Abstract
Selective expression of retinal cone opsin genes is essential for color vision, but the mechanism mediating this process is poorly understood. Both vertebrate rod and medium wavelength-sensitive (M) cone photoreceptors differentiate by repression of a short wavelength-sensitive (S)-cone differentiation program. We show that Pias3 acts in mouse cone photoreceptors to activate expression of M-opsin and repress expression of S-opsin, with the transcription factors Trβ2 and Rxrγ mediating preferential expression of Pias3 in M-cones. Finally, we observe that Pias3 directly regulates M- and S-cone opsin expression by modulating the cone-enriched transcription factors Rxrγ Rorα, and Trβ1. This study reveals that Pias3-dependent SUMOylation of photoreceptor-specific transcription factors is a common mechanism that controls both rod and cone photoreceptor subtype specification, regulating distinct molecular targets in the two cell types.
Introduction
Vertebrate image-forming retinal photoreceptors are comprised of rod and cone subtypes. Rod photoreceptors are activated by dim light, while cone photoreceptors are activated by bright light, and the two photoreceptor subtypes differ considerably in function and gene expression. Most vertebrates possess multiple cone opsin genes, whose maximum spectral absorbance are tuned to different wavelengths and are expressed in different cone photoreceptor cells. Correct expression of cone opsin subtypes is critical for normal color vision, which is essential for many ecologically-important behaviors 1-4. Mice have two cone opsin genes; maximally sensitive to short (S) and middle (M) wavelengths of light, which are expressed in opposing, partially mosaic gradients along the dorsoventral axis of the retina. Three major subtypes of cone photoreceptors are found in the mouse: S-dominant, M-dominant and mixed cones that coexpress both opsin genes 5-8.
The molecular pathways that guide rod and cone cell fate specification differ considerably. Although the homeodomain factors Otx2 and Crx are required for differentiation of both rods and cones 9, 10, the two photoreceptor classes largely use mutually exclusive sets of transcription factors to guide their differentiation. In developing rods, the rod-specific transcription factors Nrl and Nr2e3 bind to both rod and cone-specific promoters and active expression of rod-specific genes while repressing expression of S-cone-specific genes 11-14. Developing M-cone photoreceptors likewise differentiate through a process of repression of S-opsin expression followed by activation of M-opsin expression. Loss of function of the nuclear hormone receptor Trβ2, which is selectively expressed in developing cone photoreceptors and at lower levels in mature cones 15, results both in a loss of M-opsin expression and a corresponding upregulation of S-opsin 16, while loss of function of Rxrγ results in ectopic expression of S-opsin in all cones without affecting M-opsin expression 17. Injection of 3,5,3′-triiodothyronine (T3), the physiological ligand for Trβ2, leads to activation of M-opsin expression and repression of S-opsin expression, and has been proposed to regulate cone opsin specification via regulation of Trβ2 activity 18. Both the ligand binding and DNA binding domains of Trβ2 are required for regulation of cone opsin expression 19, 20. Furthermore, Trβ2 is bound to the promoters of both M- and S-cone opsins in vivo, implying that unidentified regulatory factors must also control Trβ2-dependent regulation of cone opsin gene expression, allowing it to directly repress S-opsin expression and activate M-opsin expression 21.
We previously found that Pias3-dependent SUMOylation of Nr2e3 by Pias3 is essential for mediating repression of S-cone-specific genes in rods 22. In this study, we find that Pias3-dependent SUMOylation plays a similar role in regulating cone photoreceptor subtype specification.
Results
Pias3 is selectively expressed in M-dominant cones
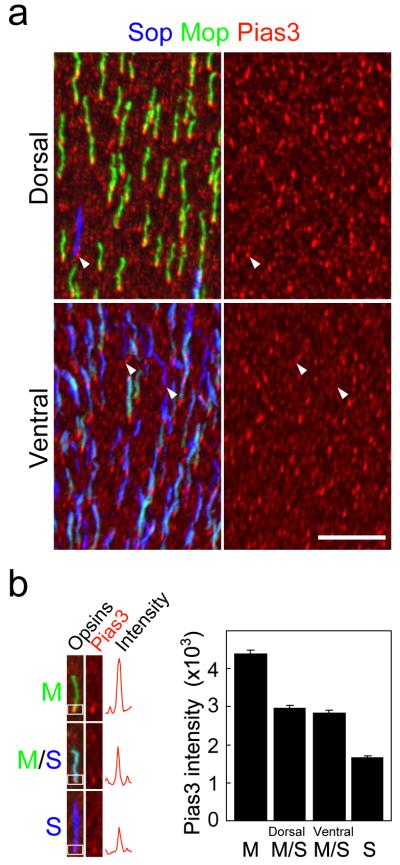
Section immunohistochemical analysis of the adult C57BL/6 mouse retina indicated that Trβ2, which is expressed in the nuclei of cone photoreceptor cells, was colocalized with Pias3 (Supplementary Fig. 1a), as previously observed for developing cones22. Antibodies for M-opsin (Mop) and S-opsin (Sop) showed that Pias3 is localized in both the nucleus and inner segment of cone photoreceptors (Supplementary Fig. 1b). Flat-mount immunohistochemistry showed that Pias3 expression was elevated in M-dominant cone photoreceptors relative to S-dominant cones in dorsal retina (Fig. 1a). In ventral retina, which lacks M-dominant cones but contains many cones that coexpress both M- and S-cone opsin, we also observe higher levels of Pias3 in M-opsin expressing cones (Fig. 1a). Quantifying Pias3 immunoreactivity revealed that Pias3 expression correlates directly with levels of M-opsin expression in cone photoreceptors, and is independent of the dorso-ventral position of these cells (Fig. 1b). Z-stack images of the flat mount images confirmed that higher levels of Pias3 expression was observed in both the nucleus and inner segment of M-opsin expressing cones when compared to S-opsin expressing cones (Supplementary Fig. 1c).
Figure 1.
Pias3 is preferentially expressed in cones expressing middle (M)-wavelength-sensitive cone opsins. (a) Horizontal view of dorsal (upper) and ventral (lower) region of a flat-mount retina from an 8wk C57BL/6 mouse immunostained with S-opsin (Sop, blue), M-opsin (Mop, green) and Pias3 (red) antibodies. Immunohistochemistry (IHC) was performed to visualize Pias3 (red channel), which is visualized separately on right panels. The arrowheads indicate cones expressing S-opsin alone that express lower levels of Pias3 than M-opsin expressing cones. Scale bar: 20 μm. (b) Quantification of Pias3 immunofluorescence by cone photoreceptor subtype. Left panel indicates the intensity from each cone subtype (M-only, M/S-coexpressing and S-only cones). Right panel represents Pias3 immunofluorescence of each cone subtype. The white boxes in the left panels represent the regions scanned in order to calculate Pias3 immunofluorescence intensity. All data are represented as mean ± s.e.m. (n = 100 for each cone subtype).
We further examined Pias3 expression in developing cone photoreceptors (Supplementary Fig. 2). Until postnatal day 4 (P4), when Pias3 is weakly expressed in cones, all cones express S-cone opsins. After P6, the onset of cone expression of Pias3 coincides with repression of S-opsin expression and the activation of M-opsin expression. To determine if enriched expression of Pias3 in long-wavelength cones was evolutionarily conserved, we identified two potential Pias3 homologues in zebrafish (Supplementary Fig. 3a). We found that these genes were coexpressed in an identical subset of developing cone photoreceptors at both 5 days and 4 months post-fertilization (Supplementary Fig. 3b, c; data not shown). Two-color in situ hybridization using cone opsin probes at 4 months post-fertilization, indicated that both Pias3 homologues were expressed in LWS (long wavelength-sensitive) cones and Rh2 (rhodopsin-like) cone subtypes, along with UV-sensitive SWS1 cones, but were not detectable in the blue-sensitive SWS2 (short wavelength-sensitive type 2) cone subtypes (Supplementary Fig. 3c). This pattern of Pias3 expression resembles that of the mouse, in that Pias3 is expressed in cones expressing the LWS opsin – the orthologue of mouse M-cone opsin – and is excluded from the short-wavelength sensitive SWS2 cone opsins. It differs in that Pias3 is expressed in SWS1-postive cones, which is the orthologue of the mouse S-cone opsin.
Pias3-dependent SUMOylation regulates M/S-opsin expression
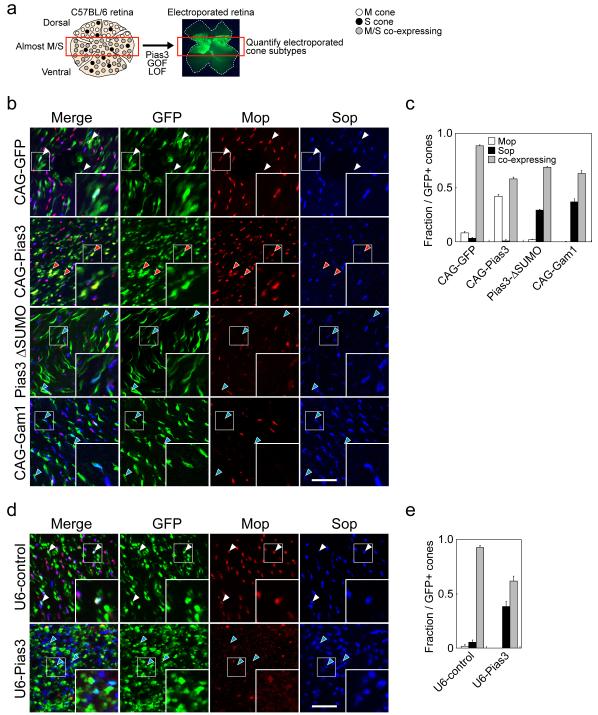
We next used in vivo electroporation to determine whether Pias3-dependent SUMOylation controlled cone opsin subtype expression 22. For these studies, we examined only electroporated cells in the central retina, where the vast majority of cone photoreceptors co-express both M- and S-cone opsins (Fig. 2a). When Pias3 is overexpressed using the ubiquitous CAG promoter, we observed that GFP-positive cones upregulate expression of M-opsin and downregulate S-opsin expression, with many cones in the central retina shifting from coexpressing both M- and S-cone opsins to expressing M-opsin exclusively (Fig. 2b, c). ShRNA-mediated knockdown of Pias3, on the other hand, resulted in an increase in GFP-positive cones that exclusively express S-opsin, while inhibiting M-opsin expression (Fig. 2d, e). Though Pias3 knockdown also induces S-opsin expression in electroporated rods 22, these cells express S-opsin at much lower levels than cones, and can easily be excluded from analysis (data not shown). To further determine whether the E3 SUMO ligase activity of Pias3 was required for these effects, we also overexpressed a SUMOylase-deficient mutant of Pias3 (Pias3ΔSUMO) 23, which was previously shown to inhibit Pias3-depedent SUMOylation of Nr2e3 22, and observed a phenotype much like that seen following shRNA-mediated Pias3 knockdown (Fig. 2b, c). Finally, to determine whether a global inhibition of SUMOylation would phenocopy these effects, we overexpressed the adenoviral protein Gam1, which selectively targets the sole cellular E1 SUMO ligase for proteolytic destruction 24, and observed that this also phenocopied the effects of Pias3 loss of function (Fig. 2b, c).
Figure 2.
Pias3-dependent SUMOylation is necessary and sufficient to direct cone opsin subtype expression. (a) Schematic diagram for the experimental procedure of GOF/LOF analyses of Pias3 function on cone photoreceptor subtype specification. Since cone opsin expression of WT mouse retinas shows opposing, partially mosaic gradients along the dorsoventral axis, we quantified electroporated cones at central region of the retina where most cones coexpress M and S opsins. Flat mount immunohistochemistry was performed to visualize S- and M-opsin expression in electroporated cones. (b) Confocal analysis of flat-mount IHC analysis of overexpression of GFP (CAG-GFP) and full-length Pias3 (CAG-Pias3), SUMOylation-deficient mutants (Pias3 ΔSUMO) and Gam1 protein (CAG-Gam1), which globally inhibits SUMOylation. (c) Composition of electroporated cone subtypes shown in Fig. 2b. (d) Control shRNA (U6-control) and shRNA-mediated knockdown (U6-Pias3). These constructs were electroporated in vivo at P0 and harvested at P14 and visualized by flat mount IHC with antibodies to Mop, Sop and GFP. Red, blue and white arrowheads indicate electroporated cones that are M-dominant, S-dominant and mixed (coexpressing), respectively. The inset in each panel is a high magnification image of electroporated cones (white box). Scale bar: 20 μm. (e) Composition of electroporated cone subtypes shown in Fig. 2c. All data are represented as mean ± s.d. (n = 3).
Trβ2 and Rxrγ directly activate Pias3 expression
Since Trβ2 and Rxrγ are both required for cone subtype specification, but are maximally expressed nearly one week prior to the onset of any differential expression of cone opsins 16, 17, we next investigated whether these transcription factors might act by directly regulating expression of Pias3. We first examined the proximal promoter of Pias3, and found a thyroid hormone response element (TRE) that was evolutionarily conserved among mammals (Fig. 3a). Since thyroid hormone receptors often bind TREs as heterodimers with Rxr family members 25, we next determined whether Trβ2 and Rxrγ could cooperate to transactivate expression of Pias3. Using HEK293 cell-based luciferase reporters, we observed that cotransfection of Trβ2 and Rxrγ led to a dose-dependent activation of transcription from the Pias3 proximal promoter (Fig. 3b).
Figure 3.
Pias3 is a transcriptional target of Trβ2 and Rxrγ. (a) Identification of an evolutionarily conserved thyroid hormone response element (TRE) in the mammalian Pias3 promoter. (b) Trβ2 and Rxrγ cooperate to activate expression of a Pias3 promoter luciferase reporter construct, and enhanced it in the presence of 9-cis retinoic acid (9CRA). Pink and green triangles represent increasing doses treatment of 9CRA (5 μM and 10 μM) and 3,5,3′-triiodothyronine (T3, 1 nM and 5 nM), respectively. ‘+’ and ‘++’ represent 15 ng and 30 ng of transfected plasmid, respectively. All data are represented as mean ± s.d. (n = 3). (c) Trβ2 and Rxrγ selectively co-occupy the Pias3 promoter. Chromatin immunoprecipitation (ChIP) assays were performed using P14 mouse retinas. Results are shown as gel images of PCR products representing promoter sequences of the Pias3 promoter. (d) Quantitative real-time PCR ChIP analysis of the Pias3 promoter for the antibodies used in Fig. 3b. Relative IP versus Input ratio is presented as percent of Input, which is calculated based on a formula: (2−ΔCt) x 100%, where ΔCt=Ct [IP] – Ct [Input]. All data are represented as mean ± s.d. (n = 3). (e) Flat mount IHC of the dorsal region of an 8wk WT and Trβ2−/− mouse retinas labeled with Sop, Mop and Pias3 antibodies. Immunostaining with Pias3 (red channel) is visualized separately on right panels. Scale bar: 20 μm. (f) Quantification of Pias3 immunofluorescence by cone photoreceptor subtype. All data are represented as mean ± s.e.m. (n = 100 cells).
In parallel, we tested the effect of ligands of these nuclear hormone receptors. 9-cis retinoic acid (9CRA), which is a physiological ligand for heterodimeric Rxrγ 26, is enriched in the dorsal neonatal mouse retina 27, while only low levels of T3 are detected. We observed that 9CRA enhanced activation of the Pias3 promoter in the presence of both Trβ2 and Rxrγ Meanwhile, neither 9CRA with Rxrγ alone nor T3 enhanced activation of Pias3 (Fig. 3b), and all-trans retinoic acid (ATRA) could not enhance the activation (data not shown). Finally, we observed that the dose-dependent transcriptional activation induced by 9CRA in cells expressing both Trβ2 and Rxrγ was blocked by the simultaneous addition of T3 (Fig. 3b). These data suggest that T3 might act to antagonize the effects of 9CRA on Pias3 expression. This finding may explain the seemingly paradoxical effects of treatment with high concentrations of T3 during the first postnatal week, which we found leads to a repression of M-opsin expression, rather than the activation of M-opsin expression that is seen at later stages (Onishi, et al. data not shown), and implies that 9CRA can only efficiently upregulate Pias3 expression when T3 levels are low.
Genetic disruption of Trβ2 preserves retinal expression of the Trβ1 splice form of the Trβ gene. Trβ1 is prominently expressed in the postnatal retina 16, and we sought to determine whether it was expressed in a similar cellular pattern to Trβ2 and could functionally substitute for Trβ2 in regulating Pias3 transcription. In situ hybridization analysis with probes specific to the first exons of Trβ1 and Trβ2 indicated that both Trβ1 and Trβ2 mRNAs were enriched in a subset of cells in the outer portion of the outer nuclear layer, where cone photoreceptor cell bodies are located. This pattern of Trβ1 expression is preserved in Trβ2−/− mice (Supplementary Fig. 4a). When Trβ1 and Rxrγ were cotransfected, a modest increase in Pias3 reporter expression was observed (Supplementary Fig. 4b). However, 9CRA produced no significant increase in reporter expression. We therefore conclude that Trβ2, in conjunction with Rxrγ and 9CRA, directly activates transcription of Pias3 in developing retina.
To confirm that Trβ2 and Rxrγ directly regulate Pias3 expression in vivo, we performed chromatin immunoprecipitation (ChIP) analysis of the Pias3 promoter, and found that Trβ2 and Rxrγ were bound to this region in developing retina (Fig. 3c, d). We next investigated whether loss of function of Trβ2 led to loss of differential expression of Pias3 in developing cones. Examining retinas of Trβ2−/− mice, along with the loss of the great majority of M-opsin expression, we observed that all cone photoreceptors expressed a uniform and relatively low level of Pias3 that is characteristic of S-dominant cones (Fig. 3e, f).
Pias3 modulates Trβ2 and T3–regulated cone opsin expression
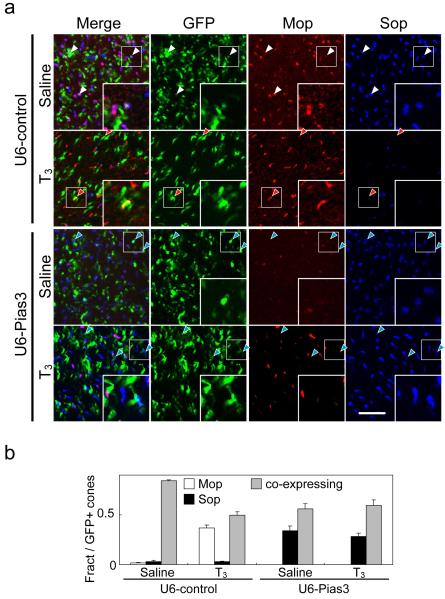
Though thyroid hormone itself did not directly regulate the Pias3 promoter luciferase construct, we investigated whether Pias3 might influence T3-dependent regulation of cone opsin subtype expression. We observed, as have others 18, 20, that injecting T3 into mice in the second postnatal week both enhances M-opsin expression and represses S-opsin expression (Fig. 4a, b). We found that this effect was blocked by shRNA-mediated reduction of Pias3 expression (Fig. 4a, b).
Figure 4.
T3-regulated cone photoreceptor subtype specification is Pias3-dependent. (a) Flat-mount IHC of the P12 mouse retinas electroporated in vivo at P0 with U6-control and U6-Pias3 labeled with antibodies to Mop, Sop, and GFP. The electroporated pups were injected subcutaneously with either saline or T3 from P7 to P12. Red, blue and white arrowheads indicate electroporated cones that are M-dominant, S-dominant and mixed (coexpressing), respectively. Inset in each panel is high magnification image of electroporated cones (white box). Scale bar: 20 μm. (b) Composition of electroporated cone subtypes is shown in Fig. 3E. All data are represented as mean ± s.d. (n = 3).
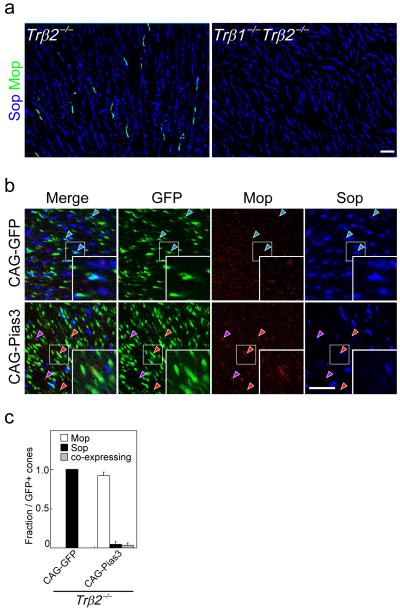
Flat-mount immunohistochemistry indicated that some cone photoreceptors in the dorsal retina still expressed M-opsin in Trβ2−/− mice, although no M-opsin expression was detected in mice lacking both the β1 and β2 splice forms of the receptor (Fig. 5a). Using isoform-specific probes, we determined that both Trβ1 and Trβ2 are expressed in a subset of cells in outer nuclear layer in a pattern consistent with that of cone photoreceptors. We observe a very similar expression pattern of Trβ1 and Trβ2 expression in cones of wildtype mice, and that Trβ1 expression was preserved in Trβ2−/− retina (Supplementary Fig. 4a). We further observed that, unlike Trβ2, Trβ1 does not efficiently cooperate with 9CRA-bound Rxrγ to activate the Pias3 promoter luciferase construct expression (Supplementary Fig. 4b; Fig. 3b), suggesting that Trβ1 might not compensate for the loss of Trβ2 in regulating Pias3 expression. In addition, Trβ1 mRNA is upregulated beginning at P10 16, 17, when M-opsin mRNA expression is first observed. We thus hypothesized that Trβ1 might contribute to regulation of cone opsin expression following the developmental onset of expression of Pias3, and the indispensable function of Trβ2 in regulating cone photoreceptor development might be mediated through transcriptional control of Pias3.
Figure 5.
Pias3-dependent regulation of cone subtype specification can occur independently of Trβ2. (a) Flat mount IHC of the dorsal region of 8wk Trβ2−/− and Trβ1−/− Trβ2−/− mouse retinas labeled with antibodies to Sop and Mop. (b) Flat-mount immunohistochemistry of the P14 Trβ2−/− mouse retinas electroporated in vivo at P0 with CAG-GFP and CAG-Pias3 labeled with antibodies to Mop, Sop, and GFP antibodies. Blue arrowheads indicate electroporated S-dominant cones. Red arrowheads indicate electroporated cones that are M-dominant, while purple arrowheads indicate M-dominant cones that do not express detectable levels of GFP. Inset in each panel is a high magnification image of electroporated cones (white box). Purple arrowheads represent non-electroporated cells expressing M-opsin. (c) Composition of electroporated cone subtypes shown in Fig. 5b. All data are represented as mean ± s.d. (n = 3). Scale bar: 20 μm.
To determine whether Pias3 overexpression could regulate cone opsin expression in the absence of Trβ2, we used in vivo electroporation to overexpress Pias3 in neonatal Trβ2−/− retina. Overexpression of Pias3 robustly induced M-opsin expression and repressed S-opsin expression in Trβ2−/− retina at P14 (Fig. 5b, c). Interestingly, Trβ2−/− retinas electroporated with CAG-Pias3 also occasionally showed expression of M-opsin in cells that were not GFP positive (Fig. 5c). The mechanism underlying this effect is unclear.
Based on these findings, we hypothesized that Pias3 might directly regulate cone opsin expression by binding and SUMOylating nuclear hormone receptors such as Trβ1 that are bound at these promoters. Using ChIP analysis, we next determined that Trβ1 and Trβ2, along with Rxrγ, were bound to the promoters of many photoreceptor-specific genes, including both M- and S-cone opsin, but were generally absent from the promoters of genes selectively expressed either in non-photoreceptor cells of the retina or in non-retinal tissues (Supplementary Fig. 5).
Pias3 directly regulates cone opsin expression
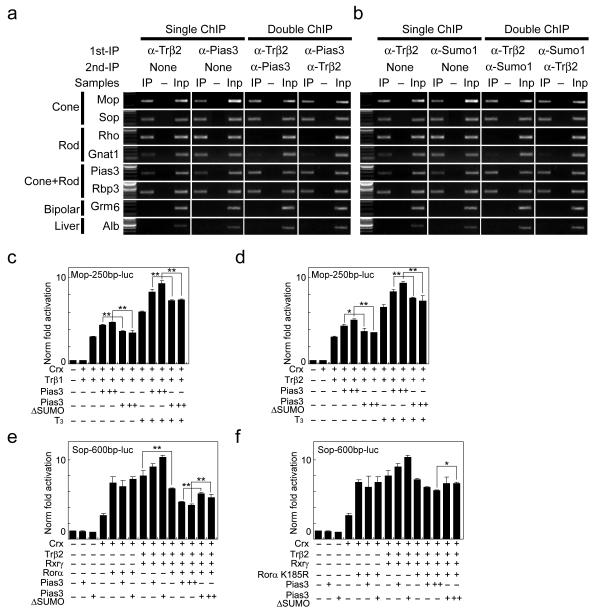
Since Trβ2 is cone photoreceptor-specific in retina 15, 16, we next performed sequential ChIP to determine whether Pias3 is present at cone opsin promoters in retinal cones, and whether these promoters are bound by hyperSUMOylated proteins, as has been previously reported for photoreceptor-specific promoters in retinal rods 22. We found that Pias3 was present at both the S- and M-cone opsin promoters in vivo, along with Trβ2 (Fig. 6a and Supplementary Fig. 5b), and that proteins bound to these promoter sequences were hyperSUMOylated (Fig. 6b and Supplementary Fig. 5c), suggesting that Pias3 may directly regulate cone opsin gene expression by SUMOylation of target proteins at these promoters. Furthermore, we also observed that the promoter of the rod-specific genes rhodopsin and Gnat1, when bound by Trβ2 in cones, were neither bound by Pias3 nor by Sumo1-conjugated proteins, unlike the promoters of M- and S-cone opsins (Fig. 6a, b). Likewise, the rhodopsin and Gnat1 promoter sequences were not bound by Pias3 or by Sumo1-conjugated proteins in Nrl−/− mice, which lack rod photoreceptors altogether (Supplementary Fig. 6).
Figure 6.
Molecular mechanism of action of Pias3 in developing cones. (a) Quantitative sequential ChIP analysis for Trβ2 and Pias3. (b) Quantitative sequential ChIP analysis for Trβ2 and Sumo1. (c and d) Pias3-dependent SUMOylation activates expression of M-cone opsin luciferase reporter constructs in the presence of either Crx or Trβ1 (c) or Trβ2 (d). SUMOylation-deficient mutants (Pias3 ΔSUMO) could not enhance reporter expression. (e and f) Rorα activates expression of S-cone opsin luciferase reporter constructs in the presence of Crx. Rorα represses expression of S-cone opsin luciferase reporter constructs in the presence of Crx, Trβ2 and Rxrγ, and Pias3-dependent SUMOylation of Rorα enhances this repression (e). SUMOylation site mutants of Rorα (K185R) showed less efficient repression (f). ‘+’ and ‘++’ in Fig. 6c-f represent 10 ng and 20 ng, respectively. All data in Fig. 6c-f are represented as mean ± s.d. (n = 3). * p < 0.05 (Student’s t-test).
In addition to Trβ1, Trβ2 and Rxrγ, another strong candidate substrate for Pias3-dependent SUMOylation in cone photoreceptors is the orphan nuclear hormone receptor Rorα. This protein, observed in cones after P3, is bound to the promoters of cone-specific genes in mouse retina and can activate expression of S-cone opsin promoter luciferase constructs 28. Furthermore, Rorα is a known substrate for Pias3-dependent SUMOylation 29. Using cell-based luciferase analysis, we tested whether Pias3-dependent SUMOylation could directly account for activation of M-opsin transcription and repression of S-opsin transcription in the presence of these factors.
We first confirmed that both Trβ1 and Trβ2 activated expression of an M-cone opsin promoter luciferase construct in the presence of Crx. Pias3 enhanced Trβ-dependent transcription in a dose-dependent manner. Trβ1-dependent transcription was further enhanced by the addition of T3, and this effect was likewise enhanced by Pias3. SUMOylation-deficient mutants of Pias3, however, failed to show any dose-dependent increase in reporter expression, either in the absence or presence of T3 (Fig. 6c, d). We conclude that Pias3-dependent SUMOylation activates M-opsin expression by enhancing T3-dependent transcriptional activation mediated by either Trβ1 or Trβ2
We further observed that Crx, Trβ2, and Rxrγ cooperate to activate S-opsin expression (Fig. 6e). Rorα in combination with Crx also activated S-opsin transcription, as previously reported 28. However, when Rorα was tested in combination with Crx, Trβ2, and Rxrγ this resulted in a significant inhibition of S-opsin promoter construct expression relative to that observed these factors when tested without Rorα. This inhibition was enhanced by Pias3-dependent SUMOylation, as E3 SUMO ligase-deficient mutants of Pias3 showed a significantly reduced ability to repress S-opsin promoter construct expression relative to wildtype Pias3. To determine if SUMOylation of Rorα by Pias3 was directly responsible for the observed repression of S-cone opsin, we tested whether repression was deficient in K185R SUMOylation site mutants of Rorα 29. This mutant showed substantially reduced inhibition of S-cone opsin reporter expression, and adding Pias3 did not result in any enhanced transcriptional repression in this mutant (Fig. 6f). Taken together, these findings suggest that Pias3-dependent SUMOylation of Rorα mediates repression of S-cone opsin expression.
We also found that Rorα expression was upregulated in the cone photoreceptors of Trβ2−/− mice (Supplementary Fig. 7). The observation suggests that upregulated Rorα in Trβ2−/− cones, which express lower levels of Pias3, enhance S-opsin expression in combination with Crx. Furthermore, overexpression of Pias3 in Trβ2−/− cones may thus result in higher levels of SUMOylated Rorα, accounting for the enhanced Pias3-dependent repression of S-opsin observed in this mutant background (Fig. 5b, c).
Discussion
These findings have a number of implications. First, they identify Pias3-dependent SUMOylation as critical for both initial patterning of cone photoreceptor subtypes in mouse retina and in maintenance of cone opsin expression. This may resolve a number of mechanistic questions regarding cone opsin patterning in mice. Though both Trβ2 and Rxrγ are necessary for regulating cone opsin patterning, maximal expression of these genes in developing cones occurs at just prior to birth, days before differential expression of S- and M-opsin is observed 30. Furthermore, both genes are uniformly expressed in all cone subtypes 15-18. Though a dorsoventral gradient of T3 has been detected in P10 retina, no such gradient is found in neonatal retina 18. The reason for this early peak in expression of Trβ2 and Rxrγ has been unclear, along with how activity of these factors is regulated in early postnatal retina.
Our data suggests that the initial expression of Pias3 in dorsal, M-dominant cone photoreceptors is directly mediated by Trβ2 and 9CRA-bound Rxrγ. 9CRA can be synthesized both directly from 9-cis retinal and by spontaneous non-enzymatic isomerization of all-trans retinoic acid. In the developing mouse retina, two retinaldehyde dehydrogenases, Aldh1a1 and Aldh1a3 are expressed in the dorsal and ventral region, respectively 31, 32. Aldh1a1 synthesizes 9CRA more effectively than Aldh1a3 33, 34. Furthermore, all-trans RA is enriched in dorsal neonatal mouse retina 27. Our data suggests that this difference in 9CRA distribution leads to higher levels of Pias3 expression in cone photoreceptors in the dorsal retina. Later in the first postnatal week, Pias3-dependent SUMOylation modulates the activity of Rorα which, in conjunction with unliganded Trβ1 and/or Trβ2 and Rxrγ,downregulates expression of S-cone opsin. Simultaneously, Pias3 enhances T3 and Trβ-dependent activation of M-cone opsin expression, with the higher levels of M-opsin expression in dorsal retina reflecting the dorsoventral gradient of T3 that is observed after the first postnatal week (Supplementary Fig. 8). Pias3 thus acts as a dual-function transcriptional coregulator to simultaneously coordinate expression of both M- and S-cone opsin genes.
Second, these data reveal an unexpected common molecular pathway linking rod and cone photoreceptor specification. Genetic data has indicated that, prior to activating expression of rod and M-cone specific genes; both developing photoreceptor subtypes must first repress expression of S-cone specific genes. However, the transcription factors that mediate this process in rods – Nrl and Nr2e3 – are not expressed in cones, while Trβ2 and Rxrγ are not expressed in rods. Thus, it has been unclear whether there is any common molecular pathway mediating this dual-specificity transcriptional regulatory activity. Our study reveals that Pias3 fulfills this function in both developing rods and cones, but performs this dual regulatory role by acting on different sets of nuclear hormone receptors in the two cell types. Furthermore, unlike in retinal rods, where the promoters of both rod and cone-specific genes are bound by both by Pias3 and by hyperSUMOylated proteins 22, in cone photoreceptors only cone-specific promoters show these modifications. This may reflect the fact that while rod photoreceptors must actively repress expression of S-cone specific genes in mice, in mammalian cone photoreceptors there is no evidence that rod-specific gene expression is actively repressed.
Third, this conserved function of Pias3 in photoreceptor subtype specification implies that the dual-specificity regulation of nuclear hormone function by Pias3-dependent SUMOylation has been employed repeatedly over the course of vertebrate evolution as a mechanism to direct development of photoreceptor subtypes away from a default short-wavelength cone fate. Phylogenetic analysis indicates that LWS opsins were the first visual opsins to diverge from non-visual ciliary opsins, with SWS opsins diverging from the LWS opsins soon thereafter 35-37. Thus, from very early stages of vertebrate retinal evolution, developing retinal photoreceptors would have needed to make a binary choice between expressing a long and a short-wavelength cone opsin 38,39. We hypothesize that the decision to keep SWS expression as a default developmental state was fixed at this stage. In mice, which possess only the SWS1 and LWS cone opsins and rhodopsin, SWS1 expression is a default developmental state for both rods and LWS-positive cones. Since the process of Pias3-dependent regulation of photoreceptor subtype specification is conserved between rods and cones, while the targets of Pias3 are not, our findings suggest that the differential Pias3-dependent SUMOylation of nuclear hormone receptors in an ancestral cone-like photoreceptor may thus have been a triggering event in the initial generation of photoreceptor subtype diversity in the vertebrate retina.
Analysis of the role of Pias3 homologues in the specification of photoreceptor subtype in tetrachromatic vertebrates, such as zebrafish, may allow analysis of whether Pias3 acts more generally to promote differentiation of long-wavelength cones at the expense of short-wavelength cones. In zebrafish, genetic data suggests that SWS1 is not a default developmental fate for retinal photoreceptors 40, and Pias3 homologues are expressed in all cone subtypes except for SWS2 (Supplementary Fig. 2b), suggesting that this short-wavelength cone subtype may instead serve as a default developmental fate in this species. Further investigation will be required to address this possibility.
Finally, it is now clear that neuronal subtype specification is often regulated by transcription factors that have opposing effects on expression of cell subtype-specific genes. Many other examples are known from both vertebrates and invertebrate development 41. Our finding that SUMOylation-dependent regulation of transcription factor activity coordinates the development of multiple different photoreceptor subtypes suggests that such a mechanism may be used more generally in the regulation of neuronal cell fate specification.
Supplementary Material
Acknowledgments
We thank D. Forrest for providing antibodies to Trβ2 and for supplying Trβ2−/− and Trβ1−/− Trβ2−/− mice. We also thank J. Nathans, T. Shimogori, W. Yap and members of the Blackshaw lab for their comments on the manuscript. This work was supported by NIH R01EY017015 to S.B and NIH RO1EY012543 to S.C. S.B. is a W. M. Keck Distinguished Young Investigator in Medical Science.
Appendix
Materials and Methods
Animals
Timed pregnant CD1 and C57BL/6 mice were purchased from Charles River Breeding Laboratories and Jackson Laboratory, respectively. Trβ2−/− and Trβ1−/−;Trβ2−/− mice were provided by D. Forrest at National Institute of Health. All experimental procedures were pre-approved by JHMI animal care and the Institutional Animal Care and Use Committee of Washington University School of Medicine, and conformed to the guidelines of the Association for Research in Vision and Ophthalmology for the use of live animals in vision research.
DNA Constructs
Most of cDNA constructs used in this study are shown in our recent report 22. The cDNA of mouse Trβ2 and Sop-600 luciferase vectors were provided by D. Forrest. A 600 bp fragment of mouse Pias3 promoter region (chr3:96,500,554-96,501,097) was generated by PCR, and cloned into pGL3 luciferase vector (Promega). cDNAs of human Rxrγ, Trβ1, and Rorα were shuttled from the entry clones of the Ultimate Human ORF Collection (Invitrogen) into pcDNA3.1/nV5-DEST (Invitrogen). The K185R mutant of Rorα was made by QuikChange Site-Directed Mutagenesis Kit (Stratagene). All DNA constructs used in this study are listed in Supplementary Table 1.
In vivo Electroporation
In vivo electroporation were performed at P0 as previously described 42. ~0.3 μl of DNA solution (5 μg/μl) was injected into the subretinal space of P0 mouse pups, and square electric pulses (100 V; five 50-ms pulses with 950-ms intervals) were applied with tweezer-type electrodes (BTX, model 522). CAG-GFP vector was coelectroporated (1 μg/μl) with both shRNA constructs (U6-control and U6-Pias3) to allow for visualization of transfected cells. The electroporated retinas were harvested at P14 for immunohistochemistry. For 3,5,3′-triiodothyronine (T3) treatment experiments, electroporated pups were injected subcutaneously with 1.5 μg of T3 (Sigma) or saline every 24 hrs from P7 to P12. When quantitative data was obtained, a minimum of 100 electroporated cone photoreceptors from each of a minimum of three different retinas were scored.
Immunohistochemistry
Whole retinas were fixed with 4 % paraformaldehyde in PB for ~60 min at room temperature. After 1 hr blocking with 10 % horse serum in PBST, the eyecups were immunostained for two days with anti-Pias3 (P0117, Sigma), M-opsin (JH492, a gift from J. Nathans) and S-opsin (sc-14363, Santa Cruz) antibodies in 1:2000, 1:5000 and 1:500 dilution, respectively. After secondary antibody reaction, the retinas were flattened on slide glasses. Images were taken on an Apotome confocal-style three-dimensional imaging microscope (Zeiss). The intensities of Pias3 signals in cone photoreceptor nuclei were measured by profile mode on AxioVision (Zeiss) software. Section immunohistochemistry was performed as previously described 22. Goat polyclonal anti-RORα antibody (sc-6062, Santa Cruz) was used at 1:1000 dilution. When quantitative data was obtained, a minimum of 100 cone photoreceptors from each of a minimum of three different retinas were scored.
Luciferase assays
Luciferase analysis was performed essentially as previously described 22. 24 hrs after transfection, 5 μM or 10 μM of 9-cis retinoic acid (9CRA, Sigma) and/or 1 nM or 5 nM of T3 were treated. The transfected cells were harvested 48 hrs after transfection, and the luciferase activity was measured. Since 293T cells express low levels of endogenous Pias3 (data not shown), 10ng of an shRNA construct selectively targeting human but not mouse Pias3 was included in each transfection to eliminate confounding effects of endogenous Pias3.
In situ hybridization
Single color in situ hybridization was performed as previously described 43. Two-color in situ hybridization was performed as described by Barroso-Chinea et al. with minor modifications 44. The following regions were used to generate probes for probes for in situ hybridization in zebrafish retinal sections: Pias3-1 chr18:14,151,257-14,151,760; Pias3-2 chr7:29,557,068-29,557,473 (July 2007 assembly, UCSC Genome Browser); LWS, NM_131175: 111-1184; SWS1, NM_131319: 62-1072; SWS2, NM_131192: 128-1192; and Rh2, NM_131253: 68-1117. The divergent 5′regions of Trβ1 (NM_001113417.1: 0 bp-469 bp) and Trβ2 (NM_009380.3: 0 bp-554 bp) were used to generate probes for detection of mouse Trβ1 and Trβ2 transcripts.
Chromatin immunoprecipitation
Chromatin immunoprecipitation has performed as described 45. The results of ChIP assays were analyzed using candidate gene-based PCR with primers spanning the promoter region of each gene (listed in Peng et al., 2005). Results shown are representative of at least three separate experiments. Controls include the use of normal rabbit/mouse IgG (Santa Cruz) in immunoprecipitation reactions (negative controls) and input (without IP) as positive controls in PCR reactions. Double chromatin immunoprecipitation assays (double ChIP) was performed as previously described 22, 46, with three new antibodies – to Trβ1, Trβ2 and Rxrγ – included in the assay. The sources of anti-Pias3, anti-Sumo1 and mouse IgG have been previously described 22. Other antibodies used included anti-Trβ1 (Millipore, 06-539), anti-Trβ2 (gift of D. Forrest), and anti-Rxrγ (Abgent, AT3749a). Primers used for PCR amplification of genomic DNA for Rho, Mop, Sop, Grm6, Thy1, and Alb have been previously described 22. Other primer sets used are listed on Supplementary Table 2.
Statistical analysis
The values in this study are represented as mean ± s.d. or s.e.m. Statistical comparisons were done using a two-tailed Student’s t-test, and p < 0.05 was considered statistically significant.
References
- 1.Solomon SG, Lennie P. The machinery of colour vision. Nat Rev Neurosci. 2007;8:276–286. doi: 10.1038/nrn2094. [DOI] [PubMed] [Google Scholar]
- 2.Osorio D, Vorobyev M. A review of the evolution of animal colour vision and visual communication signals. Vision Res. 2008;48:2042–2051. doi: 10.1016/j.visres.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 3.Collin SP, Trezise AE. The origins of colour vision in vertebrates. Clin Exp Optom. 2004;87:217–223. doi: 10.1111/j.1444-0938.2004.tb05051.x. [DOI] [PubMed] [Google Scholar]
- 4.Pichaud F, Briscoe A, Desplan C. Evolution of color vision. Curr Opin Neurobiol. 1999;9:622–627. doi: 10.1016/S0959-4388(99)00014-8. [DOI] [PubMed] [Google Scholar]
- 5.Peichl L. Diversity of mammalian photoreceptor properties: adaptations to habitat and lifestyle? Anat Rec A Discov Mol Cell Evol Biol. 2005;287:1001–1012. doi: 10.1002/ar.a.20262. [DOI] [PubMed] [Google Scholar]
- 6.Rohlich P, van Veen T, Szel A. Two different visual pigments in one retinal cone cell. Neuron. 1994;13:1159–1166. doi: 10.1016/0896-6273(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 7.Lukats A, Szabo A, Rohlich P, Vigh B, Szel A. Photopigment coexpression in mammals: comparative and developmental aspects. Histol Histopathol. 2005;20:551–574. doi: 10.14670/HH-20.551. [DOI] [PubMed] [Google Scholar]
- 8.Applebury ML, et al. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–523. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 9.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 10.Nishida A, et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 11.Akhmedov NB, et al. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci U S A. 2000;97:5551–5556. doi: 10.1073/pnas.97.10.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haider NB, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 13.Mears AJ, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 14.Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 15.Ng L, Ma M, Curran T, Forrest D. Developmental expression of thyroid hormone receptor beta2 protein in cone photoreceptors in the mouse. Neuroreport. 2009 doi: 10.1097/WNR.0b013e32832a2c63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng L, et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 17.Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci. 2005;46:2897–2904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- 18.Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G, Reh TA. Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc Natl Acad Sci U S A. 2006;103:6218–6223. doi: 10.1073/pnas.0509981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Applebury ML, et al. Transient expression of thyroid hormone nuclear receptor TRbeta2 sets S opsin patterning during cone photoreceptor genesis. Dev Dyn. 2007;236:1203–1212. doi: 10.1002/dvdy.21155. [DOI] [PubMed] [Google Scholar]
- 20.Pessoa CN, et al. Thyroid hormone action is required for normal cone opsin expression during mouse retinal development. Invest Ophthalmol Vis Sci. 2008;49:2039–2045. doi: 10.1167/iovs.07-0908. [DOI] [PubMed] [Google Scholar]
- 21.Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onishi A, et al. Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron. 2009;61:234–246. doi: 10.1016/j.neuron.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long J, Wang G, Matsuura I, He D, Liu F. Activation of Smad transcriptional activity by protein inhibitor of activated STAT3 (PIAS3) Proc Natl Acad Sci U S A. 2004;101:99–104. doi: 10.1073/pnas.0307598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Kliewer SA, Umesono K, Mangelsdorf DJ, Evans RM. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature. 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, Li T, Wang F, Tian H, Samuels HH. Functional evidence for retinoid X receptor (RXR) as a nonsilent partner in the thyroid hormone receptor/RXR heterodimer. Mol Cell Biol. 2002;22:5782–5792. doi: 10.1128/MCB.22.16.5782-5792.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaffrery P, Posch KC, Napoli JL, Gudas L, Drager UC. Changing patterns of the retinoic acid system in the developing retina. Dev Biol. 1993;158:390–399. doi: 10.1006/dbio.1993.1197. [DOI] [PubMed] [Google Scholar]
- 28.Fujieda H, Bremner R, Mears AJ, Sasaki H. Retinoic acid receptor-related orphan receptor alpha regulates a subset of cone genes during mouse retinal development. J Neurochem. 2009;108:91–101. doi: 10.1111/j.1471-4159.2008.05739.x. [DOI] [PubMed] [Google Scholar]
- 29.Hwang EJ, et al. SUMOylation of RORalpha potentiates transcriptional activation function. Biochem Biophys Res Commun. 2009;378:513–517. doi: 10.1016/j.bbrc.2008.11.072. [DOI] [PubMed] [Google Scholar]
- 30.Szel A, Rohlich P, Mieziewska K, Aguirre G, van Veen T. Spatial and temporal differences between the expression of short- and middle-wave sensitive cone pigments in the mouse retina: a developmental study. J Comp Neurol. 1993;331:564–577. doi: 10.1002/cne.903310411. [DOI] [PubMed] [Google Scholar]
- 31.McCaffery P, Wagner E, O’Neil J, Petkovich M, Drager UC. Dorsal and ventral retinal territories defined by retinoic acid synthesis, break-down and nuclear receptor expression. Mech Dev. 1999;82:119–130. doi: 10.1016/s0925-4773(99)00022-2. [DOI] [PubMed] [Google Scholar]
- 32.Li H, et al. A retinoic acid synthesizing enzyme in ventral retina and telencephalon of the embryonic mouse. Mech Dev. 2000;95:283–289. doi: 10.1016/s0925-4773(00)00352-x. [DOI] [PubMed] [Google Scholar]
- 33.el Akawi Z, Napoli JL. Rat liver cytosolic retinal dehydrogenase: comparison of 13-cis-, 9-cis-, and all-trans-retinal as substrates and effects of cellular retinoid-binding proteins and retinoic acid on activity. Biochemistry. 1994;33:1938–1943. doi: 10.1021/bi00173a042. [DOI] [PubMed] [Google Scholar]
- 34.Lin M, Zhang M, Abraham M, Smith SM, Napoli JL. Mouse retinal dehydrogenase 4 (RALDH4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J Biol Chem. 2003;278:9856–9861. doi: 10.1074/jbc.M211417200. [DOI] [PubMed] [Google Scholar]
- 35.Lamb TD. Evolution of vertebrate retinal photoreception. Philos Trans R Soc Lond B Biol Sci. 2009;364:2911–2924. doi: 10.1098/rstb.2009.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shichida Y, Matsuyama T. Evolution of opsins and phototransduction. Philos Trans R Soc Lond B Biol Sci. 2009;364:2881–2895. doi: 10.1098/rstb.2009.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terakita A. The opsins. Genome Biol. 2005;6:213. doi: 10.1186/gb-2005-6-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowmaker JK. Evolution of vertebrate visual pigments. Vision Res. 2008;48:2022–2041. doi: 10.1016/j.visres.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 39.Lamb TD, Collin SP, Pugh EN., Jr. Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat Rev Neurosci. 2007;8:960–976. doi: 10.1038/nrn2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alvarez-Delfin K, et al. Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proc Natl Acad Sci U S A. 2009;106:2023–2028. doi: 10.1073/pnas.0809439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- 42.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blackshaw S, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barroso-Chinea P, et al. Detection of two different mRNAs in a single section by dual in situ hybridization: a comparison between colorimetric and fluorescent detection. J Neurosci Methods. 2007;162:119–128. doi: 10.1016/j.jneumeth.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 45.Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- 46.Geisberg JV, Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.