Abstract
Over time, organisms developed various strategies to adapt to their environment. Circadian clocks are thought to have evolved to adjust to the predictable rhythms of the light-dark cycle caused by the rotation of the Earth around its own axis. The rhythms these clocks generate persist even in the absence of environmental cues with a period of about 24 hours. To tick in time, they continuously synchronize themselves to the prevailing photoperiod by appropriate phase shifts. In this study, we disrupted two molecular components of the mammalian circadian oscillator, Rev-Erbα and Period1 (Per1). We found that mice lacking these genes displayed robust circadian rhythms with significantly shorter periods under constant darkness conditions. Strikingly, they showed high amplitude resetting in response to a brief light pulse at the end of their subjective night phase, which is rare in mammals. Surprisingly, Cry1, a clock component not inducible by light in mammals, became slightly inducible in these mice. Taken together, Rev-Erbα and Per1 may be part of a mechanism preventing drastic phase shifts in mammals.
Introduction
In mammals, many physiological and behavioural parameters are subjected to daily variations. For instance, blood pressure, heart rate, body temperature, or sleep-wake cycles change considerably during the course of a day. The central circadian (Latin: “around a day”) clock, which governs these rhythms, is located in the suprachiasmatic nuclei (SCN) of the ventral hypothalamus [1], [2]. Since this internal pacemaker can tell time only approximately, it has to be reset every day by environmental signals. The strongest synchronizer for the endogenous clock is light, which signals directly to the SCN via the retinohypothalamic tract [3]. To reset peripheral oscillators, rhythms generated in the SCN are sent to the rest of the body via both neuronal and humoral signals.
Much of the understanding concerning the functioning of the circadian clock has come from extensive work done in model organisms mutant for one or several clock genes. In mammals, it was found that the molecular clockwork is based on cell-autonomous interlocked positive and negative transcriptional/translational feedback loop (TTL) [4]. The positive limb of the core loop is formed by the transcription factors CLOCK and BMAL1. Heterodimers of these proteins bind to E-box motifs in the promoters of Per, Cry, Rev-Erbα and other genes controlled by the clock. In the cytoplasm, the PER proteins form complexes with CRY, which then enter the nucleus. Once the complexes reach their destination, CRY inhibits the CLOCK/BMAL1 driven transcription, thereby establishing a negative feedback loop [5]–[7]. The suppressed E-box driven transcription leads to a decline in PER, CRY and REV-ERBα protein levels, which finally re-activates CLOCK-BMAL1 driven transcription. Due to the decrease in REV-ERBα, Bmal1 transcription is no longer inhibited [8], [9]. Hence RORα can activate Bmal1 expression by binding to the now vacant retinoic acid related orphan receptor response elements (RORE) in its promoter [10]–[12].
The TTL described above can be modulated by external signals. As mentioned previously, the strongest Zeitgeber (literally, “time giver” in German) for the mammalian clock is light. To better study the impact of light on the circadian system animals kept under constant darkness (DD) are subjected to a brief light pulse at different time points. During the subjective day, light has little or no effect on the resetting of the clock. By contrast, light exposure during the subjective night is capable of shifting the pacemaker. Depending on whether the light is administered during the early or late subjective night, the animal either delays or advances its clock, respectively [13]–[15].
Several studies attempted to identify the molecular mechanisms underlying resetting in response to a light pulse. However, in mammals only little is understood compared to Drosophila and zebrafish [16]–[18]. Since both Rev-Erbα−/− and Per1Brdm1 single mutant mice are characterized by an aberrant light response during late night [9], [19], we were interested in analyzing the resetting phenotype of double mutants. We monitored their wheel-running behaviour under constant conditions and established a phase response curve (PRC). We show that Rev-Erbα−/−Per1Brdm1 double mutant mice maintain a functional circadian clock although they display a shorter period with higher variance between individuals. Moreover, clock gene expression and their light induction are altered in mice deficient for both Rev-Erbα and Per1. Strikingly, they show high amplitude resetting and Cry1 becomes slightly light inducible in these mice after a light pulse delivered at the end of the subjective night. We conclude that Rev-Erbα and Per1 may participate in a mechanism leading to moderate light evoked behavioral responses typically observed in mammals.
Results
Generation of Rev-Erbα−/−Per1Brdm1 double mutant mice
Since Rev-Erbα and Per1 mutant mice both show shortened period length in constant darkness [9], [20], we wanted to test circadian parameters in mice bearing both mutations. To this end, Rev-Erbα [9] and Per1 [20] single mutants were crossed. Double mutant offspring were intercrossed to produce both wild-type and mutant animals. Genotyping of these mice (Fig. S1) revealed that all genotypes were obtained at the expected Mendelian ratios (Fig. S2A). Moreover, they were all morphologically indistinguishable from their wild-type littermates. In addition, Rev-Erbα−/−Per1Brdm1 double mutants were fertile and gave birth to normally sized litters at the expected intervals (Fig. S2B, S2C).
Rev-Erbα−/−Per1Brdm1 double mutant mice display a shorter circadian period in DD
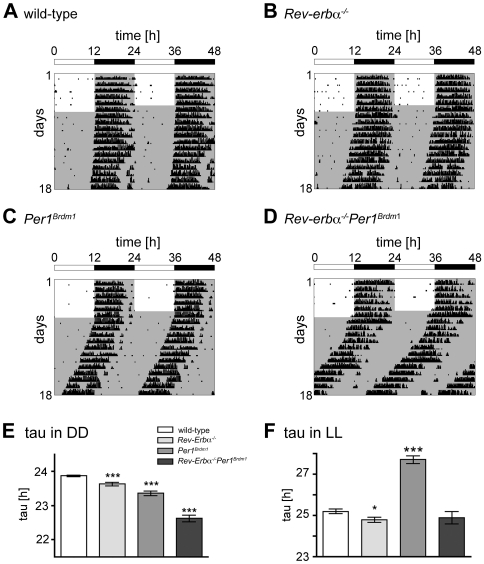
To study the effect of absent Rev-Erbα and Per1 on locomotor activity, mice were housed individually and their wheel-running activity was monitored [21]. Initially, animals were kept in a 12-h light:12-h dark cycle (LD 12∶12 h) for several days until they were stably entrained. Subsequently, they were released into constant darkness (DD). Under LD conditions, Rev-Erbα−/−Per1Brdm1 double mutants displayed an activity profile comparable to that of wild-type mice (Fig. S3). Upon release into DD, double mutants remained rhythmic and ran with a circadian period of 22.62±0.1 h (mean ± SEM; N = 34) which is significantly shorter than that of the wild-type (23.86±0.02 h; N = 38) and Rev-Erbα −/− (23.62±0.05 h; N = 27) or Per1Brdm1 (23.35±0.06 h; N = 22) single mutant mice (Fig. 1A–E). This might indicate that the absence of these two clock genes has a cumulative effect on circadian period in DD.
Figure 1. Representative wheel-running activity plots.
(A–D) Representative locomotor activity records of wild-type (A), Rev-Erbα−/− (B), Per1Brdm1 (C), and Rev-Erbα−/−Per1Brdm1 double mutant mice (D) under both light/dark (white and grey shaded, respectively) and constant darkness (grey shaded) conditions. Black bars represent wheel-revolutions. The actograms are double-plotted with the activity of the following light/dark cycle plotted to the right and below the previous light/dark cycle. The white and black bar on the top of each actogram indicates light and darkness, respectively. (E) Average free-running period (tau) in constant darkness: Rev-Erbα−/− (23.62±0.05 h; N = 27), Per1Brdm1 (23.35±0.06 h; N = 22), Rev-Erbα−/−Per1Brdm1 mutant (22.62±0.1 h; N = 34), wild-type (23.86±0.02 h; N = 38). (F) Average free-running period (tau) in constant light. Rev-Erbα−/− (24.78±0.14 h; N = 8), Per1Brdm1 (27.70±0.19 h; N = 7), Rev-Erbα−/−Per1Brdm1 mutant (24.88±0.3 h; N = 11), wild-type (25.18±0.11 h; N = 12). Data are represented as mean ± SEM. Unpaired t-test was performed to compare the free-running period lengths. * p<0.05, *** p<0.0001 (compared to wild-type).
Under constant light conditions, double mutant mice display a period, which is comparable to the one of wild-type and Rev-Erbα −/− animals (Fig. 1F). Interestingly, removal of Rev-Erbα in Per1Brdm1 mutants normalizes the free-running period in LL to a level comparable to wild-type animals. Hence, it appears that knocking out both genes rescues the Per1Brdm1 phenotype in LL.
Rev-Erbα−/−Per1Brdm1 double mutant mice display reduced amplitudes in clock gene expression
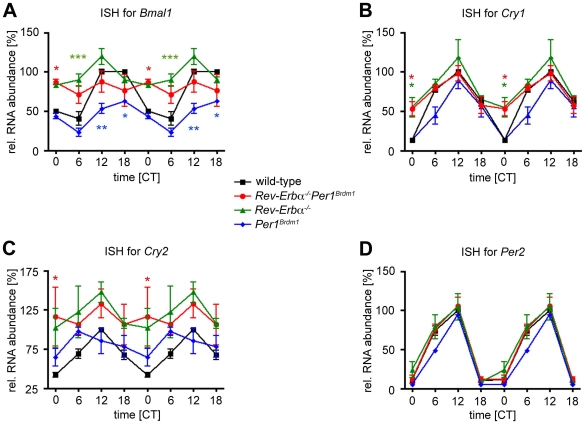
In situ hybridization analysis was used to extend our investigations to the molecular level. Clock gene expression patterns in the SCN were determined on coronal brain sections of mice kept in DD for six days. Bmal1 mRNA accumulation in Rev-Erbα−/−Per1Brdm1 double mutant mice was constantly high (Fig. 2A). However, levels were slightly lower than those reported for Rev-Erbα−/− single mutants [9]. This in combination with the observation that the increase in Bmal1 mRNA was significantly truncated in Per1Brdm1 single mutant mice could indicate that PER1 may be involved in activating Bmal1 expression.
Figure 2. In situ hybridization (ISH) profiles of cycling clock gene expression in the SCN.
Relative expression of Bmal1 (A), Cry1 (B), Cry2 (C), and Per2 (D) mRNA in the SCN under constant darkness conditions of wild-type (black line), Rev-Erbα−/−Per1Brdm1 (red line), Rev-Erbα−/− (green line), and Per1Brdm1 (blue line) mice. Values are double plotted and represented as mean ± SEM (N = 3). Two-way ANOVA with Bonferroni post-test was performed to determine significant differences between genotypes. * p<0.05; ** p<0.01; *** p<0.001.
The expression of Cry1 was rhythmic with similar peak levels in all four genotypes (Fig. 2B). By contrast, the amplitude in Cry1 mRNA accumulation was dampened in Rev-Erbα−/− single and Rev-Erbα−/−Per1Brdm1 double mutant mice due to an elevated trough at circadian time 0 (CT0). Expression of Cry2 mRNA was slightly higher in Rev-Erbα−/− single and Rev-Erbα−/−Per1Brdm1 double mutant mice compared to Per1Brdm1 single mutant and wild-type animals (Fig. 2C). However, these differences were only statistically significant at CT0 for the double mutant animals compared to the wild-type. Interestingly, disruption of Per1, Rev-Erbα, or both did not alter Per2 expression patterns (Fig. 2D). Our observations confirmed the previous findings that disrupting Rev-Erbα considerably affected the expression of Bmal1 and Cry1 but had only little or no consequences on Cry2 and Per2 transcript levels [9]. While Bmal1 expression seemed to be less changed in Rev-Erbα−/−Per1Brdm1 double mutant mice compared to Rev-Erbα−/− single mutants, Cry1 expression was unchanged between these two mutants.
With the exception of the Per2 mRNA, overall clock gene expression patterns display lower amplitudes in Rev-Erbα−/−Per1Brdm1 double mutant mice. This indicates that the oscillator in these animals is probably less robust (low amplitude), which might lead to high amplitude resetting (see below).
Rev-Erbα−/−Per1Brdm1 double mutant mice display high amplitude resetting
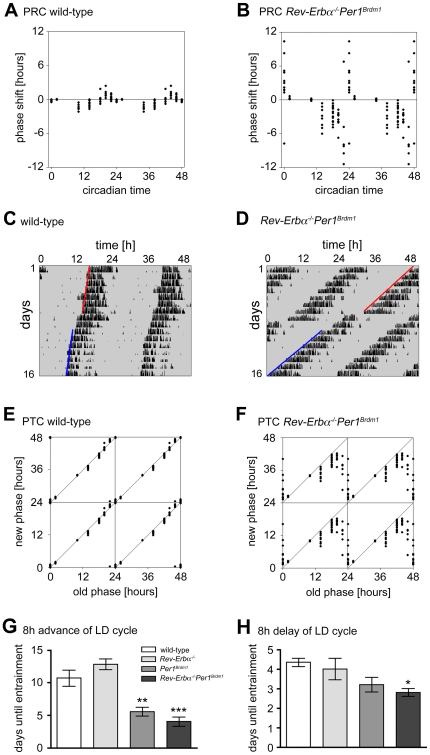
The impact of the combined absence of Rev-Erbα and Per1 on the clock-resetting response of mice was studied. A single 15 min light pulse was administered at various circadian times (CT) and the change of clock phase was quantified [22]. While wild-type mice displayed the expected low amplitude (Type I) phase response curve (PRC), Rev-Erbα−/−Per1Brdm1 double mutants showed high amplitude (Type 0-like) resetting (Fig. 3A–D) [23], [24]. For double mutant animals, the so-called “breakpoint” (transition from phase delays to phase advances) was observed between CT22 and CT24. At this time point it was not possible to distinguish whether the animals make huge phase advances or delays. Interestingly, some mice became transiently arrhythmic after a light pulse given at CT22 for some days (data not shown). The difference between weak and strong resetting can be visualized more clearly when data are plotted as a phase transition curve (PTC). In this type of visualization, the time a light pulse was given is compared to the time the phase is shifted to after the light pulse (i.e. “old” versus “new phase”). In wild-type mice, all data points were found to be close to the diagonal of the square, which has a slope of 1 (Fig. 3E). Hence, the new phase was almost identical with the old one and they displayed a so-called Type 1 resetting. By contrast, the average slope for Rev-Erbα−/−Per1Brdm1 double mutants deviated significantly from 1 (Fig. 3F).
Figure 3. Phase shifting responses to light pulses and rescheduled light/dark cycles.
(A/B) Phase response curve (PRC) of wild-type (A; N = 4–15) and Rev-Erb−/−Per1Brdm1 (B; N = 8−15) double mutant mice to 15 min light pulses. The x-axis indicates the circadian time the light pulse was administered and the y-axis represents the phase shift produced by the light pulse. Each dot represents the phase shift of one animal. (C) Representative actogram of a phase-advance response to a light pulse at CT22 of a wild-type animal. (D) Representative actogram of a phase-advance response to a light pulse at CT22 of a Rev-Erbα−/−Per1Brdm1 double mutant animal. Red lines: old phase, blue lines: new phase. (E/F) Phase transition curve (PTC) of wild-type (E) and Rev-Erb−/−Per1Brdm1 (F) double mutant mice to 15 min light pulses. Data of A and B are replotted to obtain E and F, respectively. The x-axis indicates the circadian time the light pulse was administered (old phase). The y-axis displays the circadian time to which locomotor activity was shifted after the light pulse (new phase). (G) Average time to adjust to an 8-hour advance of the light/dark (LD) cycle. Data are represented as mean ± SEM. Unpaired t-test was performed to compare the days until re-entrainment of Rev-Erbα−/− (12.80±0.80 days; N = 5), Per1Brdm1 (5.50±0.67 days; N = 6), and Rev-Erbα−/−Per1Brdm1 (4.00±0.69; N = 7) mutant to wild-type (10.67±1.26 days; N = 6) mice. ** p<0.01; *** p<0.001. (H) Average time to adjust to an 8-hour delay of the light/dark (LD) cycle. Data are represented as mean ± SEM. Unpaired t-test was performed to compare the days until re-entrainment of Rev-Erbα−/− (4.00±0.55 days; N = 5), Per1Brdm1 (3.20±0.37 days; N = 5), and Rev-Erbα−/−Per1Brdm1 (2.80±0.20 days; N = 5) mutant to wild-type (4.33±0.21 days; N = 6) mice. * p<0.05.
Due to the strong resetting phenotype of the double mutant animals we were curious to see, how fast they would adapt to an experimental jet lag. Hence we entrained them to LD 12∶12 h before they were subjected to an 8 h advance of the lighting schedule. Rev-Erbα−/−Per1Brdm1 double mutant mice achieved stable re-entrainment already after 4.00±0.69 days (mean ± SEM; N = 7) while it took two times longer for wild-type animals (10.67±1.26 days; N = 6; Fig. 3G). Likewise, Rev-Erbα−/−Per1Brdm1 (2.80±0.20 days; N = 5) double mutants adapted faster to an 8 h delay in the photoschedule (Fig. 3H) compared to wild-type mice (4.33±0.21 days; N = 6).
Cry1 is slightly light inducible in Rev-Erbα−/−Per1Brdm1 double mutant mice
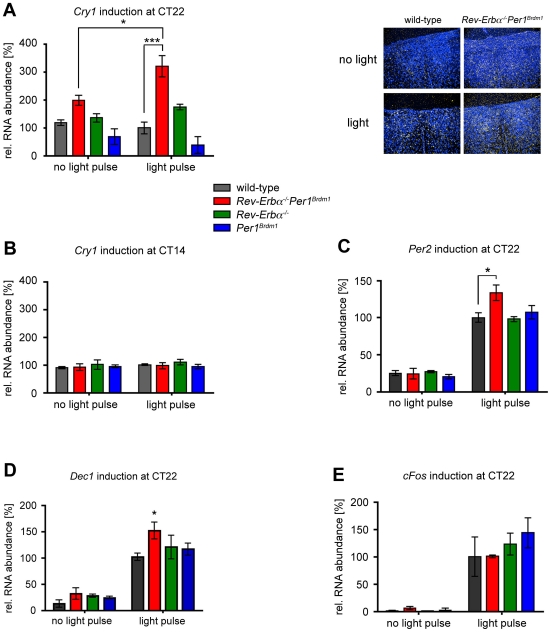
To find a possible explanation for the high amplitude resetting observed in mutants lacking both Rev-Erbα and Per1, we examined induction of clock genes in response to light. Surprisingly, Cry1 a clock gene not inducible by light in wild-type mice, was slightly increased in its expression after a light pulse at CT22 in double mutant animals relative to dark controls (Fig. 4A). As expected, wild-type and single mutant mice did not show any Cry1 induction neither at CT22 nor at CT14 (Fig. 4B). In contrast to CT22, light did not induce Cry1 expression in Rev-Erbα−/−Per1Brdm1 double mutant mice at CT14 (Fig. 4B). In addition, Per2 was induced to a higher extent in double mutant mice compared to the three other genotypes at CT22 (Fig. 4C) but not at CT14 (data not shown). Expression of Dec1, another known light inducible clock gene, was increased stronger in double mutants relative to the other genotypes at CT22 (Fig. 4D) and CT14 (data not shown). The signal intensity for cFos was similar between all genotypes after a light pulse at CT22 (Fig. 4E) indicating that the light-signalling pathway is intact. Hence the PRC observed in our double mutants is probably caused by changes in the circadian pacemaker.
Figure 4. Light induced mRNA accumulation in the SCN.
(A) Cry1 induction after a light pulse at CT22 (N = 3); (B) Cry1 induction after a light pulse at CT14 (N = 3); (C) Per2 induction after a light pulse at CT22 (N = 3−5); (D) Dec1 induction after a light pulse at CT22 (N = 3); (E) cFos induction after a light pulse at CT22 (N = 3) in wild-type (black), Rev-Erbα−/−Per1Brdm1 (red), Rev-Erbα−/− (green), and Per1Brdm1 (blue) mice. SCN were collected 45 minutes after the end of the 15-minute light pulse or mock treatment. Values are represented as mean ± SEM. Two-way ANOVA with Bonferroni post-test was performed to determine significant differences between genotypes. Unpaired t-test was used to determine significant differences between light pulsed and dark control animals. * p<0.05; *** p<0.001.
To sum up Rev-Erbα−/−Per1Brdm1 double mutant mice were fertile and morphologically indistinguishable from wild-type animals. Although they displayed a shorter circadian period in DD, their molecular oscillator seemed to be still functional. However, their response to brief light pulses was altered and they displayed high amplitude resetting. Interestingly, Cry1 became slightly light inducible if the light signal was presented at the end of the subjective night (CT22).
Absence of Cry1 affects suppression of daytime activity in Rev-Erbα−/−Per1Brdm1Cry1−/− triple mutant mice
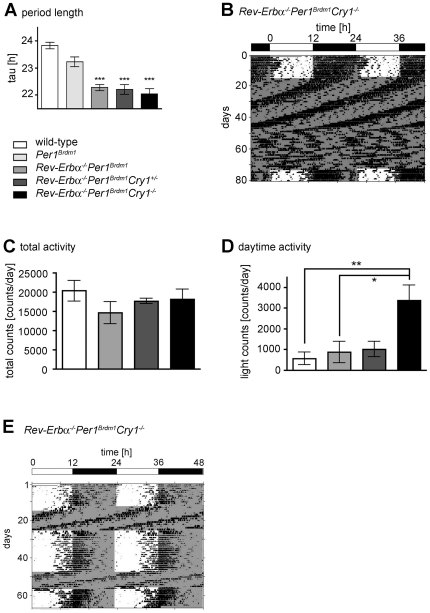
In order to unravel whether a change in Cry1 light induction is related to the observed high amplitude resetting in our double mutant mice, we generated Rev-Erbα−/−Per1Brdm1Cry1−/− triple mutant animals (Fig. S4). First, we investigated the circadian phenotype of these mice under both DD and LD conditions. In constant darkness, the circadian period lengths of Rev-Erbα−/−Per1Brdm1Cry1−/− (22.04±0.19 h; N = 8), heterozygous Rev-Erbα−/−Per1Brdm1Cry1+/− (22.20±0.18 h; N = 12) and Rev-Erbα−/−Per1Brdm1 mice (22.27±0.11 h; N = 16) carrying the same genetic background were indistinguishable from each other (Fig. 5A). In contrast to Rev-Erbα−/−Per1Brdm1 double mutant animals, triple mutants become arrhythmic after about 1 month in DD (Fig. 5B).
Figure 5. Wheel-running behaviour of Rev-Erbα−/−Per1Brdm1Cry1−/− mice.
(A) Average free-running period (tau) in constant darkness. Data are represented as mean ± SEM. One-way ANOVA was performed to compare the free-running period length of Per1Brdm1 (23.22±0.19 h; N = 5), Rev-Erbα−/−Per1Brdm1 (22.27±0.11 h; N = 16), Rev-Erbα−/−Per1Brdm1Cry1+/− (22.20±0.18 h; N = 12) and Rev-Erbα−/−Per1Brdm1Cry1−/− (22.04±0.19 h; N = 8) mutant to wild-type mice (23.82±0.12 h; N = 6). *** p<0.001. (B) Arrhythmicity of Rev-Erbα−/−Per1Brdm1Cry1−/− triple mutant in constant darkness DD. Locomotor activity record under both light/dark (LD) and DD conditions. Most animals become arrhythmic after about 1 month in DD. Black bars represent wheel-revolutions. The actogram is double-plotted with the activity of the following light/dark cycle plotted to the right and below the previous light/dark cycle. The white and black bar on the top of the actogram indicates light and darkness, respectively. (C) There is no difference in total activity between the genotypes. Data are represented as mean ± SEM. (D) Rev-Erbα−/−Per1Brdm1Cry1−/− triple mutant mice display increased daytime activity. Data are represented as mean ± SEM. Unpaired t-test was performed to determine the significant differences between the genotypes. * p = 0.0334, ** p = 0.0087. (E) Masking defect. Typical actogram of a Rev-Erbα−/−Per1Brdm1Cry1−/− triple mutant mouse under LD and DD conditions. One can clearly see that the mouse displays increased daytime activity prior to lights off under LD conditions.
Under LD conditions Rev-Erbα−/−Per1Brdm1Cry1−/− triple mutants showed comparable total activity (Fig. 5C) but significantly elevated daytime activity compared to the other genotypes (Fig. 5D). In most cases, daytime activity was clustered before dark onset (Fig. 5E) and in a few mice the dark activity extended into the light phase (Fig. 5B). This indicates reduced masking by light in which Cry1 may play an important role. This correlates with the observed advance in the phase angle of these animals (Fig. 5E).
Given our findings described above, light resetting was difficult to assess, since the triple mutants become arrhythmic in DD before establishing a stable free running rhythm. Under LD conditions activity onset can't be defined precisely (Fig. 5B, 5E), due to impaired masking. Hence, it is not possible to determine accurately whether the slight light induction of the Cry1 gene in Rev-Erbα−/−Per1Brdm1 mice plays a role in the high amplitude resetting observed in these animals.
Discussion
Here we show that mice deficient for both Rev-Erbα−/− and Per1Brdm1 display a dramatically shorter circadian period in DD compared to single mutants and wild-type animals (Fig. 1A–E). As reported previously, Rev-Erbα−/− and Per1Brdm1 single mutant mice exhibited significantly shorter circadian periods relative to wild-type animals [9], [20]. Hence the shorter period observed in the double mutants might indicate that the concomitant absence of these two clock genes has a cumulative effect on circadian period in DD.
Resetting studies revealed that Rev-Erbα−/−Per1Brdm1 double mutants show high amplitude resetting (Fig. 3A–F). A similar phenotype has been reported by Vitaterna et al. [25] for heterozygous Clock mutant mice. These mice exhibit a Type 0 PRC in response to 6-h light pulses. Interestingly, light induction of both Per1 and Per2 was not affected in the Clock+/− mutants after 6-hour light exposure. However, basal expression levels of Per genes were significantly blunted in both Clock homo- and heterozygous mice. In contrast to the observations by Vitaterna et al. [25] we could not detect any dampening in Per2 expression in our animals. In situ hybridization profiles for Per2 were completely identical between Rev-Erbα−/−Per1Brdm1 double mutant and wild-type animals (Fig. 2D). Moreover, Vitaterna et al. [25] could not detect any differences in light induction of either Per1 or Per2 between Clock heterozygous and wild-type mice [25]. By contrast, we found that Per2 induction was stronger in Rev-Erbα−/−Per1Brdm1 double mutants compared to wild-types (Fig. 4C). The differences in the two studies might stem from the protocol used for light exposure. Whereas Vitaterna et al. [25] used light pulses of 6-hour duration; our protocol is based on short light exposures of 15 minutes. Hence, the mechanisms studied in the two reports are probably different. However, consistent with the results in Clock mutants, the Rev-Erbα−/−Per1Brdm1 double mutant animals show dampened oscillation of several clock genes in the SCN (Fig. 2) supporting the concept of decreased pacemaker amplitude leading to high amplitude phase shifts.
To test functional consequences of a Type 0-like PRC, we performed jet-lag experiments, one advancing the LD cycle by 8 hours, the other delaying it by 8 hours. We find that Rev-Erbα−/−Per1Brdm1 double mutants adapt significantly faster in both paradigms (Fig. 3G, 3H). This is consistent with the findings described above, that the double mutants display larger responses to a light pulse. Hence, larger responses lead to a faster alignment between the clock phase and the LD cycle. Our findings suggest that the actual adaptation to a new time would be fast but is slowed down by components of the circadian clock, which affect various signalling pathways. This is in line with previous findings where it was demonstrated that rapid phase resetting in the liver is counteracted by active signalling pathways [26].
Whether Type 0 or a Type 1 resetting is exhibited in an animal depends not only on species but also on the strength of the stimulus. Increasing the light dose can convert a Type 1 into a Type 0 PRC [27]. In this study we used light pulses of 400 lux and we did not vary intensities. We rather modulated the receiving end of the light input by genetic mutations [27]. However, light seems to reach the SCN in all genotypes, because the immediate early gene cFos was induced comparably at CT22 (Fig. 4E). Hence, it is likely, that alterations in core clock components involved in the interpretation of the light signal are the underlying reason for the observed PRC in Rev-Erbα−/−Per1Brdm1 double mutants (Fig. 4A, 4C). However, we can't exclude differences between the genotypes in terms of light sensitivity.
Surprisingly, we observed that Cry1 became slightly light inducible in Rev-Erbα−/−Per1Brdm1 double mutant mice at CT22 but not at CT14 (Fig. 4A, 4B). So far, it has never been reported that Cry1 can be induced by light in mammals [28], [29]. In lower vertebrates like zebrafish (Danio rerio), however, Cry1a acts as a light-signalling molecule [18]. Its induction as well as that of Per2 was found to be critical for light-induced phase shifts in this species. Interestingly, we observed an induction of Per2 in our double mutants at CT22 (Fig. 4C) [30], [31]. Our findings in terms of clock gene induction in the Rev-Erbα−/−Per1Brdm1 double mutant mice parallel the ones observed in zebrafish. Interestingly, zebrafish cell lines show high amplitude phase shifts in response to light [18], [32] and zebrafish tissues are directly light responsive [33], [34]. Therefore, we tested whether tissues and cells of Rev-Erbα−/−Per1Brdm1 double mutant mice would respond directly to light. However, our experiments indicate that this is not the case (Fig. S5, S6).
Taken together it appears that in the course of evolution complex mechanisms have evolved to adapt land living vertebrates to their environment. In particular, the effects of strong light exposure had to be controlled. This might have led to new functions of Rev-erbα and Per1 keeping behavioral light responses at a moderate level. Our results suggest that loss of Rev-Erbα and Per1 results in a gain of function phenotype in mice and light responses become elevated. This leads to faster adaptation to light stimuli. Under natural conditions (e.g. nocturnal lightning) this would result in jumping of clock phase, which in turn could have adverse effects on the organism. Rev-Erbα and Per1 are probably involved together in attenuating responses to unexpected brief single light pulses to maintain coherence in the circadian system of mammals.
Materials and Methods
Animals
All animal work was performed in accordance with the guidelines of the Schweizer Tierschutzgesetz (TSchG, SR455, Abschnitt 2: Art. 5+7, Abschnitt 5: Art. 11 and Abschnitt 6: Art.12–19) and was approved by the state veterinarian of Fribourg (permit FR 101/07) and in accordance with the declaration of Helsinki. Male and female mice were used in this study in equal ratios for each genotype. Animals were raised in LD 12∶12 h and were not in a different cycle before the DD or LL experiments.
The Rev-Erbα−/−Per1Brdm1 double mutant mice used in this study were generated by crossing Rev-Erbα−/− [9] and Per1Brdm1 [20] single mutants. Intercrossing double heterozygous animals produced Rev-Erbα−/−, Per1Brdm1, Rev-Erbα−/−Per1Brdm1 and wild-type animals with a matching genetic background (mixed strain 129Sv/C57Bl/6). Homozygous breeding pairs were established using F2 offspring. To minimize genetic drift, matings were kept together as long as possible. The animals were regularly backcrossed to minimize epigenetic effects.
To obtain Rev-Erbα−/−Per1Brdm1Cry1−/− triple mutant mice, animals double mutant for Rev-Erbα−/−Per1Brdm1 or Per1Brdm1Cry1−/− [35] were crossed with each other. The Rev-Erbα+/−Per1Brdm1Cry1+/− offspring were then intercrossed to obtain the F2 generation. To get triple mutant animals, two mating types were set-up using the F2 mice: in the first case both parents were Rev-Erbα−/−Per1Brdm1Cry1+/−, in the second case one parent was Rev-Erbα−/−Per1Brdm1Cry1+/− while the other was Rev-Erbα+/−Per1Brdm1Cry1−/−. For the experiments we used mice of the F2 (Per1Brdm1, Rev-Erbα−/−Per1Brdm1, Rev-Erbα−/−Per1Brdm1Cry1+/−) and F3 (Rev-Erbα−/−Per1Brdm1, Rev-Erbα−/−Per1Brdm1Cry1+/−, Rev-Erbα+/−Per1Brdm1Cry1−/−) generation.
The genotype of the offspring was determined by PCR. The PCR protocol for Rev-Erbα was according to Preitner et al. 2002 [9]. The following primers were used:
Cry1_1: 5′-GCA TGA CCC CTC TGT CTG AT-3′
Cry1_2: 5′-TGA ATG AAC TGC AGG ACG AG-3′
Cry1_3: 5′-AAC ACG CAG ATG CAG TCG-3′
Per1_1: 5′-ACA AAC TCA CAG AGC CCA TCC-3′
Per1_2: 5′-ATA TTC CTG GTT AGC TGT AGG-3′
Per1_3: 5′-CGC ATG CTC CAG ACT GCC TTG-3′
Rev-Erbα_1: 5′-CAC CTT ACA CAG TAG CAC CAT GCC ATT CA-3′
Rev-Erbα_2: 5′-AAA CCA GGC AAA GCG CCA TTC GCC ATT CA-3′
Rev-Erbα_3: 5′-CCA GGA AGT CTA CAA GTG GCC ATG GAA GA-3′
The final primer concentration was 1.0 µM for Cry1 and 0.5 µM for Per1 and Rev-Erbα. The dNTP (Roche) concentration was 0.4 mM for Cry1 and 0.2 mM for Per1 and Rev-Erbα. The final MgCl2 concentration was 3.0 mM for Cry1, 1.5 mM for Per1 and 3.5 mM for Rev-Erbα. To improve annealing, 6 nM (NH4)2SO4 was added to the PCR reaction mix for Cry1 and Per1 or 12 nM for Rev-Erbα. 2.5 U Taq DNA polymerase (Qiagen) were used per 50 µl reaction for Per1 and 1.25 U per 25 µl reaction for Rev-Erbα and Cry1. A final concentration of 0.25x and 0.2x Q-solution was used to increase PCR specificity of the Cry1 and Per1 reaction, respectively. An initial denaturation was done at 94°C for 2 min. Subsequent denaturation was done at 94°C for 30 s followed by an annealing step of 30 s. The annealing temperature was 56°C for Per1 and 62°C for Cry1 and Rev-Erbα. For Per1 and Rev-Erbα, the elongation step was performed at 72°C for 1 min. For Cry1, the elongation step at 72°C took 7 min. After 34 (Per1), 35 (Cry1) or 36 (Rev-Erbα) cycles, the PCR was ended with a final extension at 72°C for either 7 (Cry1) or 10 min (Per1, Rev-Erbα). From time to time, Southern blot analysis was used to confirm the PCR results obtained for Per1 [20], [36] and Cry1 [37].
Locomotor activity monitoring and circadian phenotype analysis
Mice housing and handling were performed as described earlier [21]. Animals were entrained in LD 12∶12 h for 7–15 days before they were released into DD or LL. Activity was assessed with a running-wheel and evaluated using the ClockLab software package (Actimetrics). Activity records were double plotted in threshold format for 6-min bins. Period length was assessed by χ2 periodogram analysis for days 4–10 in DD or LL. To determine light induced phase shifts, an Aschoff Type I protocol was used [22]. Animals were allowed to stabilize their free-running rhythm for at least 2 months prior to the light pulse. The circadian time (CT) at the beginning of the light pulse was calculated for every mouse individually. The phase response curve was established administering 15 min light pulses at CT0 (N [wild-type/Rev-Erbα−/−Per1Brdm1] = 8/11), CT2 (N = 11/4), CT10 (N = 8/8), CT14 (N = 15/8), CT18 (N = 14/15), CT20 (N = 8/10), and CT22 (N = 13/6).
Mice subjected to experimental jet lag were entrained to LD 12∶12 h. This cycle was then advanced or delayed by 8 hours. Animals were allowed to re-entrain to the new phase for 1 month. The onsets between the two LD phases were determined. The number of days it took the animal to shift from the old onset to the new onset was counted.
In situ hybridization
Locomotor activity was monitored for each mouse to properly determine activity onsets, which is necessary to calculate CT values. To determine circadian gene expression, adult mice were first anesthetized with Attane™ Isoflurane (Provet AG) and then sacrificed on day 6 in DD. For light induction experiments, animals were kept in DD for about 2 months before they were exposed to a 15 min light pulse (400 lux) at different CTs. 45 min after the end of the light pulse, the mice were sacrificed. Control animals were sacrificed without prior light exposure.
Specimen preparation and in situ hybridization were carried out as described previously [30]. Briefly, the 35S-UTP (1250 Ci/mmol, PerkinElmer) labelled riboprobes were synthesized using the RNAMaxx™ High Yield transcription kit (Stratagene) according to manufacturer's protocol. The Bmal1 probe was made from a cDNA corresponding to nucleotides (nt) 654–1290 (accession no. AF015953; [38]), Cry1 corresponds to nt 190–771 (AB000777; [38]), Cry2 to nt 231–945 (AF156987), Per2 to nt 229–768 (AF036893; [30]), cFos to nt 237–332 [30]. 7 µm thick paraffin sections were dewaxed, rehydrated and fixed in 4% paraformaldehyde. Sections were then permeabilized using a proteinase K (Roche) digestion before they were fixed again and acetylated. After serial dehydration, hybridization was performed over-night at 55°C in a humid chamber. Stringency washes were carried out at 63°C. Slides were subjected to a ribonuclease A (Sigma) digestion and then dehydrated in graded ethanol series. Quantification was performed by densitometric analysis (GS-700 or GS-800, BioRad) of autoradiography films (Amersham Hyperfilm) using the Quantity One software (BioRad). Data from the SCN were normalized subtracting the optical density measured in the lateral hypothalamus next to the SCN. For each experiment at least 3 animals per genotype were used and 4 to 9 adjacent SCN sections per animal were analyzed. Relative RNA abundance values were calculated by defining the highest wild-type mean value of each experiment as 100%. For statistical analysis, all normalized values obtained for one brain were averaged to obtain one final value per animal.
Statistical analysis
Significant differences were determined using GraphPad Prism 4 software. Depending on the type of data, either unpaired t-test, one- or two-way ANOVA with Bonferroni post-test was performed. Values were considered significantly different with p<0.05 (*), p<0.01 (**), or p<0.001 (***).
For supplemental data see supplemental materials and methods (Methods S1).
Supporting Information
Supplemental Materials and Methods
(0.04 MB DOC)
PCR analysis of mouse tail DNA for Rev-Erbα and Per1. Primers for Rev-Erbα amplify a fragment of about 340 nucleotides (nt) on the wild-type allele and of about 200 nt on the mutant allele. The PCR for Per1 amplifies a fragment of about 290 nt on the mutant allele and of about 450 nt on the wild-type (wt) allele.
(0.42 MB TIF)
Breeding statistics. (A) Distribution of genotypes in the F2 generation born to Rev-Erbα+/−Per1+/Brdm1 double heterozygous parents. Black bars represent the expected Mendelian distribution while white bars display the observed ratios. The x-axis indicates the genotype of the F2 offspring. The percent of the total mouse number displaying a certain genotype is plotted on the y-axis. (B) Litter frequency of wild-type (white), Rev-Erbα−/−Per1Brdm1 double homozygous (dark grey), and Rev-Erbα+/−Per1+/Brdm1 double heterozygous (light grey) breeding pairs. Data are represented as mean ± SEM. On average, wild-type matings gave birth to a new litter every 32.22±2.06 days (N = 41), Rev-Erbα−/−Per1Brdm1 double homozygous breeding pairs every 32.12±2.33 days (N = 33), and Rev-Erbα/Per1 double heterozygous couples every 26.42±1.04 days (N = 60). One-way ANOVA with Bonferroni's multiple comparison was performed to compare the litter frequency of the three genotypes. * p<0.05. (C) Number of pups born to wild-type (white), Rev-Erbα−/−Per1Brdm1 double homozygous (dark grey), and Rev-Erbα+/−Per1+/Brdm1 double heterozygous (light grey) breeding pairs. Data are represented as mean ± SEM. On average, wild-type couples gave birth to 6.76±0.51 pups (N = 38), Rev-Erbα−/−Per1Brdm1 double homozygous breeding pairs to 6.18±0.43 pups (N = 33), and Rev-Erbα/Per1 double heterozygous couples to 6.94±0.31 pups (N = 69). One-way ANOVA with Bonferroni's multiple comparison did not reveal any significant differences in litter size between the three genotypes.
(1.63 MB TIF)
Total and daytime wheel-running activity. (A) Total wheel-running activity of mice kept under LD 12∶12 h (7 days). On average, wild-type mice make 17′738±808 wheel-revolutions per day (N = 32), Rev-Erbα−/−Per1Brdm1 double mutants 16′307±929 (N = 29), Rev-Erbα−/− single mutants 14′551±1420 (N = 17), and Per1Brdm1 single mutants 17′718±1195 (N = 16). (B) Rest (rho) phase activity of mice kept under LD 12∶12 h (7 days). Wild-type mice spend 5.98±1.18% (N = 32) of their total activity during the day, Rev-Erbα−/−Per1Brdm1 double mutants 6.74±1.4% (N = 29), Rev-Erbα−/− single mutants 4.6±1.4% (N = 17), and Per1Brdm1 single mutants 6.76±2.3% (N = 16). Data are represented as mean ± SEM. One-way ANOVA with Bonferroni's multiple comparison did not reveal any significant difference between genotypes for both total and rho phase activity. (C) 24-h average distribution of activity for wild-type (black; N = 32), Rev-Erbα−/−Per1Brdm1 double mutant (red; N = 29), Rev-Erbα−/− single mutant (green; N = 17), and Per1Brdm1 single mutant mice (blue; N = 15) kept in LD 12∶12 h. The black and white bar on the bottom depicts the light and dark phase, respectively. Data are represented as mean only. The x-axis indicates the phase angle while the y-axis displays the activity as average counts/minute over a ten-minute interval. Statistical analysis did not reveal any difference between the genotypes.
(4.80 MB TIF)
Characterization of Rev-Erbα−/−Per1Brdm1Cry1−/− triple mutant mice by PCR. PCR analysis of mouse tail DNA for Cry1. Fragments of 2.3 and 3.1 kbp were amplified on the wild-type and mutant allele, respectively.
(2.65 MB TIF)
Ear cells of Rev-Erbα−/−Per1Brdm1 double mutant mice are not directly light responsive. (A) Cry1 and (B) Per2 induction in the ear after a light pulse at ZT22 measured in wild-type (black), Rev-Erbα−/−Per1Brdm1 (red), Rev-Erbα−/− (green), and Per1Brdm1 (blue) mice. Mice were sacrificed at ZT22. The ears were kept in cell culture medium and immediately subjected to a 30 min light pulse. Another 30 min later the tissue was harvested for further analysis. Values are represented as mean ± SEM (N = 3). Two-way ANOVA with Bonferroni post-test did not detect significant differences between genotypes.
(0.77 MB TIF)
Mouse dermal fibroblasts of Rev-Erbα−/−Per1Brdm1 double mutant mice are not directly light responsive. Immortalized MDFs were subjected to a 30 min light pulse. A UV-filter was between the cells and the light source. The induction of Cry1 (A, B), Per2 (C, D) and Bmal1 (E, F) were assessed using real-time PCR during 8 h. Rev-Erbα−/−Per1Brdm1 double mutant MDFs are plotted on the left side (A, C, E) while the wild-type cells are on the right side (B, D, F). Data are represented as mean ± SD (N = 2 except for wild-type time-point 0 where N = 1). Two-way ANOVA with Bonferroni post-test did not reveal any significant differences between the light pulse samples and the dark controls. Values were normalized to the highest mean level within an experiment.
(1.34 MB TIF)
Acknowledgments
The authors thank Prof. Russell Foster for discussions and Prof. Ueli Schibler for Rev-erbα ko mice. Moreover they thank S. Baeriswyl and A. Sargsyan for technical assistance and Dr. J.A. Ripperger and Dr. I. Schmutz for critically reading the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work has been supported by the Swiss National Science Foundation, the European Union FP6 Integrated Project EUCLOCK and the state of Fribourg (Switzerland). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 3.Moore RY, Lenn NJ. A retinohypothalamic projection in the rat. J Comp Neurol. 1972;146:1–14. doi: 10.1002/cne.901460102. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 6.Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, et al. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 7.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 8.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 9.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 10.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 11.Giguere V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- 12.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Aschoff J. Desynchronization and resynchronization of human circadian rhythms. Aerosp Med. 1969;40:844–849. [PubMed] [Google Scholar]
- 14.Daan S, Pittendrigh CS. A functional analysis of circadian pacemakers in rodents. II. The variability of phase response curves. J Comp Physiol. 1976;106:253–266. [Google Scholar]
- 15.Pittendrigh CS. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–184. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Lin FJ, Song W, Meyer-Bernstein E, Naidoo N, Sehgal A. Photic signaling by cryptochrome in the Drosophila circadian system. Mol Cell Biol. 2001;21:7287–7294. doi: 10.1128/MCB.21.21.7287-7294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamai TK, Young LC, Whitmore D. Light signaling to the zebrafish circadian clock by Cryptochrome 1a. Proc Natl Acad Sci U S A. 2007;104:14712–14717. doi: 10.1073/pnas.0704588104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. MPer1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 20.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 21.Jud C, Schmutz I, Hampp G, Oster H, Albrecht U. A guideline for analyzing circadian wheel-running behavior in rodents under different lighting conditions. Biol Proced Online. 2005;7:101–116. doi: 10.1251/bpo109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aschoff J. Response curves in circadian periodicity. In: Aschoff J, editor. Circadian Clocks. Amsterdam: North-Holland Publishing Co; 1965. pp. 95–111. [Google Scholar]
- 23.Winfree AT. Integrated view of resetting a circadian clock. J Theor Biol. 1970;28:327–374. doi: 10.1016/0022-5193(70)90075-5. [DOI] [PubMed] [Google Scholar]
- 24.Winfree AT. Biomathematics. New York: Springer; 1980. The geometry of biological time. [Google Scholar]
- 25.Vitaterna MH, Ko CH, Chang AM, Buhr ED, Fruechte EM, et al. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci U S A. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson CH. Phase response curves: what can they tell us about circadian clocks? In: Hiroshige T, Honma K, editors. Circadian clocks from cell to human. Sapporo: Hokkaido University Press; 1992. pp. 209–249. [Google Scholar]
- 28.Field MD, Maywood ES, O'Brien JA, Weaver DR, Reppert SM, et al. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 29.Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, et al. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 30.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 31.Yan L, Silver R. Differential induction and localization of mPer1 and mPer2 during advancing and delaying phase shifts. Eur J Neurosci. 2002;16:1531–1540. doi: 10.1046/j.1460-9568.2002.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallone D, Gondi SB, Whitmore D, Foulkes NS. E-box function in a period gene repressed by light. Proc Natl Acad Sci U S A. 2004;101:4106–4111. doi: 10.1073/pnas.0305436101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature. 2000;404:87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- 34.Whitmore D, Foulkes NS, Strahle U, Sassone-Corsi P. Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci. 1998;1:701–707. doi: 10.1038/3703. [DOI] [PubMed] [Google Scholar]
- 35.Oster H, Baeriswyl S, Van Der Horst GT, Albrecht U. Loss of circadian rhythmicity in aging mPer1-/-mCry2-/- mutant mice. Genes Dev. 2003;17:1366–1379. doi: 10.1101/gad.256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez-Solis R, Zheng H, Whiting J, Krumlauf R, Bradley A. Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell. 1993;73:279–294. doi: 10.1016/0092-8674(93)90229-j. [DOI] [PubMed] [Google Scholar]
- 37.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 38.Oster H, Yasui A, van der Horst GT, Albrecht U. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 2002;16:2633–2638. doi: 10.1101/gad.233702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods
(0.04 MB DOC)
PCR analysis of mouse tail DNA for Rev-Erbα and Per1. Primers for Rev-Erbα amplify a fragment of about 340 nucleotides (nt) on the wild-type allele and of about 200 nt on the mutant allele. The PCR for Per1 amplifies a fragment of about 290 nt on the mutant allele and of about 450 nt on the wild-type (wt) allele.
(0.42 MB TIF)
Breeding statistics. (A) Distribution of genotypes in the F2 generation born to Rev-Erbα+/−Per1+/Brdm1 double heterozygous parents. Black bars represent the expected Mendelian distribution while white bars display the observed ratios. The x-axis indicates the genotype of the F2 offspring. The percent of the total mouse number displaying a certain genotype is plotted on the y-axis. (B) Litter frequency of wild-type (white), Rev-Erbα−/−Per1Brdm1 double homozygous (dark grey), and Rev-Erbα+/−Per1+/Brdm1 double heterozygous (light grey) breeding pairs. Data are represented as mean ± SEM. On average, wild-type matings gave birth to a new litter every 32.22±2.06 days (N = 41), Rev-Erbα−/−Per1Brdm1 double homozygous breeding pairs every 32.12±2.33 days (N = 33), and Rev-Erbα/Per1 double heterozygous couples every 26.42±1.04 days (N = 60). One-way ANOVA with Bonferroni's multiple comparison was performed to compare the litter frequency of the three genotypes. * p<0.05. (C) Number of pups born to wild-type (white), Rev-Erbα−/−Per1Brdm1 double homozygous (dark grey), and Rev-Erbα+/−Per1+/Brdm1 double heterozygous (light grey) breeding pairs. Data are represented as mean ± SEM. On average, wild-type couples gave birth to 6.76±0.51 pups (N = 38), Rev-Erbα−/−Per1Brdm1 double homozygous breeding pairs to 6.18±0.43 pups (N = 33), and Rev-Erbα/Per1 double heterozygous couples to 6.94±0.31 pups (N = 69). One-way ANOVA with Bonferroni's multiple comparison did not reveal any significant differences in litter size between the three genotypes.
(1.63 MB TIF)
Total and daytime wheel-running activity. (A) Total wheel-running activity of mice kept under LD 12∶12 h (7 days). On average, wild-type mice make 17′738±808 wheel-revolutions per day (N = 32), Rev-Erbα−/−Per1Brdm1 double mutants 16′307±929 (N = 29), Rev-Erbα−/− single mutants 14′551±1420 (N = 17), and Per1Brdm1 single mutants 17′718±1195 (N = 16). (B) Rest (rho) phase activity of mice kept under LD 12∶12 h (7 days). Wild-type mice spend 5.98±1.18% (N = 32) of their total activity during the day, Rev-Erbα−/−Per1Brdm1 double mutants 6.74±1.4% (N = 29), Rev-Erbα−/− single mutants 4.6±1.4% (N = 17), and Per1Brdm1 single mutants 6.76±2.3% (N = 16). Data are represented as mean ± SEM. One-way ANOVA with Bonferroni's multiple comparison did not reveal any significant difference between genotypes for both total and rho phase activity. (C) 24-h average distribution of activity for wild-type (black; N = 32), Rev-Erbα−/−Per1Brdm1 double mutant (red; N = 29), Rev-Erbα−/− single mutant (green; N = 17), and Per1Brdm1 single mutant mice (blue; N = 15) kept in LD 12∶12 h. The black and white bar on the bottom depicts the light and dark phase, respectively. Data are represented as mean only. The x-axis indicates the phase angle while the y-axis displays the activity as average counts/minute over a ten-minute interval. Statistical analysis did not reveal any difference between the genotypes.
(4.80 MB TIF)
Characterization of Rev-Erbα−/−Per1Brdm1Cry1−/− triple mutant mice by PCR. PCR analysis of mouse tail DNA for Cry1. Fragments of 2.3 and 3.1 kbp were amplified on the wild-type and mutant allele, respectively.
(2.65 MB TIF)
Ear cells of Rev-Erbα−/−Per1Brdm1 double mutant mice are not directly light responsive. (A) Cry1 and (B) Per2 induction in the ear after a light pulse at ZT22 measured in wild-type (black), Rev-Erbα−/−Per1Brdm1 (red), Rev-Erbα−/− (green), and Per1Brdm1 (blue) mice. Mice were sacrificed at ZT22. The ears were kept in cell culture medium and immediately subjected to a 30 min light pulse. Another 30 min later the tissue was harvested for further analysis. Values are represented as mean ± SEM (N = 3). Two-way ANOVA with Bonferroni post-test did not detect significant differences between genotypes.
(0.77 MB TIF)
Mouse dermal fibroblasts of Rev-Erbα−/−Per1Brdm1 double mutant mice are not directly light responsive. Immortalized MDFs were subjected to a 30 min light pulse. A UV-filter was between the cells and the light source. The induction of Cry1 (A, B), Per2 (C, D) and Bmal1 (E, F) were assessed using real-time PCR during 8 h. Rev-Erbα−/−Per1Brdm1 double mutant MDFs are plotted on the left side (A, C, E) while the wild-type cells are on the right side (B, D, F). Data are represented as mean ± SD (N = 2 except for wild-type time-point 0 where N = 1). Two-way ANOVA with Bonferroni post-test did not reveal any significant differences between the light pulse samples and the dark controls. Values were normalized to the highest mean level within an experiment.
(1.34 MB TIF)