Abstract
The ubiquitous nature of protein phosphorylation makes it challenging to map kinase-substrate relationships, which is a necessary step toward defining signaling network architecture. To trace the activity of individual kinases, we developed a semisynthetic reaction scheme, which results in the affinity tagging of substrates of the kinase in question. First, a kinase, engineered to use a bio-orthogonal ATPγS analog, catalyzes thiophosphorylation of its direct substrates. Second, alkylation of thiophosphorylated serine, threonine or tyrosine residues creates an epitope for thiophosphate ester–specific antibodies. We demonstrated the generality of semisynthetic epitope construction with 13 diverse kinases: JNK1, p38α MAPK, Erk1, Erk2, Akt1, PKCδ, PKCε, Cdk1/cyclinB, CK1, Cdc5, GSK3β, Src and Abl. Application of this approach, in cells isolated from a mouse that expressed endogenous levels of an analog-specific (AS) kinase (Erk2), allowed purification of a direct Erk2 substrate.
Kinase-substrate interactions transduce extracellular signals into appropriate intracellular responses, and mapping these relationships is fundamental to understanding how signaling-network connectivity results in distinct biological outcomes. Yet owing to a paucity of techniques that permit association of an individual kinase with its direct substrates, a great many connections remain to be defined1,2. Shared enzymology among protein kinase family members makes it difficult to follow the activity of a single kinase in the presence of all other cellular kinases. Protein chips3 circumvent this challenge by isolating a kinase and potential substrates from cellular complexity. But cellular components that impede substrate identification can also impose specificity, as kinase fidelity is often enforced through scaffolds4, cofactors and priming of nearby residues by phosphorylation5. Our goal is to develop bio-orthogonal chemical reactions, unique from the natural repertoire of cellular enzymology, to allow individual kinase substrates to be traced in the presence of signaling components that contribute to physiological specificity.
Specific kinase substrate labeling is achieved by engineering the kinase of interest to accept bio-orthogonal ATP analogs that are not used by the remainder of the kinome6. For example, AS kinases use bulky [γ-32P]ATP analogs7 to produce radiolabeled substrates of a single kinase. Application of this strategy to yeast glutathione S-transferase–tagged or tandem affinity purification (TAP)-tagged libraries has yielded hundreds of Cdc28 (ref. 8) and Pho85 (ref. 9) substrate pairs. In these screens AS kinases had been added to yeast extracts containing candidate substrates, which can be purified by virtue of their genetically encoded affinity handles. But such libraries are not available for mammalian proteins and, without the aid of an affinity handle, identification of mammalian AS kinase substrates faces the same challenges as other phosphoproteomic studies, including the dynamic range of protein abundance and substoichiometric substrate phosphorylation10. In studies of mammalian AS kinase substrates this challenge has typically been addressed by immunoprecipitating the kinase followed by identification of bound substrates. For example, AS Erk2 substrates can be identified by performing kinase assays on immune complexes11, which provides a necessary pre-enrichment of Erk2 binding proteins. Many kinase-substrate interactions are of low affinity, however, and are not amenable to this approach. Further, in all previous studies of AS kinase substrates, the kinase was either overexpressed or purified and added to cell extracts. Here we report two key advances toward the goal of unbiased identification of endogenous kinase substrates: substrate labeling in cells derived from a ‘gene knock-in’ AS kinase mouse12-14 and development of a method for affinity purification of AS kinase substrates.
To convert the phosphate, delivered specifically by an AS kinase into an affinity handle, we developed a semisynthetic reaction scheme. First, an AS kinase uses an N6-alkylated ATPγS (A*TPγS) to thiophosphorylate its substrates. Because all other kinases will instead transfer phosphate to their substrates, substituting a reactive sulfur in place of oxygen provides a unique starting point from which to differentiate AS kinase substrates from all other phosphoproteins. Reactions that can functionalize thiophosphate-containing proteins, facilitating their purification, will also modify other cellular nucleophiles. For example, alkylation of thiophosphate with the electrophilic tag p-nitrobenzylmesylate (PNBM) is concomitant with formation of undesired alkylation products resulting from derivatization of cysteine thiols. To specifically detect and purify thiophosphate reaction products, we developed polyclonal and monoclonal antibodies that discriminate thiophosphate esters from cysteine alkylation products (thioethers).
In our first report15, we had used this approach to detect substrates of the serine-threonine kinase, Cdk1. To further develop this strategy and expand the portion of the kinome tractable with this approach, we analyzed each of the reaction steps (Fig. 1a) for specificity and applicability to many kinase-substrate pairs. We screened diverse kinases for the ability to thiophosphorylate their protein substrates and found that the vast majority of kinases used ATPγS as a phosphodonor. We synthesized orthogonal A*TPγS analogs and found them to be preferred substrates for AS kinases, permitting delivery of thiophosphate to individual kinase substrates. The resulting labeled kinase substrates, which contained modified serine, threonine and tyrosine residues in the context of diverse kinase consensus motifs, were all recognized by new thiophosphate ester–specific antibodies. Our initial hapten design presented the thiophosphate ester modification on a threonine backbone15; the elicited hapten-specific antibodies, however, suffered from either low yield of specific antibody or low affinity, and were not expected to recognize modified tyrosine residues. Here we designed a new hapten (Fig. 1b) containing only the minimal desired epitope in an effort to focus the immune response on the thiophosphate ester moiety. The elicited rabbit polyclonal and monoclonal antibodies are thiosphosphate ester–specific, context-independent and capable of immunoprecipitation. Application of this semisynthetic immunoaffinity approach allowed us to purify the direct substrates of an AS kinase (Erk2), which was expressed at endogenous levels.
Figure 1.
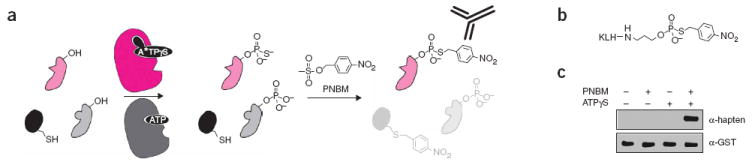
Strategy for labeling individual kinase substrates. (a) Reaction sequence for affinity tagging AS kinase substrates. First, an AS kinase (magenta) uses N6-alkylated ATPγS (A*TPγS) to thiophosphorylate its substrates (pale magenta). In a second step, alkylation with PNBM yields thiophosphate esters and thioethers. Only AS kinase substrates are recognized by α-hapten–IgG. (b) Structure of the hapten conjugate used to elicit thiophosphate ester specific antibodies. (c) JNK1 was incubated with c-Jun–GST and combinations of ATPγS and PNBM as indicated; reaction products were analyzed by western blot with α-hapten–IgG. α-GST–IgG confirms equal loading. Full-length blots are available in Supplementary Figure 1b.
RESULTS
Generation of thiophosphate ester–specific IgG
We elicited rabbit α-hapten–immunoglobulin gamma (IgG) with a hapten conjugate, consisting of a p-nitrobenzylthiophosphate ester with a three-carbon linker to keyhole limpet hemocyanin (KLH; Fig. 1b). Affinity-purified polyclonal antibodies only recognized c-Jun–GST containing both thiophosphorylation and PNBM modifications (Fig. 1c), verifying that the antibodies discriminate thiophosphate alkylation products from all other possible alkylation products. In contrast, antibodies raised in chicken (α-hapten–IgY) recognized PNBM alkylated proteins whether or not they had been thiophosphorylated (Supplementary Fig. 1 online) and were not used in any further experiments. α-hapten–IgG comprised approximately 3% of total serum IgG, yielding milligrams of antibody for western blot and kinetic experiments. Meanwhile, we used cells from this rabbit to generate hybridomas. Forty eight hybridoma supernatants were immunoreactive to a BSA-hapten conjugate, and forty had modest to excellent specificity for thiophosphate esters over thioethers (Supplementary Fig. 2a online). Several of these clones immunoprecipitated affinity-tagged histone H1 (Supplementary Fig. 2b). Clones with the strongest signal also specifically recognized thiophosphate alkylation products in the context of a whole cell lysate (Supplementary Fig. 2c); we subcoloned each of these hybridomas. We used the subclone with the strongest signal in these assays (51-8) for large-scale rabbit monoclonal antibody (RmAb) production.
Specificity and yield of PNBM alkylation
Proteins contain several functional groups that could potentially react with PNBM, and hinder automated database searching and identification of AS kinase substrates from mass spectrometric data. To determine the extent of PNBM alkylation of proteogenic amino acids, we thiophosphorylated and alkylated c-Jun–GST (as in Fig. 1c), digested it with trypsin and analyzed it by tandem mass spectrometry. After standard MASCOT-based sequencing-software analysis, c-Jun–GST was readily identified with 56% sequence coverage (Fig. 2a), indicating PNBM does not modify alcohol, amine or carboxylic acid functional groups. Manual sequencing of unassigned tandem mass spectra yielded peptides that contained alkylated cysteine and thiophosphorylated serine residues. We localized sites of cysteine alkylation by inspection of the y-ion series (Fig. 2b). Initial tandem mass spectrometry spectra of thiophosphate ester–containing peptides contained few sequence ions; lowering the collision-induced disassociation energy produced the more informative spectra (Fig. 2c). We observed a prominent 169-Da mass shift, corresponding to elimination of p-nitrobenzyl thiol, from the molecular ion, which produced an additional y-ion series.
Figure 2.
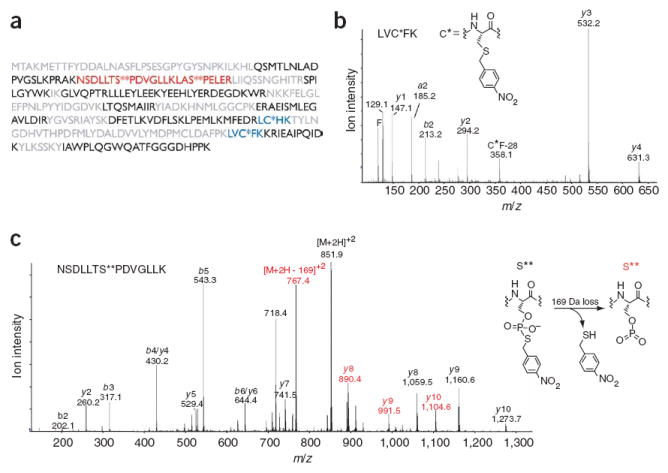
Mass spectrometric analysis of thiophosphorylated and alkylated c-Jun–GST. (a) Sequence coverage of c-Jun-GST. Residues in black were identified by automated database searching of the unmodified peptides, blue indicates peptides that contain a modified cysteine, red indicates thiophosphate ester–containing peptides, and residues in gray were not observed. (b) Tandem mass spectrum of a thioether–containing peptide. C* indicates a nitrobenzyl-modified cysteine. (c) Tandem mass spectrum of a peptide containing a modified serine (S**), which corresponds to a site of JNK1 phosphorylation. A 169-Da loss, consistent with the depicted elimination, was observed from the doubly charged molecular ion and also gave rise to an additional y-ion series. Ions that have undergone this loss are labeled in red.
To determine the yield of thiophosphate alkylation, we synthesized a model thiophosphopeptide and monitored PNBM alkylation by high-performance liquid chromatography (HPLC; Supplementary Fig. 3a–c online). The reaction was complete after a 1-h treatment with 2.5 mM PNBM. We observed only one product in the HPLC chromatogram, and mass spectrometric analysis confirmed this to be the monoalkylated peptide (Supplementary Fig. 3d).
Generality of ATPγS use and antibody recognition
To determine the general applicability of this labeling strategy, we tested each of the reaction steps with a diverse set of kinase substrate pairs. Several kinases have been reported to use ATPγS as a phosphodonor (for a partial list, see ref. 15), although often with a reduced catalytic rate, kcat (ref. 16). However, the generality of thiophosphate transfer among kinases remains unclear. We screened 15 kinases for the ability to thiophosphorylate protein substrates, and all but two (GRK2 and JNK2; data not shown) accepted ATPγS as a phosphodonor. We alkylated the thiophosphorylated substrates from the remaining 13 kinases with PNBM and analyzed them for antibody recognition in the context of different kinase consensus phosphorylation motifs17 (Figs. 3 and 4). The polyclonal antibodies recognized each labeled substrate regardless of the modified residue (serine, threonine or tyrosine) and the amino acid residues surrounding the site of phosphorylation. When the monoclonal antibody became available, we tested a diverse subset of labeled kinase substrates (PKCδ, JNK1, GSK3β, Src and CK1), and in every case the 51-8 antibody selectively recognized the modified substrate (Supplementary Fig. 2).
Figure 3.

α-hapten-IgG detection of thiophosphate-esterified kinase substrates. Each blot is labeled with the corresponding kinase and its preferred phosphorylation consensus motif. A description of the conditions for each kinase reaction is available in the Supplementary Methods; full-length blots are available in Supplementary Figure 1c.
Figure 4.

A*TPγS analog orthogonality and acceptance by AS kinases. (a) Chemical structure of A*TPγS, R indicates the site of N6 modification. (b) Kinase reactions with ATPγS and A*TPγS analogs. Following PNBM alkylation, the labeled substrates were detected with α-hapten-IgG western blot analysis, full-length blots are available in Supplementary Figure 1d. WT, wild type.
A*TPγS synthesis and use by AS kinases
A*TPγS contains two permutations from the natural kinase substrate ATP: an N6 appendage and substitution of the terminal oxygen of the γ-phosphate with sulfur (Fig. 4a). The N6 modification was introduced to the adenosine riboside, followed by diphosphorylation18 to afford N6-alkylated ADPs. We synthesized A*TPγS analogs by condensation of the N6-alkylated ADPs with an S-protected phosphodiester19, to ensure that the sulfur on the γ-phosphate is non-bridging. Previous work in our laboratory has demonstrated that N6-benzyl and N6-phenethyl ATP are consistently selected, from a diverse panel of N6–modified ATP analogs, as efficient substrates for AS kinases. Based on these data we synthesized N6-benzyl and N6-phenethyl ATPγS.
We asked if these A*TPγS analogs were bio-orthogonal (not accepted by wild-type enzymes) and if the unnatural analogs could be accepted by AS kinases. We tested wild type and AS alleles of kinases with diverse roles in cellular signaling for their ability to utilize ATPγS and A*TPγS as thiophosphate donors: PKCδ, a second messenger activated kinase20, Cdc5, the yeast polo-kinase which regulates cell division21, and JNK1, a stress activated MAP kinase22. The assay for thiophosphorylation was based on alkylating the in vitro kinase reactions with PNBM, followed by western blot analysis (Fig. 4b). Notably, wild-type kinases accepted ATPγS and could not utilize A*TPγS analogs. Each of the AS kinases were able to utilize ATPγS and one or both of the A*TPγS analogs: AS PKCδ favored N6-benzyl ATPγS, AS Cdc5 favored N6-phenethyl ATPγS, and AS JNK1 used both analogs to a similar extent. Western blot analysis provides a rapid assessment of the preferred nucleotides but does not allow quantitative analysis of the thiophosphorylation reaction. To obtain relative kinetic parameters for unnatural A*TPγS analog usage, we developed a disassociation-enhanced lanthanide fluorescence immunoassay (DELFIA)23, using europium-labeled α-hapten–IgG (Supplementary Fig. 4a,b online). The AS kinases typically had a lower specificity (kcat/Km) for ATPγS than the corresponding wild-type kinase, but we observed an increase in kcat/Km when the AS kinase used its preferred A*TPγS analog (Supplementary Fig. 4c). In each case the magnitude of kcat/Km correlated with the intensity of the corresponding band on the western blot.
Immunoprecipitation and identification of Erk2 substrates
To explore the utility of this technique to affinity label and immunoprecipitate kinase substrates in a cellular context, we used cells expressing endogenous amounts of AS Erk2. We prepared mouse embryonic fibroblasts (MEFs) from 13.5-day-old embryos of Erk1−/−Erk2+/+ and Erk1−/−Erk2AS/AS mice. Erk2 was immunoprecipitated from each of these cell lines and assayed with A*TPγS analogs; N6-phenethyl ATPγS was a preferred nucleotide substrate for AS Erk2 and was not accepted by wild-type Erk2 (data not shown). We optimized concentrations of A*TPγS and GTP, a competitor for any general nucleotidases or nonspecific phophotransferases, that yielded specifically labeled AS Erk2 substrates by adding varying concentrations of the nucleotides to digitonin-permeabilized MEFs. Alkylation and western blot analysis of cells permeabilized in the presence of 100 μM A*TPγS and 1 mM GTP revealed several proteins that were selectively labeled in the AS Erk2-expressing cells (Fig. 5a). Immunoprecipitation of these samples with the 51-8 monoclonal antibody yielded many specifi-cally enriched proteins as determined by western blot analysis (Fig. 5b). Silver staining revealed a set of bands at 200 kDa (Fig. 5c). We cut the entire length of the gel (both wild-type and AS lanes) into approximately 1-mm slices and digested them with trypsin. Mass spectrometry analysis and database searching identified each of the high-molecular-weight bands as the nucleoporin translocated promoter region (Tpr), which is a reported Erk2 substrate11. We sequenced forty-five peptides, corresponding to Tpr, from liquid chromatography–tandem mass spectrometry analysis of these gels bands, one of which is shown in Figure 5d. None of the other selectively immunoprecipitated substrates were present at a level sufficient for mass-spectrometric identification.
Figure 5.
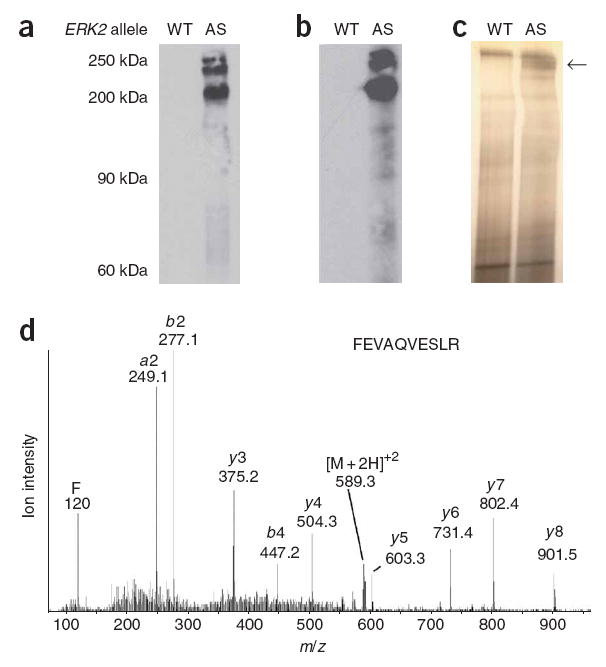
Labeling, immunoprecipitation and identification of AS kinase substrates. (a) MEFs expressing either wild-type (WT) or AS Erk2 were permeabilized, incubated with A*TPγS, alkylated and analyzed by western blot (RmAb α-hapten–IgG). (b) Western blot analysis of the same samples as in a after immunoprecipitation. (c) Silver stain gel of immunoprecipitates in b. (d) Liquid chromatography–tandem mass spectrometry spectrum of a tryptic peptide, FEVAQVESLR, from the indicated band in c. Full-length blots and silver-stained gels are available in Supplementary Figure 1e.
DISCUSSION
Presently 40 kinases have been rendered analog specific (Supplementary Table 1 online), and it is expected that most protein kinases will be amenable to this approach24. The use of AS kinases in combination with bio-orthogonal ATP analogs has permitted the identification of several AS kinase substrates, yet the vast majority of such pairs remain to be identified. Here we report a unified technique for AS kinase substrate labeling and phosphoprotein affinity purification, which is applicable to all three common types of phosphorylation. Additionally, the suite of immunoassays described here allow for the detection, quantification and purification of wild-type kinase substrates.
Another advance toward our goal of physiologically relevant assays of individual kinases is the description of a method to label substrates in gently permeabilized cells expressing endogenous levels of an AS kinase, Erk2. Immunoprecipitation of these samples, prepared from a modest amount of material (15 × 106 cells), enriched a sufficient quantity of Tpr for mass-spectrometric identification. Presently we are extending this study to identify additional kinase substrates in tissues isolated from AS kinase–expressing mice. We are also exploring the possibility of immunoprecipitating modified peptides to identify the sites of phosphorylation; this approach recently has been applied to identify tyrosine phosphorylation sites25.
High-affinity serine and threonine phosphospecific antibodies typically rely on residues or motifs26 flanking the site of phosphorylation, whereas phosphotyrosine antibodies can recognize tyrosine phosphorylated sites independent of context25. Using this semi-synthetic approach we were able to convert each of these phospho–amino acids into an epitope that can be recognized by a single antibody—a new concept in the area of modification-specific antibodies. A key feature of the epitope construction strategy is discrimination of closely related reaction products, that is, specificity for thioesters over thioethers. The ability to provide a second filter that distinguishes desired from undesired reaction products rather than relying on a single completely bio-orthogonal reaction, such as click chemistry, oxime formation or the Staudinger ligation27, may prove to be a general approach to broaden the scope of chemical functionalities and modification strategies used in bio-orthogonal tagging schemes.
METHODS
Antibody generation and purification
We conjugated hapten (Supplementary Fig. 5 online) to succinylated KLH, by reaction with N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide HCl (EDCI) and N-hydroxysulfosuccinimide (sulfo-NHS) overnight at pH 8. We purified the product on a PD-10 gel filtration column (Amersham) and used it to immunize rabbits (Epitomics) and chickens (Aves Labs). We purified polyclonal antibodies on an affinity column (diaminodipropyl amine gel; Pierce) containing immobilized hapten. After elution, we concentrated the antibodies using a 10-kDa molecular weight cut-off Amicon ultra centrifugal filter. For DELFIA, we labeled the antibodies with Eu3+ according the manufacturer’s instructions (Perkin Elmer). Monoclonal antibodies were produced by Epitomics, and screened in enzyme-linked immunosorbent assay as well as western blot and immunoprecipitation assays as described in Supplementary Methods online.
A*TPγS analog synthesis
The overall synthetic scheme is illustrated in Supplementary Figure 6 online. We converted N6-modified ribosides28 to the N6-modified ADP molecules by reaction with POCl3, followed by phosphoric acid and 1,8-diaza-bicyclo[5.4.0]undec-7-ene (DBU)18. We quenched the reactions with 0.1 M triethylammonium bicarbonate (TEAB) and purified the products with strong cation exchange chromatography (0.1 M to 1 M of TEAB) on a Hi-Prep QFF column (Amersham Biosciences). We pooled fractions containing the diphosphates, lyophilized them and analyzed them by mass spectrometry. Yields were 25–30%. We synthesized A*TPγS analogs as described for ATPγS19, and purified and analyzed them as described above for the ADP analogs. We obtained the lyophilized solids in 6–8% yield.
In vitro kinase reactions followed by antibody detection
We incubated kinases with their respective substrates in appropriate kinase buffers (see Supplementary Methods). For screens of analog preference and orthogonality using western blot analysis, we used ATPγS or A*TPγS analogs at a concentration of 1 mM. For kinetic measurements, ATPγS or A*TPγS analog concentration varied from 0.1 μM to 250 μM. We alkylated proteins with 2.5 mM PNBM for 2 h at room temperature (18–22 °C) and analyzed the products by western blotting or DELFIA. For western blotting, we diluted the antibodies 1:15,000 in TBS (pH 8.0) containing 0.5% Tween 20 (TBST) and 5% milk. We rocked the blots overnight at 4 °C, then incubated them with goat anti-IgG horseradish peroxidase (Promega) or rabbit anti-IgY horseradish peroxidase (Sigma), and imaged them (chemiluminescence on film).
Mice
All experiments involving live animals were approved by The University of California San Diego Institutional Animal Care and Use Committee (IACUC). We produced Erk2AS/AS knock-in mice using standard techniques of gene targeting by homologous recombination. A 2.7 kb short arm including exon 2 and a 6.4 kb long arm containing exon 3 were cloned into pfloxΔTK. We modified exon 3 by site-directed mutagenesis to change the CAG glutamine codon at position 102 to GGC glycine. This eliminated an Eco0109t restriction site that we used for screening. We sent the targeting vector to the University of California San Diego Cancer Center Transgenic facility to produce transfected R1 mouse embryonic stem cells. Two clones that tested position for homologous recombination and retention of the codon mutation were used to produce chimeric mice by blastocyst injection. Upon breeding, the targeted allele was transmitted to offspring and further transmitted at a Mendelian frequency.
We bred Erk2AS/AS mice retaining the neomycin-resistance gene to EIIA-cre mice29 and selected offspring for successful recombination. We then crossed Erk2AS/AS mice with Erk1−/− mice. We intercrossed the resulting mice and selected them for an Erk1−/−Erk2+/+ or Erk1−/−Erk2AS/AS genotype. We derived embryonic fibroblasts as described in the Supplementary Methods.
Cell permeablization and Erk2 substrate labeling
We trypsinized 15 × 106 wild-type or AS Erk2 MEFs cells, pelleted (200g at 4 °C) and resuspended them in DMEM to 5 × 106 cells/ml. We added phorbol 12-myristate 13-acetate (PMA) (20 ng/ml) and ionomycin (1 μM) for 5 min at 37 °C and then pelleted the cells. Permeabilization proceeded for 5 min on ice in 1× Dulbecco’s phosphate buffered saline and 1× kinase buffer (Cell Signaling) containing complete protease inhibitor cocktail (Roche), phosphatase inhibitor cocktails I and II (Calbiochem) and 50 μg/ml digitonin (Sigma). We pelleted and resuspended cells in the same buffer but without digitonin, and with 100 μM N6-phenethyl ATPγS and 1 mM GTP. The kinase reaction proceeded at 30 °C for 30 min with gentle rocking. We then pelleted and lysed the cells on ice for 15 min in 0.5 ml RIPA buffer (50 mM Tris-HCl (pH 8), 150 mM NaCl, 1.0% NP-40 and 0.1% SDS) containing 25 μM EDTA. We cleared the lysates by centrifugation, alkylated them and stored them at −80 °C.
Immunoprecipitation of Erk2 substrates with 51-8 antibody
We removed PNBM, which potently inhibits immunoprecipitation, by size-exclusion chromatography. We equilibrated PD-10 columns with RIPA buffer, applied 0.5 ml of alkylated proteins to the column and eluted them with RIPA buffer containing protease inhibitors. We collected 0.5-ml fractions; we pooled the protein-containing fractions (7–9) and pre-cleared them with 100 μl of 50% rProtG agarose (Invitrogen) for 5 h at 4 °C. Meanwhile 20 μg of 51-8 RmAb was bound to 100 μl of 50% rProtG agarose in 1.0 ml of RIPA buffer containing 0.5 mg/ml BSA for 5 h at 4 °C. After removal of the beads from the lysate, we added the antibody-bound beads. Immunoprecipitation proceeded overnight at 4 °C, and then we washed the samples 4× with 1.0 ml of RIPA buffer. We eluted bound proteins with SDS-PAGE sample buffer and analyzed them by western blotting and silver staining.
Mass spectrometry
We excised bands from either a coomassie-stained gel (c-Jun–GST) or a silver-stained gel (Tpr), and subjected them to in-gel tryptic digestion. We separated peptides with nano-HPLC and sequenced them by tandem mass spectrometry on a QSTAR-XL (Applied Biosystems). The spectra were submitted to a Mascot search to identify unmodified peptides, and we manually sequenced PNBM-modified peptides.
Additional methods
Detailed descriptions of all experiments are available in the Supplementary Methods.
Supplementary Material
Acknowledgments
We thank D. Randle and D. Morgan for wild-type Cdc5, D. Kenski for performing GRK2 kinase assays, H. Luecke for helpful suggestions, D. Maly for helpful comments on the manuscript, and V. Ohman for excellent administrative assistance. Erk1−/− mice were generously provided by G. Pages and J. Pouyssegur (Centre National de la Recherche Scientifique, Nice, France). HeLa cells were provided by the National Cell Culture Center. This work was supported by the US National Institutes of Health (NIH) National Institute of Biomedical Imaging and BioEngineering R01 EB001987, and in part by NIH grant AA013588 and funds provided by the State of California for medical research on alcohol and substance abuse through the University of California at San Francisco (to R.O.M.). J.J.A. was funded by an Achievement Rewards for College Scientists fellowship, Genentech, and the Sandler Foundation. Mass spectrometry was made possible by NIH grants NCRR RR015804 and NCRR RR001614.
Footnotes
AUTHOR CONTRIBUTIONS J.J.A. and K.M.S. wrote the manuscript. J.J.A. performed all experiments except those mentioned below. M.L. performed Erk2 substrate labeling reactions, C.S.B. performed mass spectrometry of modified peptides, J.L.P. expressed Cdc5, D.W. provided technical assistance for PKCδ experiments, A.H. constructed JNK1 plasmids, C.D.K. constructed the AS Erk2 mouse, W.C. constructed PKCδ plasmids, A.L.B. advised C.S.B., R.O.M. advised D.W. and W.C., and assisted in manuscript revision, R.J.D. advised A.H., S.M.H. advised M.L. and C.D.K. and K.M.S. advised J.J.A.
COMPETING INTERESTS STATEMENT The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/naturemethods.
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Berwick DC, Tavare JM. Identifying protein kinase substrates: hunting for the organ-grinder’s monkeys. Trends Biochem Sci. 2004;29:227–232. doi: 10.1016/j.tibs.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Ptacek J, Snyder M. Charging it up: global analysis of protein phosphorylation. Trends Genet. 2006;22:545–554. doi: 10.1016/j.tig.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 4.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 5.Biondi RM, Nebreda AR. Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions. Biochem J. 2003;372:1–13. doi: 10.1042/BJ20021641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah K, et al. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci USA. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witucki LA, et al. Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phospho-acceptor specificity of the wild-type enzyme. Chem Biol. 2002;9:25–33. doi: 10.1016/s1074-5521(02)00091-1. [DOI] [PubMed] [Google Scholar]
- 8.Ubersax JA, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 9.Dephoure N, et al. Combining chemical genetics and proteomics to identify protein kinase substrates. Proc Natl Acad Sci USA. 2005;102:17940–17945. doi: 10.1073/pnas.0509080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee WC, Lee KH. Applications of affinity chromatography in proteomics. Anal Biochem. 2004;324:1–10. doi: 10.1016/j.ab.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 11.Eblen ST, et al. Identification of novel ERK2 substrates through use of an engineered kinase and ATP analogs. J Biol Chem. 2003;278:14926–14935. doi: 10.1074/jbc.M300485200. [DOI] [PubMed] [Google Scholar]
- 12.Jaeschke A, et al. JNK2 is a positive regulator of the cJun transcription factor. Mol Cell. 2006;23:899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, et al. Inducible protein knockout reveals temporal requirement of CaMKII reactivation for memory consolidation in the brain. Proc Natl Acad Sci USA. 2003;100:4287–4292. doi: 10.1073/pnas.0636870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, et al. A chemical-genetic approach to studying neurotrophin signaling. Neuron. 2005;46:13–21. doi: 10.1016/j.neuron.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Allen JJ, Lazerwith SE, Shokat KM. Bio-orthogonal affinity purification of direct kinase substrates. J Am Chem Soc. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sondhi D, et al. Peptide and protein phosphorylation by protein tyrosine kinase Csk: insights into specificity and mechanism. Biochemistry. 1998;37:165–172. doi: 10.1021/bi9722960. [DOI] [PubMed] [Google Scholar]
- 17.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman C, Genieser HG, Veron M, Jastorff B. Novel synthesis of nucleoside 5′-polyphosphates. Bioorg Med Chem Lett. 1996;6:2571–2574. [Google Scholar]
- 19.Goody RS, Eckstein F. Thiophosphate analogs of nucleoside di- and triphosphates. J Am Chem Soc. 1971;93:6252–6257. [Google Scholar]
- 20.Kikkawa U, Matsuzaki H, Yamamoto T. Protein kinase C delta (PKC delta): activation mechanisms and functions. J Biochem. 2002;132:831–839. doi: 10.1093/oxfordjournals.jbchem.a003294. [DOI] [PubMed] [Google Scholar]
- 21.Lee KS, et al. Yeast polo-like kinases: functionally conserved multitask mitotic regulators. Oncogene. 2005;24:217–229. doi: 10.1038/sj.onc.1208271. [DOI] [PubMed] [Google Scholar]
- 22.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Gaarde WA, Hunter T, Brady H, Murray BW, Goldman ME. Development of a nonradioactive, time-resolved fluorescence assay for the measurement of Jun N-terminal kinase activity. J Biomol Screen. 1997;2:213–223. [Google Scholar]
- 24.Zhang C, et al. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat Methods. 2005;2:435–441. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]
- 25.Rush J, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, et al. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- 27.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 28.Kikugawa K, Iizuka K, Ichino M. Platelet aggregation inhibitors. 4. N6-substituted adenosines. J Med Chem. 1973;16:358–364. doi: 10.1021/jm00262a011. [DOI] [PubMed] [Google Scholar]
- 29.Holzenberger M, et al. Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucleic Acids Res. 2000;28:E92. doi: 10.1093/nar/28.21.e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


