Abstract
Tobacco smoke and its metabolites are carcinogens that increase tissue oxidative stress and induce target tissue inflammation. We hypothesized that genetic variation of inflammatory pathway genes plays a role in tobacco-related carcinogenesis and is modified by tobacco smoking. We evaluated the association of 12 single nucleotide polymorphisms of eight inflammation-related genes with tobacco-related cancers (lung, oropharynx, larynx, esophagus, stomach, liver, bladder, and kidney) using three case-control studies from: Los Angeles (population-based; 611 lung and 553 upper aero-digestive tract cancer cases and 1,040 controls); Taixing, China (population-based; 218 esophagus, 206 stomach, 204 liver cancer cases, and 415 controls); and Memorial Sloan-Kettering Cancer Center (hospital-based; 227 bladder cancer cases and 211 controls). After adjusting for age, education, ethnicity, gender, and tobacco smoking, IL10 rs1800871 was inversely associated with oropharyngeal cancer (CT+TT versus CC adjusted odds ratio [aOR]: 0.69, 95% confidence interval [CI]: 0.50–0.95), and among never smokers was positively associated with lung cancer (TT versus CT+CC aOR: 2.5, 95% CI: 1.3–5.1) and inversely with oropharyngeal cancer (CT+TT versus CC aOR: 0.63, 95% CI: 0.41–0.95). Among all pooled never smokers (588 cases and 816 controls), TNF rs1799964 was inversely associated with smoking-related cancer (CC versus CT+TT aOR: 0.36, 95% CI: 0.17–0.77). Bayesian correction for multiple comparisons suggests that chance is unlikely to explain our findings (although epigenetic mechanisms may be in effect), which support our hypotheses, suggesting that IL10 rs1800871 is a susceptibility marker for oropharyngeal and lung cancers, and that TNF rs1799964 is associated with smoking-related cancers among never smokers.
Keywords: IL10, TNF, single nucleotide polymorphisms, inflammation, tobacco-related cancer
Introduction
Tobacco smoking is a major risk factor for malignancies of the lung, upper aero-digestive tract (UADT), stomach, pancreas, liver, kidney, urinary tract, uterine cervix, and bone marrow. Epithelial cells in many of these organs are repeatedly exposed to components and metabolites of tobacco smoke, which are carcinogenic and potent inducers of inflammation. The high concentration of free radicals contained in and generated by tobacco smoke can lead to cancer through oxidative DNA damage mediated by inflammation-associated production of reactive oxygen species.
Chronic inflammation, characterized in part by altered cytokine levels, is believed to play a role in tumor initiation and promotion. Inflammatory conditions such as ulcerative colitis and inflammatory bowel disease have well-established associations with colorectal cancer,1 and lung cancer has been connected to inflammatory diseases such as tuberculosis and pneumonia.2, 3 Additionally, cancers of the stomach, liver, and esophagus have been attributed to chronic inflammation as a result of persistent infection.4 An individual’s cancer risk may be affected by genetic variations in essential cell regulatory pathways5, 6 and inflammatory responses,7, 8 and tobacco smoking may modify these effects,9, 10 suggesting that genetic variation may be important susceptibility markers for tobacco-related cancers.
In the current study, we use a pathway-based approach using data from three case-control studies (Los Angeles [LA] County, Taixing, China, and Memorial Sloan-Kettering Cancer Center [MSKCC]) to test the hypotheses that single nucleotide polymorphisms (SNPs) in inflammation-related genes are associated with smoking-related cancers of the lung, oropharynx, larynx, esophagus, stomach, liver, bladder, and kidney, and that their effects are modified by tobacco smoking.
Material and methods
Study design and participants
Detailed descriptions of the three case-control studies reported in this manuscript have been published for the LA,11 Taixing,12 and MSKCC13 and studies, which are briefly described below. All study participants provided written consent, and study protocols were approved by appropriate review boards. Subjects not meeting study-specific inclusion criteria were excluded from enrollment.
The LA study was population-based, consisting of histologically confirmed incident lung (n = 611) and UADT (n = 553) cancer cases obtained from the LA County cancer registry administered through the Cancer Surveillance Program at the University of Southern California; 1,040 controls without a history of lung or UADT cancer were matched on gender, age, and residential neighborhood, using an algorithm to identify eligible controls from a census of each case’s neighborhood. To be eligible, subjects had to be (a) 18–65 years of age during 1999–2004; (b) a resident of LA County at time of diagnosis (cases) or recruitment (controls); and (c) able to speak English or Spanish, or have a translator available at home. Recruitment rates among eligible cases were 39% and 46% for lung and UADT cases, respectively, and 79% for controls. Buccal cell samples were obtained at the end of interviews for DNA analysis.
The Taixing study was also population-based and conducted in 2000. Newly diagnosed and pathologically or clinically confirmed cases of stomach (n = 206), liver (n = 204), and esophageal (n = 218) cancers were obtained from the Taixing Tumor Registry operated through the Taixing Center for Disease Control and Prevention. A common group of healthy controls from the general population registry (n = 415) was frequency matched by age, gender, and village. Eligibility criteria required that all subjects were (a) at least 20 years old; (b) in stable medical condition; and (c) living in Taixing for at least ten years. Recruitment rates were 67%, 65%, and 57% for eligible esophagus, stomach, and liver cancer cases, respectively, and 89% for controls. Blood samples were collected at the end of interviews for DNA analysis.
The MSKCC study, conducted during 1993–1997, was hospital-based and consisted of 227 pathologically confirmed bladder cancer cases sampled from the MSKCC who had recently been diagnosed or undergone bladder surgery. Two-hundred–eleven (211) controls who had resided in the United States for at least a year were recruited from the MSKCC blood bank or from MSKCC patients who did not have cancer diagnoses and were in stable medical condition. Ninety-five percent of cases and 92% of controls agreed to participate; blood samples were collected at the end of interviews for DNA analysis.
Standardized questionnaires appropriate for each of the three studies were administered in person by trained staff. Data collected across all three studies included demographic information; detailed behavioral factors such as diet, alcohol use, and exposure to tobacco smoke; other environmental and occupational exposures; personal and family medical histories; and other exposures considered known or possible risk factors for cancers specific to each study. Between-study variation among the common demographic variables was greatest for race/ethnicity, which was most heterogeneous in the LA study (59.3% White, 17.0% Hispanic, 11.9% African-American, 8.7% Asian-American, and 3.0% other) and least in the Taixing study (100% Chinese); less than 5% of MSKCC study participants were non-White.
SNP selection and analysis
We focused on functional and potentially functional SNPs (such as amino acid-changing polymorphisms) and SNPs located in regions regulating gene transcription (such as promoter areas). We decided a priori to exclude SNPs from analysis that did not meet the following criteria among the study-specific control groups: (a) minor allele frequency (MAF) ≥ 5%; (b) Hardy-Weinberg equilibrium (HWE) P-value > Bonferroni-adjusted P-value; and (c) genotyping call rate ≥ 80% for SNPlex and ≥ 95% for TaqMan.
The majority of SNPs that violated HWE in our initial pool of SNPs were excluded from analysis due to low genotyping rates or minor allele frequencies. After using a Bonferroni-adjusted cutpoint among the study-specific control groups to assess deviations from HWE, one SNP was excluded from the LA study (TNF rs1799724 HWE P value < 0.0001), another SNP was dropped from the Taixing study (IL13 rs20541 HWE P value < 0.0001), and one more from the MSKCC study (IL13 HWE P value = 0.00018). The final pool of SNPs included in our study had allele frequencies that were in the expected range of normal variation, and none had HWE P < 0.05 among the study-specific control groups. Six SNPs met these criteria across all three studies (IL10 rs1800871; TNF rs1799964 and rs1800629; LTA rs909253; IFNGR1 rs11914; and IFNG rs2069705) while another six met inclusion criteria for at least one of the studies (IL10 rs1800872 and rs1800896; IL1A rs17561; IL1B rs1143627 and rs16944; and IL6 rs1800796). These twelve SNPs and details of their inclusion criteria are reported in Supplementary Table S1. Because population substructure can affect a SNP’s distribution, we also examined HWE by ethnicity in the LA study. At an alpha level of 0.05, deviation from HWE was suggested for several SNPs (IL1B rs1143627 among Hispanics, IL10 rs1800871 in Asian-Americans, and LTA rs909253 for African-Americans), but none exceeded the Bonferroni-adjusted cut-point of 0.05/12 = 0.0042. In instances where two or more SNPs are in strong linkage disequilibrium (i.e., r2 ≥ 0.9), the SNP with the more reliable signal is presented. Although three SNPs in the IL10 promoter region were within 0.5 kb of each other (rs1800896, rs1800871, and rs1800872), haplotype analysis was not conducted since rs1800896 was not genotyped in the LA and MSKCC studies, and because initial analysis suggested a block size of only two SNPs in the Taixing study.
Laboratory analysis
DNA was isolated using a modified phenol-chloroform method. SNPs were genotyped using the SNPlex assay (Applied Biosystems [ABI], Foster City, CA); two were also genotyped with ABI’s TaqMan assay (IL10 rs1800871 and IFNGR1 rs11914). Briefly, DNA aliquots from cases and controls were randomized onto PCR plates with the appropriate reaction mix. For SNPlex reactions, allele-specific oligonucleotide probes were hybridized to target genomic sequences. The hybridization products were purified of excess probe, amplified by PCR, and captured in streptavidin-coated microtiter plates. Fluorescently labeled ZipChute probes were then hybridized to the streptavidin-bound amplicons and detected by capillary electrophoresis. In the TaqMan assay, fluorescently-labeled sequence-specific primers were used in PCR reactions in a total volume of 5 microliters with the following modified protocol: denaturation at 92°C for ten minutes followed by 60 cycles at 92°C for 15 seconds and extension at 62°C for 80 seconds. Genotype detection for SNPlex and TaqMan assays was performed using an ABI 3730 DNA Analyzer with ABI Genemapper 4.0 software and an ABI 7900 machine with SDS 2.3 software, respectively. Call rates were >85% for SNPlex and >97% for TaqMan. Reproducibility was 0.978 for the SNPlex assay (based on re-genotyping a 3% random sample), and 0.997 for the TaqMan assay (using a 5% random sample). Concordance for SNPs genotyped on both platforms was 0.943 for IL10 rs1800871 and 0.996 for IFNGR1 rs11914.
Statistical Analysis
Fisher’s exact test was used to assess deviation from HWE, and all data were analyzed with SAS version 9.1.3. The odds ratio (OR) and 95% confidence interval (CI) for each SNP-cancer association of each study and within smoking strata were estimated using unconditional logistic regression. To test the hypothesis that an increasing number of risk alleles is associated with cancer (i.e., monotonic-response model), we tested for linear trend of the odds ratio by treating the number of risk alleles as a continuous variable in logistic regression models, designating the homozygous ancestral genotype the as the reference group based on SNP-specific information at the National Center for Biotechnology Information’s dbSNP database.14 We used these cancer site-specific genotype-cancer associations for each SNP to determine the appropriateness of selecting an inheritance model (e.g., dominant, recessive). Covariates in the LA study included race/ethnicity (non-Hispanic White, Hispanic, African-American, Asian-American, and other), pack-years of tobacco smoking (continuous), drink-years of alcohol consumption for UADT cancers (continuous), and years of education (continuous). To minimize confounding, age was divided into fine categories (29–34, 35–36, 37–38, 39–40, 41–42, 43–44, 45–46, 47–48, 49–50, 51–52, 53–54, 55–56, 57–58, and 59–62), and controls who were more than three years older than the oldest case or more than three years younger than the youngest case were excluded. This resulted in the exclusion of 11 controls from lung cancer analyses; no controls were excluded from UADT analyses. Analyses in the Taixing study were adjusted for gender, alcohol drinking (four-level ordinal), education (four-level ordinal), age (continuous), pack-years (continuous), hepatitis B surface antigen for liver cancer, and Helicobacter pylori infection status for stomach cancer. Regression models in the MSKCC study included race (White versus non-White), gender, smoking status (ever versus never), age (continuous), and years of education (continuous). Stratified analyses by smoking status (ever and never smokers) were limited to instances for which there were at least 75 cases to ensure adequate precision, and pack-years of smoking was included in regression models to address residual confounding among ever smokers. We tested the effects of the SNPs and tobacco smoking for departure from multiplicativity by fitting a model with smoking (ever/never), genotype, and their product term, and calculating the ratio of odds ratios (ROR) by taking the natural antilog of the estimated coefficient of the product term. Potentialconfounders, including ethnicity, age, gender, and pack-years of smoking were included inlogistic regression models, and individuals with the non-risk genotype who had never smoked were designated as the reference group.
For SNPs genotyped across all three studies that had point estimates consistent in direction and magnitude over all cancer sites, we investigated whether those SNPs were associated with smoking-related cancer by pooling all cases and controls across all three studies and the different tumor sites. We adjusted for study location, gender, race/ethnicity, age, and tobacco smoking (ever vs. never). Because race/ethnicity is highly correlated with study location, we tested for the presence of multicollinearity by assessing the variance inflation factor and by testing for heterogeneity between models with and without terms for ethnicity. Neither method suggested that multicollinearity was an important factor, so categories for race/ethnicity and study location were combined into one variable and included as a new covariate in adjusted logistic regression analyses for pooled cancer sites.
Given the number of comparisons made in this study, we addressed the possibility of chance findings using two Bayesian approaches. The false positive report probability (FPRP) facilitates identifying noteworthy observations when the probability of a false positive is below an investigator-predetermined threshold.15 The Bayesian false discovery probability (BFDP) is a progression of FPRP and considers the ratio of the cost of missing a true association to the cost of a false discovery.16 In both methods, an observation from our study was viewed in light of prior knowledge (i.e., prior probability) to assess the posterior probability that the association is not null. The value of the prior probability is subjective and guided by epidemiologic data and existing knowledge of the gene and related SNPs. We considered prior probabilities ranging from 0.10 to 0.01, a main effect OR of 1.5 (or 0.67), a smoking-stratified OR of 2.5 (or 0.40), and a FPRP cut-point of 0.4 and a BFDP threshold of 0.8 to determine noteworthy findings.
Results
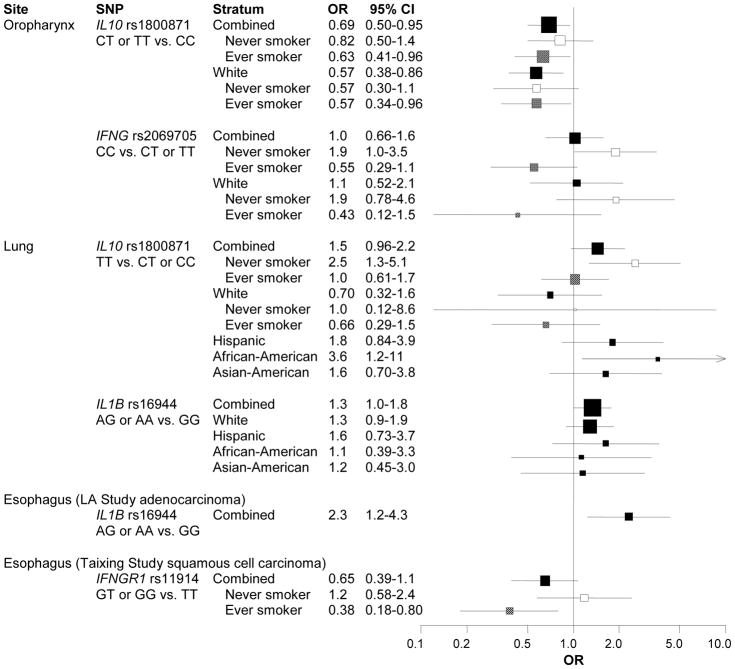
Characteristics of our study population of 2,049 smoking-related cancer cases and 1,666 controls are presented in Table 1. Compared to the LA and MSKCC studies, the majority of Taixing participants had completed less than 12 years of education and was less likely to have reported ever drinking alcohol. Controls were noticeably younger than cases in the MSKCC study. The estimated effects of the 12 SNPs stratified by cancer site are reported in Tables S2 and S3; selected results are summarized in Figure 1A. Oropharyngeal cancer was inversely associated with IL10 rs1800871 among CT heterozygotes (adjusted OR [aOR]: 0.67, 95% CI: 0.48–0.94) and possibly among TT homozygotes (aOR: 0.78, 95% CI: 0.44–1.4), suggesting a dominant inheritance model (aOR: 0.69, 95% CI: 0.50–0.95). An inverse association was also observed between IFNG rs2069705 and oropharyngeal cancer (CT and TT versus CC aOR: 0.72, 95% CI: 0.52–1.0). Compared to IL1B rs16944 GG homozygotes, the A allele was more common among lung cancer cases (aOR: 1.3, 95% CI: 1.0–1.8). This association, which was not modified by smoking status, appeared to be consistent across ethnicity.
Table 1.
Demographic and behavioral characteristics of cancer cases and controls, by study location.
| All studies pooled |
Los Angeles |
Taixing |
MSKCC |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case* N = 2,049 | Control N = 1,666 | Lung N = 611 | UADT N = 553 | Control N = 1,040 | Esophagus N = 218 | Stomach N = 206 | Liver N = 204 | Control N = 415 | Bladder N = 227 | Control N = 211 | ||||||||||||
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | |
| Age | ||||||||||||||||||||||
| 35< | 49 | 2.4 | 133 | 8.0 | 4 | 0.7 | 25 | 4.5 | 51 | 4.9 | 1 | 0.5 | 1 | 0.5 | 16 | 7.8 | 19 | 4.6 | 2 | 0.9 | 63 | 30.0 |
| 35–44 | 185 | 9.0 | 278 | 16.7 | 57 | 9.3 | 65 | 11.8 | 171 | 16.4 | 5 | 2.3 | 12 | 5.8 | 37 | 18.1 | 41 | 9.9 | 6 | 2.6 | 66 | 31.3 |
| 45–54 | 760 | 37.1 | 641 | 38.5 | 301 | 49.3 | 249 | 45.0 | 499 | 48.0 | 66 | 30.3 | 38 | 18.5 | 63 | 30.9 | 98 | 23.6 | 35 | 15.4 | 44 | 20.9 |
| 55+ | 1,054 | 51.4 | 614 | 36.9 | 249 | 40.8 | 214 | 38.7 | 319 | 30.7 | 145 | 66.5 | 155 | 75.2 | 88 | 43.1 | 257 | 61.9 | 184 | 81.1 | 38 | 18.0 |
| Missing | 1 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Gender | ||||||||||||||||||||||
| Male | 1,371 | 66.9 | 1,062 | 63.7 | 303 | 49.6 | 420 | 76.0 | 623 | 59.9 | 141 | 64.7 | 138 | 67.0 | 159 | 77.9 | 287 | 69.2 | 189 | 83.3 | 152 | 72.0 |
| Female | 678 | 33.1 | 591 | 35.5 | 308 | 50.4 | 133 | 24.1 | 417 | 40.1 | 77 | 35.3 | 68 | 33.0 | 45 | 22.1 | 128 | 30.8 | 38 | 16.7 | 46 | 21.8 |
| Missing | 0 | 0 | 13 | 0.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 6.2 |
| Ethnicity | ||||||||||||||||||||||
| White | 920 | 44.9 | 825 | 49.5 | 359 | 58.8 | 331 | 59.9 | 634 | 61.0 | 206 | 90.8 | 191 | 90.5 | ||||||||
| Hispanic | 176 | 8.6 | 206 | 12.4 | 70 | 11.5 | 100 | 18.1 | 204 | 19.6 | 6 | 2.6 | 2 | 1.0 | ||||||||
| African-American | 174 | 8.5 | 104 | 6.2 | 96 | 15.7 | 66 | 11.9 | 102 | 9.8 | 7 | 3.8 | 2 | 1.0 | ||||||||
| Asian-American | 113 | 5.5 | 64 | 3.8 | 70 | 11.5 | 41 | 7.4 | 62 | 6.0 | 1 | 0.4 | 1 | 0.5 | ||||||||
| Other | 30 | 1.5 | 38 | 2.3 | 15 | 2.5 | 13 | 2.4 | 37 | 3.6 | 2 | 0.9 | 1 | 0.5 | ||||||||
| Han Chinese | 628 | 30.7 | 415 | 24.9 | 218 | 100 | 206 | 100 | 204 | 100 | 415 | 100 | ||||||||||
| Missing | 8 | 0.4 | 15 | 0.9 | 1 | 0.2 | 2 | 0.4 | 1 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 2.2 | 14 | 6.6 |
| Education (year) | ||||||||||||||||||||||
| < 12 | 851 | 41.5 | 459 | 27.6 | 107 | 17.5 | 438 | 79.2 | 116 | 11.2 | 212 | 97.2 | 203 | 98.5 | 191 | 93.6 | 339 | 81.7 | 19 | 8.4 | 3 | 1.4 |
| 12 or more | 1,195 | 58.3 | 1,207 | 72.4 | 504 | 82.5 | 115 | 20.8 | 923 | 88.8 | 3 | 1.4 | 3 | 1.5 | 13 | 6.4 | 76 | 18.3 | 208 | 91.6 | 208 | 98.6 |
| Missing | 3 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.1 | 3 | 1.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tobacco (pack-years) | ||||||||||||||||||||||
| Never smoker | 588 | 28.7 | 816 | 49.0 | 110 | 18.0 | 159 | 28.8 | 492 | 47.3 | 94 | 43.1 | 92 | 44.7 | 85 | 41.7 | 217 | 52.3 | 38 | 16.7 | 107 | 50.7 |
| >0–20 | 416 | 20.3 | 493 | 29.6 | 102 | 16.7 | 132 | 23.9 | 353 | 33.9 | 45 | 20.6 | 48 | 23.3 | 58 | 28.4 | 91 | 21.9 | 29 | 12.8 | 49 | 23.2 |
| >20–40 | 507 | 24.7 | 236 | 14.2 | 202 | 33.1 | 139 | 25.1 | 136 | 13.1 | 48 | 22.0 | 41 | 19.9 | 38 | 18.6 | 83 | 20.0 | 31 | 13.7 | 17 | 8.1 |
| >40 | 487 | 23.8 | 94 | 5.6 | 197 | 32.2 | 123 | 22.2 | 58 | 5.6 | 24 | 11.0 | 20 | 9.7 | 11 | 5.4 | 23 | 5.5 | 107 | 47.1 | 13 | 6.2 |
| Missing | 51 | 2.5 | 27 | 1.6 | 0 | 0 | 0 | 0 | 1 | 0.1 | 7 | 3.2 | 5 | 2.4 | 12 | 5.9 | 1 | 0.2 | 22 | 9.7 | 25 | 11.9 |
| Alcohol | ||||||||||||||||||||||
| Never | 618 | 30.2 | 507 | 30.4 | 170 | 27.8 | 94 | 17.0 | 264 | 25.4 | 116 | 53.2 | 111 | 53.9 | 87 | 42.7 | 207 | 49.9 | 31 | 13.7 | 36 | 17.1 |
| Ever | 1,395 | 68.1 | 1,138 | 68.3 | 440 | 72.0 | 457 | 82.6 | 772 | 74.2 | 95 | 43.6 | 90 | 43.7 | 105 | 51.5 | 205 | 49.4 | 187 | 82.4 | 161 | 76.3 |
| Missing | 36 | 1.8 | 21 | 1.3 | 1 | 0.2 | 2 | 0.4 | 4 | 0.4 | 7 | 3.2 | 5 | 2.4 | 12 | 5.9 | 3 | 0.7 | 9 | 4.0 | 14 | 6.6 |
| H. pylori infection | 71 | 34.5 | 114 | 27.5 | ||||||||||||||||||
| Chronic hepatitis B | 132 | 64.7 | 102 | 24.6 | ||||||||||||||||||
MSKCC: Memorial Sloan-Kettering Cancer Center; UADT: upper aero-digestive tract.
Also includes 30 kidney cancer cases from MSKCC.
Figure 1.
Because tobacco smoking is a strong risk factor for our outcomes, we re-examined the SNP-cancer associations, stratifying by ever/never smoking status. These results are reported in Table S4 and summarized in Figure 1A. Lung cancer was associated with IL10 rs1800871 among never smokers (TT versus CC or CT aOR: 2.5, 95% CI: 1.3–5.1) but not among ever smokers, though there was some heterogeneity across ethnicity. While the point estimate for Whites was less than 1.0, estimates were greater than 1.0 for the other ethnicities, though they tended to be imprecise. Oropharyngeal cancer was inversely associated with IL10 rs1800871 among ever smokers (CT or TT versus CC aOR: 0.63, 95% CI: 0.41–0.95) and positively associated with IFNG rs2069705 among never smokers (CC versus CT or TT aOR: 1.9, 95% CI: 1.0–3.5). Esophageal cancer in the Taixing study was less common among ever smokers who had at least one variant G allele for IL6 rs1800796 (aOR: 0.54, 95% CI: 0.31–0.95). Risk estimates for SNP-cancer associations after restricting to current smokers were in the same direction as smoking stratified results but greater in magnitude (i.e., further from the null) and less precise.
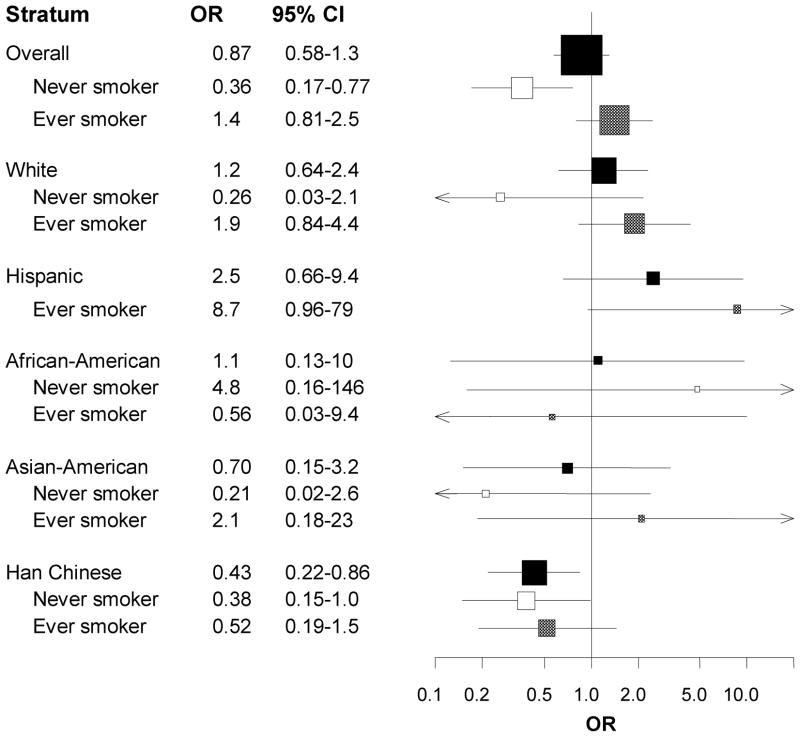
Of the six SNPs that were genotyped across all three studies, the most consistent SNP-cancer estimates were observed for TNF rs1799964 among never smokers. Smoking-related cancers were much less common among never smokers homozygous for the variant TNF rs1799964 C allele compared to never smokers with at least one T allele (aOR: 0.36, 95% CI: 0.17–0.77). This inverse association was not observed among ever smokers (Figure 1B and Table S5). When stratified by ethnicity, the relative rarity of cancers among never smokers compared to ever smokers tended to persist, although smoking-stratified estimates for some ethnicities were imprecise.
The estimated effects of genotype-smoking-status combinations for selected SNPs are shown in Table 2. In site-stratified analysis, we observed that the estimated joint effect of recessive IL10 rs1800871 genotype and tobacco smoking was less than what would be expected under the null hypothesis of multiplicativity of effects among lung cancer cases and controls (aROR: 0.31, 95% CI: 0.14–0.69), which persisted across ethnicity, although adjusting for pack-years attenuated the estimate. There was also an indication of departure from multiplicative effects for IFNGR1 rs11914 esophageal cancer in the Taixing study (aROR: 0.37, 95% CI: 0.13–1.0). In our pooled analysis, there was evidence of greater than multiplicative interaction among individuals recessive for TNF rs1799964 (aROR: 3.4, 95% CI: 1.4–8.6), which was fairly consistent across ethnicity with the exception of African-Americans, whose unstable estimates were a reflection of that group having the least number of individuals homozygous for the rare genotype among controls.
Table 2.
Modification of selected SNP-specific odds ratios for tobacco-related cancers pooled and stratified by ethnicity, smoking status, and cancer site.
| SNP and cancer site | Ever smoker | Genotype | Case | Control | cOR | 95% CI | aOR | 95% CI | FPRP prior probability |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.10 | 0.05 | 0.01 | |||||||||
| TNF rs1799964 | |||||||||||
| All cancer sites pooled | No | TT + CT | 434 | 633 | 1.0 | 1.0 | |||||
| No | CC | 10 | 35 | 0.42 | 0.20–0.85 | 0.41 | 0.19–0.85 | 0.22 | 0.37 | 0.76 | |
| Yes | TT + CT | 1,044 | 650 | 2.3 | 2.0–2.7 | 2.4 | 2.0–2.8 | <0.01 | <0.01 | <0.01 | |
| Yes | CC | 45 | 20 | 3.3 | 1.9–5.6 | 3.3 | 1.9–5.8 | <0.01 | <0.01 | 0.02 | |
| ROR | 3.4 | 1.4–8.2 | 3.4 | 1.4–8.6 | 0.25 | 0.41 | 0.78 | ||||
| White | No | TT + CT | 11 | 329 | 1.0 | 1.0 | |||||
| No | CC | 1 | 12 | 0.25 | 0.03–1.9 | 0.26 | 0.03–2.1 | ||||
| Yes | TT + CT | 528 | 314 | 5.0 | 3.9–6.4 | 3.6 | 2.8–4.8 | <0.01 | <0.01 | <0.01 | |
| Yes | CC | 25 | 9 | 8.2 | 3.73–18 | 6.7 | 2.9–15 | 0.01 | 0.01 | 0.07 | |
| ROR | 6.7 | 0.74–60 | 7.1 | 0.76–66 | |||||||
| Hispanic | No | TT + CT | 43 | 75 | 1.0 | 1.0 | |||||
| No | CC | 0 | 3 | ||||||||
| Yes | TT + CT | 77 | 102 | 1.3 | 0.82–2.1 | 1.3 | 0.80–2.2 | ||||
| Yes | CC | 6 | 1 | 10 | 1.2–90 | 11 | 1.3–97 | 0.75 | 0.87 | 0.87 | |
| ROR | |||||||||||
| African-American | No | TT + CT | 19 | 29 | 1.0 | 1.0 | |||||
| No | CC | 1 | 1 | 1.5 | 0.11–32 | 2.7 | 0.15–49 | ||||
| Yes | TT + CT | 103 | 43 | 3.7 | 1.9–7.2 | 3.5 | 1.8–7.1 | 0.02 | 0.05 | 0.20 | |
| Yes | CC | 1 | 1 | 1.5 | 0.09–26 | 1.8 | 0.10–31 | ||||
| ROR | 0.28 | 0.01–15 | 0.18 | 0.004–11 | |||||||
| Asian-American | No | TT + CT | 36 | 26 | 1.0 | 1.0 | |||||
| No | CC | 1 | 3 | 0.23 | 0.02–2.4 | 0.24 | 0.02–2.5 | ||||
| Yes | TT + CT | 38 | 21 | 1.3 | 0.60–2.6 | 1.3 | 0.63–2.7 | ||||
| Yes | CC | 3 | 1 | 2.1 | 0.20–22 | 2.2 | 0.21–22 | ||||
| ROR | 6.8 | 0.25–181 | 10.0 | 0.31–332 | |||||||
| Han Chinese | No | TT + CT | 221 | 164 | 1.0 | 1.0 | |||||
| No | CC | 7 | 14 | 0.37 | 0.15–0.94 | 0.38 | 0.14–0.98 | 0.47 | 0.65 | 0.91 | |
| Yes | TT + CT | 283 | 151 | 1.4 | 1.1–1.8 | 1.5 | 1.1–2.2 | 0.18 | 0.31 | 0.70 | |
| Yes | CC | 8 | 8 | 0.74 | 0.27–2.0 | 0.78 | 0.27–2.2 | ||||
| ROR | 1.4 | 0.37–5.6 | 1.4 | 0.33–5.4 | |||||||
| IL10 rs1800871 | |||||||||||
| Lung | No | CC + CT | 63 | 387 | 1.0 | 1.0 | |||||
| No | TT | 28 | 33 | 5.2 | 3.0–9.2 | 3.1 | 1.6–5.9 | 0.02 | 0.04 | 0.19 | |
| Yes | CC + CT | 387 | 424 | 5.6 | 4.2–7.6 | 1.2 | 0.82–1.8 | ||||
| Yes | TT | 47 | 61 | 4.7 | 3.0–7.5 | 1.2 | 0.68–2.1 | ||||
| ROR | 0.16 | 0.08–0.33 | 0.31 | 0.14–0.69 | 0.12 | 0.23 | 0.60 | ||||
| White | No | CC + CT | 24 | 263 | 1.0 | 1.0 | |||||
| No | TT | 1 | 10 | 1.1 | 0.13–8.9 | 1.0 | 0.12–8.3 | ||||
| Yes | CC + CT | 270 | 266 | 11 | 7.1–17 | 1.8 | 1.0–3.2 | 0.29 | 0.46 | 0.82 | |
| Yes | TT | 15 | 22 | 7.5 | 3.4–16 | 1.2 | 0.45–3.2 | ||||
| ROR | 0.61 | 0.07–5.6 | 0.67 | 0.07–6.5 | |||||||
| Hispanic | No | CC + CT | 19 | 71 | 1.0 | 1.0 | |||||
| No | TT | 9 | 10 | 3.4 | 1.2–9.5 | 3.8 | 1.2–12 | 0.43 | 0.61 | 0.89 | |
| Yes | CC + CT | 30 | 87 | 1.3 | 0.67–2.5 | 0.72 | 0.31–1.7 | ||||
| Yes | TT | 5 | 20 | 0.93 | 0.31–2.8 | 0.70 | 0.21–2.3 | ||||
| ROR | 0.21 | 0.05–0.93 | 0.25 | 0.05–1.2 | |||||||
| African-American | No | CC + CT | 8 | 28 | 1.0 | 1.0 | |||||
| No | TT | 0 | 1 | ||||||||
| Yes | CC + CT | 58 | 42 | 4.8 | 2.0–12 | 1.1 | 0.33–3.8 | ||||
| Yes | TT | 17 | 6 | 9.9 | 2.9–34 | 4.7 | 1.1–21 | 0.65 | 0.80 | 0.95 | |
| ROR | |||||||||||
| Asian-American | No | CC + CT | 9 | 16 | 1.0 | 1.0 | |||||
| No | TT | 18 | 11 | 2.9 | 0.96–8.8 | 3.7 | 1.1–12 | 0.52 | 0.70 | 0.92 | |
| Yes | CC + CT | 18 | 10 | 3.2 | 1.0–10 | 0.76 | 0.15–3.9 | ||||
| Yes | TT | 10 | 12 | 1.5 | 0.46–4.8 | 0.48 | 0.10–2.4 | ||||
| ROR | 0.17 | 0.03–0.84 | 0.18 | 0.03–1.1 | |||||||
| IFNG rs2069705 | |||||||||||
| Oropharynx | No | TT+CT | 60 | 339 | 1.0 | 1.0 | |||||
| No | CC | 21 | 67 | 1.8 | 1.0–3.1 | 1.9 | 1.0–3.4 | 0.30 | 0.47 | 0.82 | |
| Yes | TT+CT | 115 | 360 | 1.8 | 1.3–2.6 | 0.82 | 0.52–1.3 | ||||
| Yes | CC | 16 | 90 | 1.0 | 0.55–1.8 | 0.48 | 0.24–0.98 | 0.36 | 0.55 | 0.86 | |
| ROR | 0.31 | 0.14–0.70 | 0.31 | 0.13–0.73 | 0.19 | 0.33 | 0.72 | ||||
| White | No | TT+CT | 44 | 233 | 1.0 | 1.0 | |||||
| No | CC | 8 | 21 | 2.0 | 0.84–4.8 | 2.0 | 0.83–5.0 | ||||
| Yes | TT+CT | 79 | 239 | 1.8 | 1.2–2.6 | 0.90 | 0.51–1.6 | ||||
| Yes | CC | 3 | 25 | 0.64 | 0.18–2.2 | 0.39 | 0.11–1.4 | ||||
| ROR | 0.18 | 0.04–0.81 | 0.21 | 0.05–1.0 | 0.68 | 0.82 | 0.96 | ||||
| IFNGR1 rs11914 | |||||||||||
| Esophagus (Taixing) | No | TT | 67 | 161 | 1.0 | 1.0 | |||||
| No | GT + GG | 17 | 36 | 1.1 | 0.60–2.2 | 1.1 | 0.55–2.2 | ||||
| Yes | TT | 96 | 141 | 1.6 | 1.1–2.4 | 2.0 | 0.97–4.0 | ||||
| Yes | GT + GG | 12 | 42 | 0.69 | 0.34–1.4 | 0.74 | 0.29–1.9 | ||||
| ROR | 0.40 | 0.15–1.0 | 0.37 | 0.13–1.0 | 0.51 | 0.68 | 0.92 | ||||
aOR: odds ratio (OR) adjusted for study location/ethnicity (pooled analysis only), ethnicity (lung analysis only), age, gender, education, tobacco smoking, and alcohol use (esophagus analysis only); CI: confidence interval; cOR: crude OR; FPRP: false positive report probability. FPRP values are the posterior probabilities of reporting a false positive result, given the statistical power to detect a smoking-stratified aOR or ROR of at least 2.5. Values of FPRP less than 0.4 are bold-faced, indicating noteworthy observations; ROR: ratio of odds ratios; SNP: single nucleotide polymorphism.
Given the many comparisons in our study, we considered the probability of chance finding (Table S6). Assuming a prior probability of 0.10 and a FPRP cut-point of 0.4, the associations between lung cancer and IL1B rs16944 and between oropharyngeal cancer and polymorphisms of IL10 rs1800871 and IFNG rs2069705 appear to be noteworthy. Under the same prior, and assuming that false nondiscovery is four times as costly as false discovery (i.e., a BFDP threshold of 0.8), the association between oropharyngeal cancer and IL10 rs1800871 may be important. Considering associations that are below both FPRP and BFDP thresholds, IL10 rs1800871 appears to be an important marker for oropharyngeal cancer risk. IL10 rs1800871 may also be associated with lung cancer among never smokers and with oropharyngeal cancer among ever smokers. Chance does not seem to be a likely explanation in our study population. The TNF rs1799964 SNP, which only appeared to be associated with stomach cancer in site-specific analyses, also seems to be associated with smoking-related cancers among never smokers, even at a 5% prior probability. Additional associations adjusted for multiple comparisons may be noteworthy for several other smoking-stratified associations at a prior probability of 0.10 but none passed FPRP and BFDP criteria using more conservative priors.
Discussion
Among never smokers in our overall study population of 2,097 cases and 1,666 controls, we found that TNF rs1799964 was associated with smoking-related cancer. Cancer site-specific analysis suggests that oropharyngeal cancer is inversely associated with IL10 rs1800871 and IFNG rs2069705. Smoking-stratified analyses suggest that some SNP-cancer associations become more apparent within strata of smoking status and that some associations may be site-specific. TNF rs1799964, for example, seems to be an important SNP among never smokers for smoking-related cancer as a whole, while the IL10 rs1800871 association with lung cancer was observed only among never smokers (aOR: 2.5, 95% CI: 1.3–5.1 versus aOR: 1.0, 95% CI: 0.61–1.7 for ever smokers). After adjustment for multiple comparisons, the associations between TNF rs1799964 and any smoking-related cancer among never smokers and between IL10 rs1800871 and cancers of the oropharynx and lung do not appear to be due to chance.
Whether IL10 polymorphisms affect lung cancer risk remains to be determined but some reports suggest an association, although differences in models of inheritance and risk estimates suggest some heterogeneity. A population-based case-cohort study estimated a 60% increase in lung cancer risk for individuals with at least one copy of the IL10 rs1800872 variant A allele (which is in high linkage disequilibrium with rs1800871), and a slightly weaker association among current smokers.17 A positive association between non-small cell lung cancer (NSCLC) and IL10 rs1800871 was estimated by Van Dyke and colleagues in a population-based case-control study among Caucasian women (aOR: 1.39, 95% CI: 0.96–2.02), and the magnitude of the association was still elevated, though less precisely, among a smaller number of African-American women (aOR: 1.32, 95% CI: 0.60–2.88).18 IL10 rs1800871 was recessively associated with NSCLC in a Chinese population (OR: 1.37, 95% CI: 1.10–2.09) but it is not clear if the estimate was adjusted for tobacco smoking, age, sex, and gender, although those characteristics seemed similarly distributed between cases and controls.19 Weaker associations between rs1800871 and lung cancer have also been reported in a Chinese population for homozygous for the rare allele (aOR: 1.38, 95% CI: 0.57–3.38)20 and among non-Hispanic Caucasians with at least one variant T allele (aOR: 1.43, 95% CI: 0.78–2.63).21 Despite differences in interpreting the association between IL10 rs1800871 and lung cancer, point estimates from published studies (i.e., OR ~1.4) are comparable to our own estimate (aOR: 1.5, 95% CI: 0.96–2.2), and the consistency between these estimates (P heterogeneity > 0.99) further suggests that IL10 rs1800871 might be associated with elevated lung cancer risk, but the magnitude is not large.
IL-10 expression in oropharyngeal squamous cell carcinoma (SCC) has been inversely associated with survival22 and tumor grade and stage.23 These studies, however, do not rule out the possibility of association through altered cytokine expression as a result of somatic mutations within the tumor. Several studies of germline mutations suggest that IL10 polymorphisms, which are associated with periodontitis,24, 25 may also be associated with tongue cancer26 and oral neoplasms.27 IL10 rs1800896, which is not in high linkage disequilibrium with rs1800871 (r2 = 0.30), was associated with oral SCC in a hospital-based case-control study (aOR: 2.65, 95% CI: 1.28–5.46).28 Another study from Sichuan, China reported that compared to the rs1800871 TT genotype, the CC genotype (which is more common in Asian populations) was associated with oral cancer (OR: 1.45, 95% CI: 0.88–2.39).29 Recalculating the estimate with the CC genotype as the referent group yields a measure of association (OR: 0.69, 95% CI: 0.42–1.14) that is nearly identical to the effect estimated in our study (aOR: 0.69, 95% CI: 0.50–0.95).
Our results suggest that IL1B rs16944 may predict lung cancer, consistent with published reports,30–32 but our association did not hold after correction for multiple comparisons. Based on a large, multi-center case-control study, IL1B rs1143627 (linkage disequilibrium with rs16944: r2 = 0.94) was not associated with lung cancer, suggesting that our finding for rs16944 may be due to chance.33 It is possible, though, that the association of lung cancer with IL1B polymorphisms could involve a pathway that is not primarily mediated by rs1143627.34, 35
The involvement of TNF polymorphisms in cancer has been reported for several malignancies, including lymphomas6, 36, 37 and lung cancer, though more null results have been reported for lung cancer18, 21, 32, 38 than non-null associations.39 The most consistent associations seem to come from gastric cancer studies. A meta-analysis of TNF polymorphisms reported positive associations for rs1800629 (summary OR [sOR]: 1.49, 95% CI: 1.11–1.99) and rs1799724 (sOR: 1.57, 95% CI: 0.91–2.70).5 We observed a similar association for rs1800629 in our gastric cancer sample (aOR: 1.3, 95% CI: 0.74–2.4). The apparent lack of published associations for rs1799964 could be because it may be most noticeable among non-smokers, for whom the strong effect of tobacco-smoking is not as important a risk factor.
The difference in effects estimated between never smokers and ever smokers in our study suggests that smoking can modify the rate ratios between some inflammation-related SNPs and smoking related-cancers. In our pooled analysis, for example, smoking-related cancers were less common among never smokers with the TNF rs1799964 CC genotype, and the variant C allele did not appear protective among ever smokers (Table 2). Less than multiplicative smoking-SNP interactions were also suggested between lung cancer and IL10 rs1800871, oropharyngeal cancer and IFNG rs2069705, and esophageal cancer and IFNGR1 rs11914.
The mechanisms underlying these observations are unknown. Genetic polymorphisms in the numerous pathways involved in carcinogenesis affect cancer risk, and tobacco smoking may modify these effects.9, 10, 40, 41 The epithelial cells from many of the organs in our study are repeatedly exposed to components of tobacco smoke or their metabolic byproducts, which are carcinogenic,42 known to cause vasoconstriction,43 inhibit cell proliferation and angiogenesis,44, 45 and are potent inducers of inflammation.46, 47 The high levels of free radicals contained in48 and generated by tobacco smoke can lead to cancer through oxidative DNA damage mediated by inflammation-associated production of reactive oxygen species.49
Promoter polymorphisms of TNF are fairly numerous and in linkage disequilibrium with each other and with nearby genes,50 which may have cooperative effects,51 potentially complicating the interpretation of single-SNP associations. One of the most commonly studied TNF SNPs is the rs1800629 G-308A polymorphism.52 The variant rs1799964 C allele, which is not in linkage disequilibrium with the G-308A polymorphism (r2 = 0.07), appears to be associated with increased TNF expression.53 TNF is an inflammatory cytokine and its expression by peripheral blood mononuclear cells has been demonstrated to increase following exposure to tobacco smoke.54 Interestingly, TNF was expressed at higher levels in cells from non-smokers than smokers at all time-points, which lends potential biologic support for our observation that smoking-related cancers were less common among never smokers with the putative high expression TNF rs1799964 CC genotype. Haplotype studies of the gene encoding the anti-inflammatory cytokine IL-10 suggest that the GCC haplotype (i.e., rs1800896 G, rs1800871 C, and rs1800872 C) is associated with high IL-10 production.55, 56 Although haplotype data were unavailable in the LA study, the rs1800871 variant T allele associated with low production appeared to be an important SNP for lung cancer among never smokers while the variant did not seem to increase risk among ever smokers.
The Van Dyke study of NSCLC among women18 reported associations for six SNPs that were also genotyped in our study (IL1B rs1143627 and rs16944; TNF rs1799964 and rs1800629; LTA rs909253; and IL10 rs1800871). The estimated magnitude and direction of association for these SNPs were similar to our own estimates, both overall and among women, although comparability of our TNF results was affected by the small number of women with the variant C allele, resulting in unstable estimates. While the majority of our SNPs do not result in amino acid substitutions, cancers of the oropharynx and lung appear to be associated with IL10 rs1800871. The SNPs may influence cancer in part through modifying transcription and/or translation. Synonymous SNPs have been demonstrated to alter protein structure by affecting RNA splicing57 and the stability58, 59 and translation rate60 of mRNA. However, further work assessing how these SNPs may affect transcription, translation, and protein conformation would help shed light on these hypothesized mechanisms.
Our study design and analytic strategy had a number of strengths and weaknesses. The associations in our study are subject to confounding and biases related to information ascertainment and subject selection. We included ethnicity as a covariate in regression models to address admixture but the effects of population stratification may still residually confound our estimates, particularly for SNPs with allele frequencies that differ greatly between ethnicities, such as IL10 rs1800871 and IFNG rs2069705. The association we observed between lung cancer and IL10 rs1800871, for example, might be partially due to uncontrolled differences in ethnicity. However, the ethnic diversity of the LA study facilitated examination of ethnicity-specific ratios of odds ratios, which were consistent in magnitude across ethnicity (except for African-Americans, for whom no never smoking lung cancer cases were observed with the risk genotype), suggesting that tobacco smoking and the recessive IL10 rs1800871 genotype may interact on a less than multiplicative scale.
Because TNF rs1799964, which appeared to be an important SNP for non-smokers in pooled analysis, does not appreciably vary across ethnic groups, it seems unlikely that population stratification was a significant problem. We attempted to minimize information bias by using strict inclusion and exclusion criteria for cases and controls, and by implementing stringent quality control measures in our laboratory. We also employed trained interviewers who used standardized questionnaires to collect detailed information on potential risk factors and related covariates to address confounding. Differential recollection of exposures for cases and controls could result in misclassification bias. Although we included pack-years of tobacco smoking in regression models to address residual confounding among ever smokers, our estimates may still be confounded by the strong effect of smoking. While we selected SNPs based on a pathway-based approach, additional markers per gene selected over a range of important regions (e.g., splice sites, promoter areas, and tagging SNPs) would have been helpful to better characterize a gene, evaluate haplotypes, and ascertain whether an observed association could be mediated through linkage with SNPs.
The inverse association estimated for the TNF rs1799964 C variant among never smokers reported (Table 2) may reflect a true gene-environment interaction (in which the TNF rs1799964 CC genotype may afford some protection only in the absence of tobacco smoking) but a number of problems need to be considered. Data were sparse for some strata across ethnicity, even among the 920 cases and 825 controls who were self-identified as White. In particular, there were few observations of never smokers with the TNF rs1799964 CC variant genotype (especially true among cases), and none of the confidence intervals for these strata excluded the null when there were less than five observations per cell. The >60-fold ratios of the upper and lower 95% confidence limits illustrate the instability of some of these genotype-smoking estimates.
Cancer is a multi-factorial process and a simple deterministic SNP-cancer association is unlikely. Given the number of comparisons that we made, our results could have been entirely due to chance. We therefore used two Bayesian approaches to account for Type I error, which suggest that our results are not purely chance findings. An inherent feature of Bayesian correction, though, is the use of subjective prior probabilities, which is susceptible to publication bias.61 Alternatively, our results may be due to other genetic and/or epigenetic mechanisms (e.g., gene amplification, translocation, loss of heterozygosity, DNA methylation, genomic imprinting, and histone modification).62, 63 These reasons underscore the importance of considering our results in the broader context of existing knowledge and studies and not to overemphasize the results of a particular study.
Differential participation of cases and controls and their willingness to donate samples for DNA analysis may have created selection bias. Of the nine smoking-related sites in our study, esophageal, liver, lung, and stomach cancers have very low five-year survival rates, reflected in the high percentage of cases in our study who died before they could be interviewed. Selection bias may exist if factors related to participation were differentially associated with exposure (e.g., SNP genotype) for cases and controls. However, because a potential participant’s genotype would have been unknown at time of recruitment, this type of bias seems unlikely. Selection bias could have occurred if a particular genotype was associated with survival. It is unknown, however, if the study was biased by this type of selection because a literature search did not yield sufficient information on the prognostic value of the SNPs. However, tumor grade and stage did not appear to vary by SNP genotype (Table S7). The relatively small number of cases and controls in the Taixing and MSKCC studies resulted in low statistical precision, and had less than 80% power to detect main effect odds ratios of 1.5 or 0.67 for many of the SNP-cancer associations. Of the four SNPs in the Taixing study with sufficiently high minor allele frequencies to have at least 80% power (IL10 rs1800871, IL10 rs1800872, LTA rs909253, and IL6 rs1800796), the 95% confidence intervals from the multivariate model included both positive and inverse associations. One SNP in the MSKCC study with sufficiently high minor allele frequency (IL1B rs1143627) was associated with bladder cancer (aOR = 4.3), but with an extremely wide confidence interval (95% CI: 1.3–14) (the association did not pass multiple comparisons correction) and was therefore not reported. The LA study had better precision due to its relatively large sample size for assessing main and smoking-stratified effects for the lung and UADT sites. The ethnic diversity in the pooled sample and the LA study also allowed us to examine ethnicity-specific SNP-cancer associations to assess admixture. Additional strengths include the ability to examine SNP-cancer associations across a number of different cancer sites; the use of a population-based study design for the LA and Taxing studies; and control for multiple comparisons using two Bayesian approaches.
Our results if valid suggest that TNF rs1799964 is inversely associated with smoking-related cancers among never smokers, and that IL10 rs1800871 is a susceptibility marker for lung cancer among never smokers, and for oropharyngeal cancer among ever smokers.
Supplementary Material
Acknowledgments
This work was supported in part by NIH National Institute of Environmental Health Sciences, National Cancer Institute, Department of Health and Human Services, Grants ES06718, ES01167, DA11386, CA90833, CA77954, CA09142, CA96134, CA 128099, GM53275, the Alper Research Center for Environmental Genomics, and The Seymour Family Gift for Innovative Investigator-Initiated Research in Bladder Cancer at UCLA’s Jonsson Comprehensive Cancer Center and UCLA Center for Occupational and Environmental Health. We would like the thank Dr. Thomas M. Mack and Dr. Wendy Cozen for their contributions for the LA population-based lung and UADT case-control study. We are indebted to the studies’ participants for their time and dedication.
Abbreviations
- aOR
adjusted odds ratio
- aROR
adjusted ratio of odds ratios
- BFDP
Bayesian false discovery probability
- CI
confidence interval
- cOR
crude odds ratio
- FPRP
false positive report probability
- HWE
Hardy-Weinberg equilibrium
- LA
Los Angeles
- MAF
minor allele frequency
- MSKCC
Memorial Sloan-Kettering Cancer Center
- NSCLC
non-small cell lung cancer
- OR
odds ratio
- ROR
ratio of odds ratios
- SCC
squamous cell carcinoma
- SNP
single nucleotide polymorphism
- sOR
summary odds ratio
- UADT
upper aero-digestive tract
Footnotes
Novelty and impact statement: We use data from three case-control studies to demonstrate that common variants in inflammation-related genes are associated with smoking-related cancers, and that the association varies by cancer site and by smoking status. These variants may be useful susceptibility markers for smoking-related cancers and improve our understanding of the role of inflammation in carcinogenesis.
References
- 1.Ekbom A, Helmick C, Zack M, Adami HO. Ulcerative colitis and colorectal cancer. A population-based study. N Engl J Med. 1990;323:1228–33. doi: 10.1056/NEJM199011013231802. [DOI] [PubMed] [Google Scholar]
- 2.Alberg AJ, Ford JG, Samet JM American College of Chest Physicians. Epidemiology of lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132:29S–55S. doi: 10.1378/chest.07-1347. [DOI] [PubMed] [Google Scholar]
- 3.Engels EA. Inflammation in the development of lung cancer: epidemiological evidence. Expert Rev Anticancer Ther. 2008;8:605–15. doi: 10.1586/14737140.8.4.605. [DOI] [PubMed] [Google Scholar]
- 4.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–44. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 5.Gorouhi F, Islami F, Bahrami H, Kamangar F. Tumour-necrosis factor-A polymorphisms and gastric cancer risk: a meta-analysis. Br J Cancer. 2008;98:1443–51. doi: 10.1038/sj.bjc.6604277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang SS, Cerhan JR, Hartge P, Davis S, Cozen W, Severson RK, Chatterjee N, Yeager M, Chanock SJ, Rothman N. Common genetic variants in proinflammatory and other immunoregulatory genes and risk for non-Hodgkin lymphoma. Cancer Res. 2006;66:9771–80. doi: 10.1158/0008-5472.CAN-06-0324. [DOI] [PubMed] [Google Scholar]
- 7.Taioli E. Gene-environment interaction in tobacco-related cancers. Carcinogenesis. 2008;29:1467–74. doi: 10.1093/carcin/bgn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Zhao H, Suk R, Christiani DC. Genetic susceptibility to tobacco-related cancer. Oncogene. 2004;23:6500–23. doi: 10.1038/sj.onc.1207811. [DOI] [PubMed] [Google Scholar]
- 9.Hussain SK, Madeleine MM, Johnson LG, Du Q, Malkki M, Wilkerson HW, Farin FM, Carter JJ, Galloway DA, Daling JR, Petersdorf EW, Schwartz SM. Cervical and vulvar cancer risk in relation to the joint effects of cigarette smoking and genetic variation in interleukin 2. Cancer Epidemiol Biomarkers Prev. 2008;17:1790–9. doi: 10.1158/1055-9965.EPI-07-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Tan W, Hao B, Miao X, Zhou G, He F, Lin D. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 2003;63:8057–61. [PubMed] [Google Scholar]
- 11.Hashibe M, Morgenstern H, Cui Y, Tashkin DP, Zhang ZF, Cozen W, Mack TM, Greenland S. Marijuana use and the risk of lung and upper aerodigestive tract cancers: results of a population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2006;15:1829–34. doi: 10.1158/1055-9965.EPI-06-0330. [DOI] [PubMed] [Google Scholar]
- 12.Mu LN, Lu QY, Yu SZ, Jiang QW, Cao W, You NC, Setiawan VW, Zhou XF, Ding BG, Wang RH, Zhao J, Cai L, et al. Green tea drinking and multigenetic index on the risk of stomach cancer in a Chinese population. Int J Cancer. 2005;116:972–83. doi: 10.1002/ijc.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao W, Cai L, Rao JY, Pantuck A, Lu ML, Dalbagni G, Reuter V, Scher H, Cordon-Cardo C, Figlin RA, Belldegrun A, Zhang ZF. Tobacco smoking, GSTP1 polymorphism, and bladder carcinoma. Cancer. 2005;104:2400–8. doi: 10.1002/cncr.21446. [DOI] [PubMed] [Google Scholar]
- 14.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–27. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel U, Christensen J, Wallin H, Friis S, Nexo BA, Raaschou-Nielsen O, Overvad K, Tjonneland A. Polymorphisms in genes involved in the inflammatory response and interaction with NSAID use or smoking in relation to lung cancer risk in a prospective study. Mutat Res. 2008;639:89–100. doi: 10.1016/j.mrfmmm.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Van Dyke AL, Cote ML, Wenzlaff AS, Chen W, Abrams J, Land S, Giroux CN, Schwartz AG. Cytokine and cytokine receptor single-nucleotide polymorphisms predict risk for non-small cell lung cancer among women. Cancer Epidemiol Biomarkers Prev. 2009;18:1829–40. doi: 10.1158/1055-9965.EPI-08-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih C-M, Lee Y-L, Chiou H-L, Hsu W-F, Chen W-E, Chou M-C, Lin L-Y. The involvement of genetic polymorphism of IL-10 promoter in non-small cell lung cancer. Lung Cancer. 2005;50:291–97. doi: 10.1016/j.lungcan.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Hosgood HD, 3rd, Menashe I, Shen M, Yeager M, Yuenger J, Rajaraman P, He X, Chatterjee N, Caporaso NE, Zhu Y, Chanock SJ, Zheng T, et al. Pathway-based evaluation of 380 candidate genes and lung cancer susceptibility suggests the importance of the cell cycle pathway. Carcinogenesis. 2008;29:1938–43. doi: 10.1093/carcin/bgn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engels EA, Wu X, Gu J, Dong Q, Liu J, Spitz MR. Systematic evaluation of genetic variants in the inflammation pathway and risk of lung cancer. Cancer Res. 2007;67:6520–7. doi: 10.1158/0008-5472.CAN-07-0370. [DOI] [PubMed] [Google Scholar]
- 22.Fujieda S, Sunaga H, Tsuzuki H, Fan GK, Saito H. IL-10 expression is associated with the expression of platelet-derived endothelial cell growth factor and prognosis in oral and oropharyngeal carcinoma. Cancer Lett. 1999;136:1–9. doi: 10.1016/s0304-3835(98)00281-x. [DOI] [PubMed] [Google Scholar]
- 23.Chandler SW, Rassekh CH, Rodman SM, Ducatman BS. Immunohistochemical localization of interleukin-10 in human oral and pharyngeal carcinomas. Laryngoscope. 2002;112:808–15. doi: 10.1097/00005537-200205000-00008. [DOI] [PubMed] [Google Scholar]
- 24.Reichert S, Machulla HK, Klapproth J, Zimmermann U, Reichert Y, Glaser CH, Schaller HG, Stein J, Schulz S. The interleukin-10 promoter haplotype ATA is a putative risk factor for aggressive periodontitis. J Periodontal Res. 2008;43:40–7. doi: 10.1111/j.1600-0765.2007.00992.x. [DOI] [PubMed] [Google Scholar]
- 25.Sumer AP, Kara N, Keles GC, Gunes S, Koprulu H, Bagci H. Association of interleukin-10 gene polymorphisms with severe generalized chronic periodontitis. J Periodontol. 2007;78:493–7. doi: 10.1902/jop.2007.060309. [DOI] [PubMed] [Google Scholar]
- 26.Tezal M, Sullivan MA, Reid ME, Marshall JR, Hyland A, Loree T, Lillis C, Hauck L, Wactawski-Wende J, Scannapieco FA. Chronic periodontitis and the risk of tongue cancer. Arch Otolaryngol Head Neck Surg. 2007;133:450–4. doi: 10.1001/archotol.133.5.450. [DOI] [PubMed] [Google Scholar]
- 27.Tezal M, Grossi SG, Genco RJ. Is periodontitis associated with oral neoplasms? J Periodontol. 2005;76:406–10. doi: 10.1902/jop.2005.76.3.406. [DOI] [PubMed] [Google Scholar]
- 28.Vairaktaris E, Yapijakis C, Serefoglou Z, Avgoustidis D, Critselis E, Spyridonidou S, Vylliotis A, Derka S, Vassiliou S, Nkenke E, Patsouris E. Gene expression polymorphisms of interleukins-1 beta, −4, −6, −8, −10, and tumor necrosis factors-alpha, -beta: regression analysis of their effect upon oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2008;134:821–32. doi: 10.1007/s00432-008-0360-z. [DOI] [PubMed] [Google Scholar]
- 29.Yao JG, Gao LB, Liu YG, Li J, Pang GF. Genetic variation in interleukin-10 gene and risk of oral cancer. Clin Chim Acta. 2008;388:84–8. doi: 10.1016/j.cca.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Asada M, Yasuda H, Ebihara S, Tomita N, Suzuki S, Sato M, Kubo H, Yamaya M. Interleukin-1beta gene polymorphisms associated with risk of lung cancer in Japanese. Lung Cancer. 2006;54:261–3. doi: 10.1016/j.lungcan.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Zienolddiny S, Ryberg D, Maggini V, Skaug V, Canzian F, Haugen A. Polymorphisms of the interleukin-1 beta gene are associated with increased risk of non-small cell lung cancer. Int J Cancer. 2004;109:353–6. doi: 10.1002/ijc.11695. [DOI] [PubMed] [Google Scholar]
- 32.Lee KM, Shen M, Chapman RS, Yeager M, Welch R, He X, Zheng T, Hosgood HD, Yang D, Berndt SI, Chanock S, Lan Q. Polymorphisms in immunoregulatory genes, smoky coal exposure and lung cancer risk in Xuan Wei, China. Carcinogenesis. 2007;28:1437–41. doi: 10.1093/carcin/bgm030. [DOI] [PubMed] [Google Scholar]
- 33.Campa D, Hung RJ, Mates D, Zaridze D, Szeszenia-Dabrowska N, Rudnai P, Lissowska J, Fabianova E, Bencko V, Foretova L, Janout V, Boffetta P, et al. Lack of association between polymorphisms in inflammatory genes and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:538–9. doi: 10.1158/1055-9965.EPI-04-0513. [DOI] [PubMed] [Google Scholar]
- 34.Hu Z, Shao M, Chen Y, Zhou J, Qian J, Xu L, Ma H, Wang X, Xu Y, Lu D, Shen H. Allele 2 of the interleukin-1 receptor antagonist gene (IL1RN*2) is associated with a decreased risk of primary lung cancer. Cancer Lett. 2006;236:269–75. doi: 10.1016/j.canlet.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 35.Lind H, Zienolddiny S, Ryberg D, Skaug V, Phillips DH, Haugen A. Interleukin 1 receptor antagonist gene polymorphism and risk of lung cancer: A possible interaction with polymorphisms in the interleukin 1 beta gene. Lung Cancer. 2005;50:285–90. doi: 10.1016/j.lungcan.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Chanudet E, Ye H, Ferry J, Bacon CM, Adam P, Muller-Hermelink HK, Radford J, Pileri SA, Ichimura K, Collins VP, Hamoudi RA, Nicholson AG, et al. A20 deletion is associated with copy number gain at the TNFA/B/C locus and occurs preferentially in translocation-negative MALT lymphoma of the ocular adnexa and salivary glands. J Pathol. 2009;217:420–30. doi: 10.1002/path.2466. [DOI] [PubMed] [Google Scholar]
- 37.Morgan GJ, Adamson PJ, Mensah FK, Spink CF, Law GR, Keen LJ, Roman E, Davies FE, Rollinson S, Child JA, Bidwell JL. Haplotypes in the tumour necrosis factor region and myeloma. Br J Haematol. 2005;129:358–65. doi: 10.1111/j.1365-2141.2005.05467.x. [DOI] [PubMed] [Google Scholar]
- 38.Seifart C, Plagens A, Dempfle A, Clostermann U, Vogelmeier C, von Wichert P, Seifart U. TNF-alpha, TNF-beta, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers. 2005;21:157–65. doi: 10.1155/2005/707131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih CM, Lee YL, Chiou HL, Chen W, Chang GC, Chou MC, Lin LY. Association of TNF-alpha polymorphism with susceptibility to and severity of non-small cell lung cancer. Lung Cancer. 2006;52:15–20. doi: 10.1016/j.lungcan.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW, Tardon A, Serra C, Carrato A, Garcia-Closas R, Lloreta J, Castano-Vinyals G, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649–59. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcus PM, Hayes RB, Vineis P, Garcia-Closas M, Caporaso NE, Autrup H, Branch RA, Brockmoller J, Ishizaki T, Karakaya AE, Ladero JM, Mommsen S, et al. Cigarette smoking, N-acetyltransferase 2 acetylation status, and bladder cancer risk: a case-series meta-analysis of a gene-environment interaction. Cancer Epidemiol Biomarkers Prev. 2000;9:461–7. [PubMed] [Google Scholar]
- 42.Engstrom PF, Clapper M, Schnoll RA, Orleans CT. Prevention of Tobacco-Related Cancers. In: Kufe DW, Pollock RE, Weichselbaum RR, Bast RC Jr, Gansler TS, editors. Cancer Medicine. 6. Hamilton, Ontario: B.C. Decker; 2003. [Google Scholar]
- 43.Kinane DF, Chestnutt IG. Smoking and periodontal disease. Crit Rev Oral Biol Med. 2000;11:356–65. doi: 10.1177/10454411000110030501. [DOI] [PubMed] [Google Scholar]
- 44.Ji L, Melkonian G, Riveles K, Talbot P. Identification of pyridine compounds in cigarette smoke solution that inhibit growth of the chick chorioallantoic membrane. Toxicol Sci. 2002;69:217–25. doi: 10.1093/toxsci/69.1.217. [DOI] [PubMed] [Google Scholar]
- 45.Melkonian G, Cheung L, Marr R, Tong C, Talbot P. Mainstream and sidestream cigarette smoke inhibit growth and angiogenesis in the day 5 chick chorioallantoic membrane. Toxicol Sci. 2002;68:237–48. doi: 10.1093/toxsci/68.1.237. [DOI] [PubMed] [Google Scholar]
- 46.Hasnis E, Bar-Shai M, Burbea Z, Reznick AZ. Mechanisms underlying cigarette smoke-induced NF-kappaB activation in human lymphocytes: the role of reactive nitrogen species. J Physiol Pharmacol. 2007;58 (Suppl 5):275–87. [PubMed] [Google Scholar]
- 47.Kitamura M, Kasai A. Cigarette smoke as a trigger for the dioxin receptor-mediated signaling pathway. Cancer Lett. 2007;252:184–94. doi: 10.1016/j.canlet.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 48.Pryor WA, Prier DG, Church DF. Electron-spin resonance study of mainstream and sidestream cigarette smoke: nature of the free radicals in gas-phase smoke and in cigarette tar. Environ Health Perspect. 1983;47:345–55. doi: 10.1289/ehp.8347345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brody JS, Spira A. State of the Art. Chronic Obstructive Pulmonary Disease, Inflammation, and Lung Cancer. Proc Am Thorac Soc. 2006;3:535–37. doi: 10.1513/pats.200603-089MS. [DOI] [PubMed] [Google Scholar]
- 50.Posch PE, Cruz I, Bradshaw D, Medhekar BA. Novel polymorphisms and the definition of promoter ‘alleles’ of the tumor necrosis factor and lymphotoxin alpha loci: inclusion in HLA haplotypes. Genes Immun. 2003;4:547–58. doi: 10.1038/sj.gene.6364023. [DOI] [PubMed] [Google Scholar]
- 51.Smith AJ, Humphries SE. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev. 2009;20:43–59. doi: 10.1016/j.cytogfr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Fargion S, Valenti L, Dongiovanni P, Fracanzani AL. TNFalpha promoter polymorphisms. Methods Mol Med. 2004;98:47–58. doi: 10.1385/1-59259-771-8:047. [DOI] [PubMed] [Google Scholar]
- 53.Higuchi T, Seki N, Kamizono S, Yamada A, Kimura A, Kato H, Itoh K. Polymorphism of the 5′-flanking region of the human tumor necrosis factor (TNF)-alpha gene in Japanese. Tissue Antigens. 1998;51:605–12. doi: 10.1111/j.1399-0039.1998.tb03002.x. [DOI] [PubMed] [Google Scholar]
- 54.Ryder MI, Saghizadeh M, Ding Y, Nguyen N, Soskolne A. Effects of tobacco smoke on the secretion of interleukin-1beta, tumor necrosis factor-alpha, and transforming growth factor-beta from peripheral blood mononuclear cells. Oral Microbiol Immunol. 2002;17:331–6. doi: 10.1034/j.1399-302x.2002.170601.x. [DOI] [PubMed] [Google Scholar]
- 55.Rady PL, Matalon R, Grady J, Smith EM, Hudnall SD, Kellner LH, Nitowsky H, Tyring SK, Hughes TK. Comprehensive Analysis of Genetic Polymorphisms in the Interleukin-10 Promoter: Implications for Immune Regulation in Specific Ethnic Populations. Genetic Testing. 2004;8:194–203. doi: 10.1089/gte.2004.8.194. [DOI] [PubMed] [Google Scholar]
- 56.Suárez A, Castro P, Alonso R, Mozo L, Gutiérrez C. Interindividual variations in constitutive interleukin-10 messenger RNA and protein levels and their association with genetic polymorphisms. Transplantation. 2003;75:711–17. doi: 10.1097/01.TP.0000055216.19866.9A. [DOI] [PubMed] [Google Scholar]
- 57.Nielsen KB, Sorensen S, Cartegni L, Corydon TJ, Doktor TK, Schroeder LD, Reinert LS, Elpeleg O, Krainer AR, Gregersen N, Kjems J, Andresen BS. Seemingly neutral polymorphic variants may confer immunity to splicing-inactivating mutations: a synonymous SNP in exon 5 of MCAD protects from deleterious mutations in a flanking exonic splicing enhancer. Am J Hum Genet. 2007;80:416–32. doi: 10.1086/511992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang D, Johnson AD, Papp AC, Kroetz DL, Sadée W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenetics and Genomics. 2005;15:693–704. [PubMed] [Google Scholar]
- 59.Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–3. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 60.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 61.Rothman KJ, Greenland S, Lash TL. Modern Epidemiology. 3. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 62.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 63.Schottenfeld D, Fraumeni JF. Cancer epidemiology and prevention. 3. Oxford; New York: Oxford University Press; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.