 loop is seen in two structures --celebrex/native ovPGHS-1 and flurbiprofen/R120Q/native ovPGHS-1—the former is active while the latter is relatively inactive. We are also unable to link the “down” conformation to time-dependent inactivation. Celebrex is reversibly bound in the celebrex/native ovPGHS-1, while flurbiprofen is bound in a manner that only slowly dissociates in the flurbiprofen/R120Q/native ovPGHS-1 complex.
We are assuming in this discussion that the crystal structures closely resemble the solution structures. If so, then changes in enzyme efficiency must result from relatively subtle changes in the relative positions of the phenolic oxygen of the Tyr385 radical and the 13proS hydrogen of substrates. Alternatively, the small structural changes may lead to electronic changes that affect the stability of the Tyr385 radical.
Because we observed a shift in residues of the side pocket (510-515) and the dimer interface (123-127) in some of the structures, we suspected that there could be alternate entry and exit routes to and from the active site. We performed tunnel calculations for our ovPGHS-1 structures using Caver and found a secondary tunnel leading out along the side pocket region. The secondary tunnel was observed in the structures that had alternate conformation of residues 510-515 (configurations Y and Z of Table 4) in the side pocket region. It is not known whether the putative polar channel is a ligand or water channel or this route into the active site exists in membrane-associated PGHS enzyme in vivo. However, it is noteworthy that His90 in the ASA/diclofenac structure and His513 in the unoccupied ovPGHS-1 structure are disordered. Because His90 resides in the MBD and His513 lies adjacent to the MBD, this suggests that residues 510-515 could control access into the active site, however, this needs to be investigated further.
Acknowledgments
Funding Information
These studies were supported by Grants from NIH-NIGMS GM68848 (W.L.S) and a postdoctoral fellowship from the Heart and Stroke Foundation of Canada (R.S.S.).
Abbreviations
- AA
arachidonic acid
- ASA
acetylsalicylic acid or aspirin
- COX-1
cyclooxygenase-1
- COX-2
cyclooxygenase-2
- DFN
diclofenac
- FBP
flurbiprofen
- PG
prostaglandin
- PGHS
prostaglandin endoperoxide H synthase
- POX
peroxidase
- MBD
membrane binding domain
- nsNSAID
non-selective nonsteroidal anti-inflammatory drugs
- hu
human
- ov
ovine
Footnotes
Data deposition Atomic coordinates and structure factors have been deposited to the Protein Data Bank, www.PDB.org (PDB ID codes 3N8V, 3N8W, 3N8X, 3N8Y, and 3N8Z).
References
- 1.Rouzer CA, Marnett LJ. Cyclooxygenases: structural and functional insights. J Lipid Res. 2009;50(Suppl):S29–34. doi: 10.1194/jlr.R800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider C, Pratt DA, Porter NA, Brash AR. Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol. 2007;14:473–488. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith WL. Nutritionally essential fatty acids and biologically indispensable cyclooxygenases. Trends Biochem Sci. 2008;33:27–37. doi: 10.1016/j.tibs.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 5.van der Donk WA, Tsai AL, Kulmacz RJ. The cyclooxygenase reaction mechanism. Biochemistry. 2002;41:15451–15458. doi: 10.1021/bi026938h. [DOI] [PubMed] [Google Scholar]
- 6.Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 7.Grosser T, Fries S, FitzGerald GA. Biological basis for the cardiovascular consequences of COX-2 inhibition: therapeutic challenges and opportunities. J Clin Invest. 2006;116:4–15. doi: 10.1172/JCI27291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laskowski RA, Hutchinson EG, Michie AD, Wallace AC, Jones ML, Thornton JM. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem Sci. 1997;22:488–490. doi: 10.1016/s0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- 9.Picot D, Loll PJ, Garavito RM. The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1. Nature. 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 10.Garavito RM, Mulichak AM. The structure of mammalian cyclooxygenases. Annu Rev Biophys Biomol Struct. 2003;32:183–206. doi: 10.1146/annurev.biophys.32.110601.141906. [DOI] [PubMed] [Google Scholar]
- 11.Malkowski MG, Ginell SL, Smith WL, Garavito RM. The productive conformation of arachidonic acid bound to prostaglandin synthase. Science. 2000;289:1933–1937. doi: 10.1126/science.289.5486.1933. [DOI] [PubMed] [Google Scholar]
- 12.Yuan C, Rieke CJ, Rimon G, Wingerd BA, Smith WL. Partnering between monomers of cyclooxygenase-2 homodimers. Proc Natl Acad Sci U S A. 2006;103:6142–6147. doi: 10.1073/pnas.0601805103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan C, Sidhu RS, Kuklev DV, Kado Y, Wada M, Song I, Smith WL. Cyclooxygenase Allosterism, Fatty Acid-mediated Cross-talk between Monomers of Cyclooxygenase Homodimers. J Biol Chem. 2009;284:10046–10055. doi: 10.1074/jbc.M808634200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prusakiewicz JJ, Duggan KC, Rouzer CA, Marnett LJ. Differential sensitivity and mechanism of inhibition of COX-2 oxygenation of arachidonic acid and 2-arachidonoylglycerol by ibuprofen and mefenamic acid. Biochemistry. 2009;48:7353–7355. doi: 10.1021/bi900999z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rimon G, Sidhu RS, Lauver DA, Lee JY, Sharma NP, Yuan C, Frieler RA, Trievel RC, Lucchesi BR, Smith WL. Coxibs interfere with the action of aspirin by binding tightly to one monomer of cyclooxygenase-1. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0909765106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulmacz RJ, Lands WE. Stoichiometry and kinetics of the interaction of prostaglandin H synthase with anti-inflammatory agents. J Biol Chem. 1985;260:12572–12578. [PubMed] [Google Scholar]
- 17.Yeates TO. Detecting and overcoming crystal twinning. Methods Enzymol. 1997;276:344–358. [PubMed] [Google Scholar]
- 18.Yeates TO, Fam BC. Protein crystals and their evil twins. Structure. 1999;7:R25–29. doi: 10.1016/S0969-2126(99)80016-1. [DOI] [PubMed] [Google Scholar]
- 19.Zwart PH, Grosse-Kunstleve RW, Lebedev AA, Murshudov GN, Adams PD. Surprises and pitfalls arising from (pseudo)symmetry. Acta Crystallogr D Biol Crystallogr. 2008;64:99–107. doi: 10.1107/S090744490705531X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harman CA, Rieke CJ, Garavito RM, Smith WL. Crystal structure of arachidonic acid bound to a mutant of prostaglandin endoperoxide H synthase-1 that forms predominantly 11-hydroperoxyeicosatetraenoic acid. J Biol Chem. 2004;279:42929–42935. doi: 10.1074/jbc.M403013200. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W, Carter Charles W., Jr. Methods in Enzymology. Academic Press; 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 22.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 23.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, G.-K. RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta K, Selinsky BS, Kaub CJ, Katz AK, Loll PJ. The 2.0 A resolution crystal structure of prostaglandin H2 synthase-1: structural insights into an unusual peroxidase. J Mol Biol. 2004;335:503–518. doi: 10.1016/j.jmb.2003.10.073. [DOI] [PubMed] [Google Scholar]
- 26.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallographica Section D-Biological Crystallography. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, A. A. V. a. E. J. D. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 30.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLano WL. MacPyMOL: A PyMOL-based Molecular Graphics Application for MacOS X. DeLano Scientific LLC; Palo Alto: 2007. [Google Scholar]
- 32.Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 34.Padilla JE, Yeates TO. A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr D Biol Crystallogr. 2003;59:1124–1130. doi: 10.1107/s0907444903007947. [DOI] [PubMed] [Google Scholar]
- 35.Loll PJ, Picot D, Garavito RM. The structural basis of aspirin activity inferred from the crystal structure of inactivated prostaglandin H2 synthase. Nat Struct Biol. 1995;2:637–643. doi: 10.1038/nsb0895-637. [DOI] [PubMed] [Google Scholar]
- 36.Rowlinson SW, Kiefer JR, Prusakiewicz JJ, Pawlitz JL, Kozak KR, Kalgutkar AS, Stallings WC, Kurumbail RG, Marnett LJ. A novel mechanism of cyclooxygenase-2 inhibition involving interactions with Ser-530 and Tyr-385. J Biol Chem. 2003;278:45763–45769. doi: 10.1074/jbc.M305481200. [DOI] [PubMed] [Google Scholar]
- 37.Vecchio AJ, Simmons DM, Malkowski MG. Structural basis of Fatty Acid substrate binding to cyclooxygenase-2. J Biol Chem. 285:22152–22163. doi: 10.1074/jbc.M110.119867. [DOI] [PMC free article] [PubMed] [Google Scholar]