Abstract
MicroRNAs are small RNAs that function as regulators of posttranscriptional gene expression. MicroRNAs are encoded by genes, and processed to form ribonucleoprotein complexes that bind to messenger RNA (mRNA) targets to repress translation or degrade mRNA transcripts. The microRNAs are particularly abundant in the brain where they serve as effectors of neuronal development and maintenance of the neuronal phenotype. They are also expressed in dendrites where they regulate spine structure and function as effectors in synaptic plasticity. MicroRNAs have been evaluated for their roles in brain ischemia, traumatic brain injury, and spinal cord injury, and in functional recovery after ischemia. They also serve as mediators in the brain's response to ischemic preconditioning that leads to endogenous neuroprotection. In addition, microRNAs are implicated in neurodegenerative disorders, including Alzheimer's, Huntington, Parkinson, and Prion disease. The discovery of microRNAs has expanded the potential for human diseases to arise from genetic mutations in microRNA genes or sequences within their target mRNAs. This review discusses microRNA discovery, biogenesis, mechanisms of gene regulation, their expression and function in the brain, and their roles in brain ischemia and injury, neuroprotection, and neurodegeneration.
Keywords: brain injury, microRNA, neurodegeneration, neuroprotection, posttranscriptional gene expression, translational repression
Introduction
Post-translational regulation of gene expression by small RNAs is one of the most important discoveries in the history of cell biology. This discovery was first noted by scientists who observed that exogenous gene sequences introduced into petunias could ‘silence' genes with similar gene sequences that were endogenous to the petunia (Napoli et al, 1990). This gene-silencing phenomenon was then observed in Ceanorhadbitis elegans by Andrew Fire and Rob Mello, who discovered its underlying mechanism, which they termed ‘RNA interference' (RNAi) (Fire et al, 1998). They proposed that RNAi is a cellular immune response to infection by double-stranded RNA (dsRNA) carried by viruses. Eukaryotic cells see exogenous dsRNA as foreign; thus infected cells process the dsRNA into short interfering RNAs (siRNAs) which bind to and cause the degradation of matching endogenous messenger RNA (mRNA) sequences (RNAi), preventing the synthesis of proteins necessary for viral replication. Fire and Mello were awarded the Nobel Prize in Physiology or Medicine in 2006 for their discovery of RNAi.
Studies in C. elegans suggested that an endogenous small RNA (lin-4) could regulate translation of lin-14, a protein required for postembryonic development, through an RNA–RNA interaction within the 3′-untranslated region (3′-UTR) of lin-14 (Lee et al, 1993). Subsequent studies supported the hypothesis that endogenous small RNAs could regulate mRNA translation, and the term ‘microRNA' was introduced in a series of articles in Science (Lagos-Quintana et al, 2001; Lau et al, 2001; Lee and Ambros, 2001). The microRNAs are members of a new family of small RNAs that are encoded by the genome, but their sequence is not translated into a protein (i.e., noncoding RNA). New members of the noncoding RNA family continue to be discovered, including the Piwi-interacting RNAs (Aravin et al, 2007) and the endogenous siRNAs (Lee et al, 2006), which silence transposable elements and protect the germ line from mutations (for reviews see Okamura and Lai, 2008; Werner and Sayer, 2009). The rapid pace of small RNA science is evidenced by the number of microRNAs in the most recent miRBase Sequence Database, Release 15, which contains 14,197 entries representing precursor microRNAs expressing 15,632 mature microRNA products in 133 species (Griffiths-Jones et al, 2008). A web resource for classified and clustered Piwi-interacting RNAs, piRNABank, underscores the large number of these RNAs that have already been discovered (Sai Lakshmi and Agrawal, 2008). Thus, as the number of distinct families of noncoding RNAs has now grown to over two dozen, the importance of these small RNAs on the regulation and/or modification of other RNAs, DNA, or proteins cannot be overstated (Burian, 2007).
MicroRNA Biogenesis
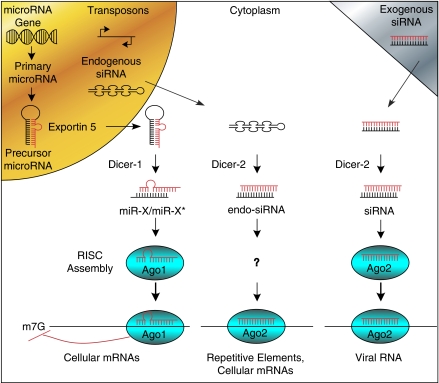
Recent reviews provide in-depth discussions of the biogenesis of microRNAs transcribed from genes (Chua et al, 2009; Davis and Hata, 2009; Winter et al, 2009), endogenous siRNAs that arise from bidirectional transcription of transposons, and exogenous siRNAs that arise from infection by dsRNA virus or experimental manipulations (Ghildiyal and Zamore, 2009; Okamura and Lai, 2008; Pontes and Pikaard, 2008). Figure 1 depicts the processing steps for microRNAs, endogenous siRNAs, and exogenous siRNAs. MicroRNA genes produce primary microRNA transcripts containing at least one ∼70 nucleotide hairpin loop. Further cleavage produces a precursor transcript that is transported into the cytoplasm by Exportin 5. Dicer then cleaves the RNA into an imperfect duplex of 20 to 25 nucleotides representing the mature microRNA and its complementary microRNA* strand. One strand of the duplex is degraded; in some cases, mature microRNAs are produced from both strands and designated as miR-X/miR-X* with the asterisk indicating the less predominantly expressed transcript. The mature microRNA bound to Dicer then binds to Argonaute (Ago) proteins to form RNA-induced silencing complexes (RISCs) (Morris, 2008; Tang, 2005; van den Berg et al, 2008). Parallel pathways form siRNAs from endogenous or exogenous dsRNAs, but use distinct isoforms of Dicer and Ago proteins.
Figure 1.
Biogenesis of microRNAs, endogenous short interfering RNAs (siRNAs), and exogenous siRNAs. Endogenous microRNAs are transcribed from nuclear genes into primary microRNA transcripts containing an ∼70 nucleotide hairpin loop structure, which are cleaved into precursor microRNA transcripts. The nuclear protein, Exportin 5, transports precursor microRNAs into the cytoplasm, where it is cleaved by Dicer to an imperfect miR-X:miR-X* duplex of ∼20 to 25 nucleotides. One strand of the duplex is degraded, and the remaining, mature microRNA binds to Dicer and Argonaute (Ago) proteins to form RNA-induced silencing complexes (RISCs). MicroRNAs target sequences within cellular messenger RNAs (mRNAs) causing repression of translation initiation and subsequent degradation of some mRNAs. Parallel processes in the cytoplasm produce siRNAs derived from endogenous transposons, which can target repetitive elements and cellular mRNAs, or from exogenous siRNAs that target viral mRNAs.
MicroRNA Regulation of Messenger RNA Targets
MicroRNAs in RISCs predominantly form Watson–Crick base pairs with sequences within the 3′-UTR of their mRNA targets (Bartel, 2004; Chan and Slack, 2006; Jackson and Standart, 2007; Pillai et al, 2007). The primary specificity determinant for mRNA target selection is a limited region of sequences within the microRNA, known as the ‘seed' (Ghildiyal and Zamore, 2009). However, interactions between proteins bound to microRNAs or their mRNA targets can also influence target selection (Forman and Coller, 2010). MicroRNAs primarily regulate posttranscriptional gene expression by translational repression or degradation of mRNA targets (Ambros, 2004; Bartel, 2004; Chen and Meister, 2005; Gebauer and Hentze, 2004; Pillai et al, 2007), but microRNAs have also been shown to activate translation during cell cycle arrest (Steitz and Vasudevan, 2009; Vasudevan and Steitz, 2007; Vasudevan et al, 2008).
Initiation of translation requires a direct interaction between eukaryotic initiation factor 4E and a 7-methylguanosine sequence on mature mRNAs (Gebauer and Hentze, 2004; Merrick, 2004; Richter and Sonenberg, 2005). When microRNAs are bound to the mRNA 3′-UTR, Ago proteins in the RISC interact with the 7-methylguanosine cap of the mRNA, which blocks eukaryotic initiation factor 4E binding to the mRNA and initiation of translation (Kiriakidou et al, 2007). This is the earliest event known to be regulated by microRNAs, but further mRNA degradation may serve to strengthen mRNA silencing (Mathonnet et al, 2007). Studies on miR-124 mRNA targets support that (1) microRNAs reduce both translation and abundance of mRNA targets, (2) translation is blocked at initiation or ribosomes preferentially drop off near the translation start site, and (3) regulation of translation and mRNA decay are correlated (Hendrickson et al, 2009). Thus, most mRNAs are not differentially targeted for either translational repression or mRNA decay.
MicroRNA function is not dependent on binding to one particular region of the mRNA, as studies show that association of a microRNA-ribonucleoprotein at any position on a target mRNA (3′- or 5′-UTR) is mechanistically sufficient to repress translation at some step downstream of initiation (Lytle et al, 2007). This finding is consistent with microRNA binding to sequences in the 5′-UTR of receptor-interacting protein 140 (Tsai et al, 2009), and with multiple independent studies showing that microRNAs target sequences in mRNA coding regions, although the effects are weaker than for sites in the 3′-UTR (Forman and Coller, 2010).
Once microRNAs and their associated RISCs are bound to an mRNA target, the whole complex can be sequestered into processing bodies (Pillai et al, 2005, 2007; Sheth and Parker, 2003). This action involves phosphorylation of Ago2 in the RISCs by p38 mitogen-activated protein kinase (Zeng et al, 2008). When a subsequent cellular stress releases the RISC-bound mRNAs from processing bodies, the freed mRNAs are recruited to ribosomes and translation can ensue (Bhattacharyya et al, 2006). Sequestration of RISC-bound mRNAs in processing bodies also occurs in synapses (Konecna et al, 2009), suggesting that this mechanism is an important regulator of mRNA translation in response to synaptic activity.
MicroRNA Target Prediction
The first open-source software for target prediction, miRanda 2005, revealed that overrepresented groups of microRNA targets include mRNAs for transcription factors, components of the microRNA machinery, and other proteins involved in translational regulation (John et al, 2004). In contrast, a large number of genes for proteins involved in basic cellular processes have very short 3′-UTRs and are specifically depleted of microRNA-binding sites (Stark et al, 2005). Vertebrate microRNAs target ∼200 mRNA transcripts each (Krek et al, 2005), but the number of predicted targets per microRNA can vary from a few to >800 transcripts. A recent extensive review discusses the main aspects of computational algorithms for microRNA gene finding, microRNA target prediction, and regulation of microRNA genes, and provides online resources for target prediction programs (Li et al, 2010). MiRanda 2005 and 2008 (Betel et al, 2008; John et al, 2004), and PicTar (Krek et al, 2005), allow combinatorial analysis of microRNAs for common targets, which is important because mRNAs targeted by multiple microRNAs show enhanced translational repression (Doench and Sharp, 2004).
Currently, the first order approach to target prediction utilizes sequence alignments between the seed region of the microRNA and sequences within the mRNA target. However, specificity can be increased by analyzing the evolutionary conservation and structural accessibility of mRNA binding sites, as well as the nucleotide composition or location of binding sites within the mRNA 3′-UTR (Alexiou et al, 2009). Databases of targets with experimental validation include Tarbase (Sethupathy et al, 2006), Ago (Shahi et al, 2006), and miRNAMAP (Hsu et al, 2006, 2008). At the present time, there is no universal standard for establishing a causal relationship between microRNAs and predicted mRNA targets, such as Koch's postulates for the relationship between a microbe and a disease. However, Kuhn et al (2008) proposed that four criteria should be met before microRNA target validation is considered confirmed: (1) the microRNA/mRNA interaction must be experimentally verified, (2) the microRNA and mRNA target must be coexpressed, (3) a given microRNA must have a predictable effect on target protein expression, and (4) microRNA-mediated regulation of target gene expression should equate to altered biological function (Kuhn et al, 2008). Thus, rapid and continued evolution of prediction tools and criteria for experimental validation are essential to establish the functional effects of microRNAs on their mRNA targets.
MicroRNA Expression and Functions in the Brain
MicroRNAs serve important roles in development and function in the brain (Bicker and Schratt, 2008; Christensen and Schratt, 2009; Schratt, 2009; Fiore et al, 2008; Zeng, 2009). Studies support that tissue-specific microRNAs serve to establish and maintain protein expression profiles underlying distinct cellular phenotypes. The discovery of seven brain-specific microRNAs (miR-9, miR-124a, miR-124b, miR-135, miR-153, miR-183, and miR-219) in mouse and human differentiating neurons implicated these microRNAs as effectors in mammalian neuronal processes (Sempere et al, 2004). Further studies showed that expression levels of the brain-specific miR-124 are >100 times higher in mouse CNS than in other organs, whereas levels of muscle-specific miR-1 are 100 to 1000 times lower in mouse CNS than in heart and skeletal muscle (Mishima et al, 2007). Transfection of brain-specific miR-124 into HeLa cells shifted the expression profile toward that of brain, whereas transfection of the heart and skeletal muscle-specific miR-1 into HeLa cells shifted the expression profile toward that of muscle (Lim et al, 2005). In addition, microarray studies of miR-124- and miR-1-transfected cells revealed reduced levels of many mRNA targets, not just their protein correlates, suggesting that miR-1 and miR-124 downregulate many of their mRNA targets.
Studies also support that anatomically specific microRNAs serve to establish and maintain protein expression profiles underlying cellular phenotypes in distinct neuroanatomical regions. For example, the expression of miR-92b, miR-146b, let-7g, miR-551b, miR-330*, and miR-384 is significantly higher in rat hippocampus than in cortex (He et al, 2007). In adult mice, expression of 44 microRNAs is enriched >3-fold in spinal cord, cerebellum, medulla oblongata, pons, hypothalamus, hippocampus, neocortex, olfactory bulb, eye, and pituitary gland (Bak et al, 2008). Analysis of microRNAs in rat amygdala, cerebellum, hippocampus, hypothalamus, and substantia nigra revealed 48 microRNAs with >3-fold enrichment between two or more brain regions, and reciprocal expression between microRNAs in the cerebellum and forebrain regions, suggesting area-specific functions for these brain microRNAs (Olsen et al, 2009). Specific development expression of miR-449 is important in the choroid plexus, the area on the brain ventricles where cerebrospinal fluid is produced, as miR-449 targets the transcription factor E2f5 which regulates cerebrospinal fluid production (Redshaw et al, 2009). Two of the most highly expressed microRNAs in adult brain, miR-124 and miR-128, are preferentially expressed in neurons, in contrast to miR-23 expression that is restricted to astrocytes, or miR-26 and miR-29 that are more strongly expressed in astrocytes than neurons (Smirnova et al, 2005). It should be noted that nearly half of all microRNA genes are located within introns, or noncoding regions, of their host genes, and intronic microRNAs can silence genes that are functionally antagonistic to the host gene to ensure expression of the host gene. For example, the apoptosis-associated tyrosine kinase gene is essential for neuronal differentiation. However, transcription of the apoptosis-associated tyrosine kinase gene also generates miR-338 from an apoptosis-associated tyrosine kinase intron, which in turn silences genes that negatively regulate neuronal differentiation (Barik, 2008).
Functions of Brain-Specific MicroRNAs
The transcriptional repressor, RE1-silencing transcription factor (REST), inhibits the expression of neuronal genes in nonneuronal cells (Chong et al, 1995; Schoenherr and Anderson, 1995), whereas the brain-specific miR-124a promotes a neuronal-like phenotype by decreasing the levels of hundreds of nonneuronal transcripts (Lim et al, 2005). REST inhibits miR-124a expression, which allows the translation of hundreds of nonneuronal transcripts, but REST is absent in mature neurons, which allows the derepression of neuronal genes, including miR-124a, and a resultant neuronal phenotype (Conaco et al, 2006).
Like REST, the small C-terminal domain phosphatase 1 is an antineural factor expressed in nonneuronal tissues, and as a target of miR-124, suppression of small C-terminal domain phosphatase 1 is critical for inducing neurogenesis during CNS development (Visvanathan et al, 2007). In the mouse subventricular zone, miR-124 repression of the SRY-box transcription factor Sox9 allows progression of the subventricular zone stem cell lineage to neurons (Cheng et al, 2009). miR-124 also represses polypyrimidine tract binding protein 1, which leads to increased expression of polypyrimidine tract binding protein 2 and correct alternative mRNA splicing that is necessary for the transition from a nonneuronal to neuronal phenotype (Makeyev et al, 2007). These studies support that miR-124 specifically targets hundreds of mRNAs for proteins that define nonneuronal phenotypes and that repression and/or degradation of ‘lingering' mRNAs by miR-124 allows the transition from nonneuronal to neuronal phenotypes during development.
Studies show that microRNAs localized to dendrites serve to regulate dendritic spine structure and size by actions on distinct mRNA targets, supporting a role for microRNAs in translational control that is necessary for synaptic plasticity and memory (Costa-Mattioli et al, 2009). For example, miR-132 expression induced neurite outgrowth in cortical neurons (Vo et al, 2005) and enhanced dendrite morphogenesis in hippocampal neurons through repressed expression of the GTPase-activating protein, p250GAP (Wayman et al, 2008). In addition, miR-134 localized to the synapto-dendritic compartment of rat hippocampal neurons negatively regulates the size of dendritic spines by inhibiting translation of LIM domain kinase 1 (Limk1), which controls spine development (Schratt et al, 2006). Exposure of neurons to brain-derived neurotrophic factor relieved the inhibition of translation by miR-134, suggesting a mechanism for neurotrophic control of dendritic spine plasticity (Schratt et al, 2006). Further, miR-138 localized in dendrites negatively regulates the size of dendritic spines, whereas inhibition of miR-138 enlarges dendritic spines, and miR-138 targets include acyl protein thioesterase 1, which regulates palmitoylation and membrane localization of synaptic proteins (Siegel et al, 2009).
Most neuronal microRNAs are detected in dendrites and distributed through the somatodendritic compartment across a nearly constant gradient (Kye et al, 2007). However, a subset of mouse forebrain microRNAs are significantly enriched or depleted in synaptoneurosomes (Smalheiser, 2008). These synaptically enriched microRNAs show structurally distinct precursor microRNA features, which may allow differential binding to proteins that mediate dendritic transport, or prevent premature processing into mature microRNAs during transport (Smalheiser, 2008). Precursor microRNAs are predominantly associated with postsynaptic densities, whereas mature microRNAs are predominantly associated with soluble components of synaptic fractions. The significant correlation between synaptic enrichment of precursor and mature microRNAs suggests that they are likely processed within dendritic spines (Lugli et al, 2008). Dicer is preferentially expressed in the Golgi-reticulum area in differentiated neurons and glia (Barbato et al, 2007), and neuronal-mediated calcium influxes in mouse forebrain neurons activate proteases such as calpain, which liberates Dicer from the postsynaptic density (Lugli et al, 2005, 2008). These studies support a model where synaptic stimulation and increased calcium within dendritic spines activates calpain, resulting in release of Dicer from the postsynaptic density and processing of local precursor microRNAs into mature microRNAs, which are then incorporated into RISCs that bind to mRNA targets in the vicinity (Smalheiser and Lugli, 2009).
MicroRNAs in Synaptic Plasticity
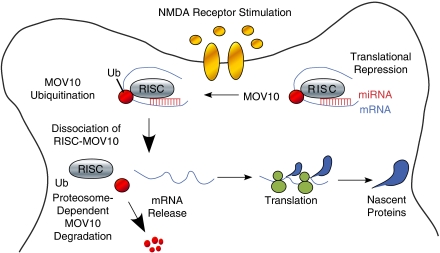
Persistent changes in synaptic strength that lead to learning and memory require both protein synthesis and protein degradation. However, recent studies on local translational regulatory mechanisms provide a model to explain these paradoxical requirements (Banerjee et al, 2009). As depicted in Figure 2, adapted from Banerjee et al (2009), N-methyl--aspartic acid receptor stimulation of neuronal synapses rapidly induced ubiquitination and proteosomal degradation of the RISC protein, MOV10 (Banerjee et al, 2009). The newly freed mRNA transcripts for Limk1, calmodulin-dependent Kinase II (α-CaMKII), and depalmitoylating enzyme lysophospholipase1 (Lypla1) could then selectively enter the polysome compartment where initiation of translation ensued. The Lypla1 mRNA was associated with brain-enriched miR-138, while the activity-dependent translation of Lypla1 and α-CaMKII was driven by their 3′-UTR sequences, and was MOV10 and proteasome dependent (Banerjee et al, 2009).
Figure 2.
Model for microRNAs as effectors of synaptic learning and memory. Synapses that contain RNA-induced silencing complex (RISC)-bound messenger RNAs (mRNAs) respond to N-methyl--aspartic acid receptor stimulation by dissociation of the translationally repressed mRNAs and ubiquitination (Ub) of the RISC-associated protein, MOV10. The freed mRNAs enter the polysome compartment where translation is initiated, and ubiquitinated MOV10 is rapidly degraded by the proteasome. This model accounts for the paradoxical requirement of both protein degradation and protein synthesis that are required for persistent changes in synaptic strength that are necessary for learning and memory. Adapted from Banerjee et al (2009).
Further studies show that synaptic activity regulates the expression of distinct microRNAs that serve as effectors in neuronal responses to stimuli which lead to synaptic plasticity. For example, expression studies revealed that several hippocampal microRNAs are altered by chemical induction of long-term potentiation and by metabotropic glutamate receptor-dependent long-term depression (Park and Tang, 2009). The majority of altered microRNAs followed a similar temporal expression profile, while some displayed distinct expression profiles. The discovery of microRNAs upregulated at specific times after long-term potentiation and long-term depression suggests that synaptic activation can elicit microRNA-mediated suppression of mRNA translation to prevent excess protein synthesis during the expression of synaptic plasticity (Park and Tang, 2009). In Aplysia californica, miR-124 is exclusively expressed in a presynaptic sensory-motor synapse and restricts serotonin-induced synaptic facilitation by regulating CREB, revealing a role for miR-124 in long-term synaptic plasticity in mature neurons (Rajasethupathy et al, 2009). In rodent cells, nicotine-induced expression of miR-140* repressed translation of dynamin 1, which is essential for synaptic endocytosis in the CNS, and thus miR-140* is likely important for neural plasticity in nicotine addiction (Huang and Li, 2009).
MicroRNAs in Brain Injury and Neuroprotection
MicroRNAs in Brain Ischemia
Brain ischemia is one of the most common causes of disability and death worldwide (Lloyd-Jones et al, 2009), and studies reveal a role for microRNAs in responses to ischemic insults. Transient focal ischemia in adult rat brain regulates the expression of microRNAs predicted to target proteins known to mediate inflammation, transcription, neuroprotection, receptor function, and ionic homeostasis in the brain (Dharap et al, 2009). The mRNA levels for proteins important to microRNA biogenesis pathways, including Drosha, Dicer, the cofactor Pasha, and the precursor microRNA transporter Exportin 5, were not altered after transient ischemia. However, transient ischemia repressed miR-145 expression, which resulted in increased translation of its mRNA target, superoxide dismutase-2, in postischemic adult rat brain (Dharap et al, 2009). It is interesting to note that in silico studies revealed eight microRNAs induced by transient ischemia with complementarity to 877 gene promoters, suggesting that microRNAs also regulate gene expression (Dharap et al, 2009). There is also specific induction of miR-497 in mouse brain after transient ischemia, and in mouse N2A neuroblastoma (N2A) cells after oxygen–glucose deprivation (Yin et al, 2010). Levels of miR-497 correlated with oxygen–glucose deprivation-induced effects on N2A cells: decreased miR-497 suppressed cell death, whereas increased miR-497 increased neuronal loss. As miR-497 directly binds to the 3′-UTR of Bcl-2/-w, the knockdown of cerebral miR-497 in mice enhanced Bcl-2/-w protein levels in the ischemic region, attenuated brain infarction, and improved neurological outcome after focal ischemia. These studies show that miR-497 promotes ischemic neuronal death by repressing expression of Bcl-2 and Bcl-w, supporting the role of apoptosis in the pathogenesis of ischemic brain injury (Yin et al, 2010).
Studies also support the potential for microRNAs as novel biomarkers for vascular injury and diseases. Expression profiling of microRNAs in ischemic rat brains revealed significant changes in several microRNAs, and some of the microRNAs highly expressed in ischemic brain were detected in blood samples (Jeyaseelan et al, 2008). Peripheral blood examined in ischemic stroke patients revealed differential expression of microRNAs implicated in endothelial cell and vascular function, erythropoiesis, angiogenesis, neural function, and hypoxia, and altered microRNAs were detectable even several months after the onset of stroke (Tan et al, 2009). Rat models of ischemia, brain hemorrhage, and kainate-induced seizures also revealed regulated expression of microRNAs in hippocampus and blood in each treatment group, many of which changed >1.5-fold in both tissues (Liu et al, 2010).
Evidence also suggests that microRNAs serve as effectors in neointimal lesion formation, and in angiogenesis in normal and injured brain. The miR-17–92 cluster is highly expressed in human endothelial cells and miR-92a, a component of this cluster, targets several mRNAs for proangiogenic proteins (Bonauer et al, 2009). Overexpression of miR-92a in endothelial cells blocked angiogenesis, and systemic administration of a miR-92a antagomir led to enhanced blood vessel growth and functional recovery of damaged tissue in mouse models of limb ischemia and myocardial infarction (Bonauer et al, 2009). In a similar vein, profiling of microRNAs in vascular walls after balloon injury revealed that miR-21 is overexpressed in injured vascular tissue, and that miR-21 depletion inhibited formation of neointimal lesions (Ji et al, 2007). Depletion of miR-21 decreased cell proliferation and increased cell apoptosis, and targets of miR-21 include the phosphatase and tensin homolog protein (PTEN) and Bcl-2 (Ji et al, 2007).
Platelets are crucial for the maintenance of hemostasis and contribute to thrombosis and vessel occlusion that underlies stroke and acute coronary syndromes. Although platelets are anucleate, they do contain mRNAs and are capable of protein synthesis (Weyrich et al, 2004). Human platelets have been shown to contain microRNAs and Dicer in Ago2 protein complexes, as well as mRNA for the P2Y purinoceptor 12 that is involved in platelet aggregation, suggesting a role for microRNAs in this system (Landry et al, 2009).
MicroRNAs in Traumatic Brain Injury
MicroRNA expression studies suggest a role for postinjury microRNAs in traumatic brain injury processes. In rat cortex and hippocampus, traumatic brain injury induced expression of several microRNAs and caused global upregulation of miR-21 after injury (Lei et al, 2009). Similarly, in rat and mouse hippocampus, controlled cortical impact injury decreased expression of 50 microRNAs and increased expression of 35 microRNAs (Redell et al, 2009). Predicted targets of validated microRNAs regulated by impact (miR-107, miR-130a, miR-223, miR-292-5p, miR-433-3p, miR-451, miR-541, and miR-711) include several proteins and pathways known to be initiated after injury, including signal transduction, transcriptional regulation, proliferation, and differentiation (Redell et al, 2009). Traumatic spinal cord injury also alters the expression of microRNAs in adult rats, and predicted targets for the spinal cord injury-regulated microRNAs include proteins involved in inflammation, oxidation, and apoptosis that are known to underlie pathogenesis of spinal cord injury (Liu et al, 2009a). Given that mechanical forces are important considerations in traumatic brain injury and spinal cord injury, it is significant that mechanical shear stresses modulate vascular homeostasis through microRNA regulation of targets (Weber et al, 2010). Human endothelial cells subjected to unidirectional shear stress increased expression of 13 microRNAs, with the greatest change observed for miR-21, which targets PTEN. Subsequent studies in human vascular endothelial cells exposed to shear stress or transfected with miR-21 showed downregulation of PTEN, whereas cells overexpressing miR-21 had decreased apoptosis, increased phosphorylation of endothelial nitric oxide synthase, and nitric oxide production (Weber et al, 2010). Together, these findings show that shear stress can regulate microRNA expression in endothelial cells, and suggests potential roles for microRNAs in mechanotransduction and injury response.
MicroRNAs in Ischemic Tolerance
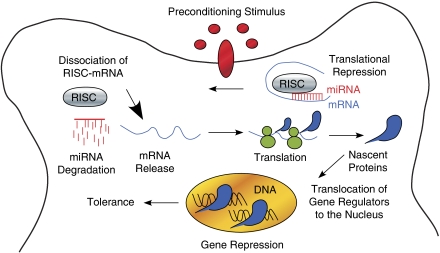
Ischemic tolerance is the response to a short duration of ischemia (preconditioning), which protects cells against a subsequent injurious duration of ischemia (Dirnagl et al, 2009, 2003; Liu et al, 2009b; Stenzel-Poore et al, 2007). Preconditioning-induced tolerance is known to require new protein synthesis (Barone et al, 1998) and exhibits a transient repression of gene expression (Stenzel-Poore et al, 2003). Studies to examine a role for microRNAs as regulators of new protein synthesis revealed that preconditioning both upregulated and downregulated microRNA expression in mouse cortex (Lusardi et al, 2010). One prominent predicted target of the decreased microRNAs was the global transcriptional regulator, methyl CpG binding protein 2 (MeCP2), which had no prior recognized role in preconditioning or tolerance. Subsequent studies in spontaneously hypertensive rats also observed that multiple microRNAs downregulated after preconditioning are predicted to target MeCP2 mRNAs (Dharap and Vemuganti et al, 2010). MeCP2 is a potent transcriptional repressor (Nan et al, 1998) and transcriptional activator (Chahrour et al, 2008). Mutations in the MeCP2 gene cause Rett syndrome (Hite et al, 2009) and several other CNS disorders including mental retardation, Angelman syndrome, and autism (Gonzales and LaSalle, 2010). Given that repressed gene expression is a feature of tolerance, it was significant that expression of MeCP2 rapidly increased in preconditioned mouse cortex with no correlating changes in mRNA expression and MeCP2 knockout mice showed increased susceptibility to ischemia (Lusardi et al, 2010), suggesting that MeCP2 may be an effector of preconditioning-induced tolerance. Figure 3 depicts a model for microRNAs in preconditioning-induced tolerance that is consistent with the known features of tolerance; new protein synthesis and repressed gene expression. In this model, the preconditioning stimulus derepresses RISC-bound mRNAs and the newly freed mRNAs are translated. Among the newly synthesized proteins are transcriptional regulators, which translocate into the nucleus and regulate gene expression. The role of MeCP2 in tolerance may be to simultaneously repress nonessential genes to conserve energy in ischemic cells where oxygen and glucose are depleted, and to activate essential genes required for cell survival. Consistent with the transient nature of tolerance, MeCP2 expression returns to normal when spared subsequent ischemic challenge. This model is similar to that for N-methyl--aspartic acid-dependent learning and memory (Figure 2; Banerjee et al, 2009), and supports that microRNAs are effectors of rapid, transient translation of synaptic proteins in response to neuronal stimuli. These models also support that the nature of the stimulus dictates the phenotype of the response, as shown for distinct preconditioning paradigms (Stenzel-Poore et al, 2007).
Figure 3.
Model for microRNAs as effectors of preconditioning-induced tolerance. Synapses that contain RNA-induced silencing complex (RISC)-bound messenger RNA (mRNAs) respond to a preconditioning stimulus by dissociation of the translationally repressed mRNAs and degradation of the microRNAs. The freed mRNAs are translated, and new proteins include transcriptional regulators that translocate into the nucleus and regulate gene expression. This model accounts for the know features of preconditioning-induced tolerance, the requirement of new protein synthesis and transcriptional repression in tolerance.
MicroRNAs in Neurodegeneration
Recent reviews underscore the roles for microRNAs in neurodegenerative disorders (Barbato et al, 2009; Eacker et al, 2009; Hebert and De Strooper, 2009; Weinberg and Wood, 2009; Yelamanchili and Fox, 2009). Here, we present an overview of studies suggesting microRNAs as effectors in the processes underlying four specific neurodegenerative diseases, which hold promise for new approaches for the treatment or prevention of these neurological disorders.
MicroRNAs in Alzheimer's Disease
Alzheimer's disease (AD) is an incurable, degenerative, and terminal condition, and is the most common form of dementia. The majority of AD cases are sporadic (not genetically inherited), yet some genes may act as risk factors, and studies implicate a role for microRNAs in sporadic AD (Maes et al, 2009) and frontotemporal lobar degeneration (Rademakers and Rovelet-Lecrux, 2009). Generation of the amyloid-β peptides that aggregate in the brain of AD patients requires two sequential cleavages of amyloid precursor protein (APP), the first of which requires β-site of APP cleaving enzyme (BACE1). The finding that microRNAs in the miR-29 gene cluster (miR-9, miR-29a, and miR-29b-1) can regulate BACE1 expression in vitro is consistent with significantly decreased expression of miR-29a and miR-29b-1 in AD patients with abnormally high levels of BACE1, and suggests that loss of specific microRNAs contributes to increased BACE1 and amyloid-β levels in sporadic AD (Hebert et al, 2008). However, human genetic studies of microRNA-binding sites within the APP or BACE1 3′-UTRs, or microRNAs in the miR-29 gene cluster, did not support a major contribution of microRNA genetic variability in AD (Bettens et al, 2009). Expression of microRNAs in the miR-20a family (miR-20a, miR-17-5p, and miR-106b) reveal a tight correlation with APP expression during brain development and in differentiating neurons, and sporadic AD patients have a significant decrease in miR-106b expression (Hebert et al, 2009). In addition, miR-107 that is predicted to target the BACE1 3′-UTR at multiple sites is significantly decreased in patients with the earliest stages of pathology (Wang et al, 2008b). The further finding that BACE1 mRNA levels increase as miR-107 levels decrease in the progression of AD suggests that miR-107 may be involved in accelerated disease progression through the regulation of BACE1 (Wang et al, 2008b).
Increased expression of BACE protein, but not mRNA, is seen in the brain by postmortem analyses of AD patients and in a mouse model of AD. The BACE1 mRNA 3′-UTR contains predicted binding sites for miR-298 and miR-328, and in vitro studies confirm the regulation of BACE1 protein expression by these microRNAs (Boissonneault et al, 2009). Further studies in AD brain revealed that increased expression of miR-146a correlates with decreased expression of the predicted mRNA target, complement factor H, an important repressor of brain inflammatory responses (Lukiw et al, 2008). Increased expression of miR-9, miR-125b, and miR-146a in AD brain is correlated with neuropathological changes (Sethi and Lukiw, 2009). Together, these studies suggest that microRNAs contribute to BACE1 and APP expression, and warrant further studies to examine the role of microRNAs in AD pathogenesis.
MicroRNAs in Huntington Disease
Huntington disease (HD) is a dominantly inherited neurodegenerative disorder that is incurable and ultimately fatal (Johnson et al, 2008). The transcription factor REST silences neuronal gene expression in nonneuronal cells, and REST is sequestered in the cytoplasm in part through binding to the Huntingtin protein. However, Huntingtin proteins that contain polyglutamine expansions cannot bind to REST, which frees REST to translocate to the nucleus where it represses neuronal gene expression. Recent studies revealed dysregulated expression of several neuronal-specific microRNAs in mouse models of HD and in human HD that likely result from REST repression (Johnson et al, 2008). The loss of microRNA expression correlates with increased expression of several mRNA targets, supporting that HD reflects a loss of neuronal identity caused in part by dysregulation of both transcriptional and posttranscriptional gene expression (Johnson et al, 2008). Further studies show that expression of several microRNAs with upstream REST-binding sites are decreased in human HD, including the brain-enriched miR-9/miR-9*, which target REST and CoREST, respectively, suggesting that bifunctional microRNAs serve as effectors of a double-negative feedback loop between the REST-silencing complex and the microRNAs that it regulates (Packer et al, 2008).
MicroRNAs in Parkinson Disease
There is also evidence for a role of microRNAs in Parkinson disease (PD), a neurodegenerative disorder caused by environmental and genetic factors. For example, miR-133b is specifically expressed in midbrain dopaminergic neurons and regulates the maturation and function of midbrain dopaminergic neurons, but miR-133b is deficient in patients with PD (Kim et al, 2007). In addition, fibroblast growth factor 20 is a risk factor for PD, and genetic analysis of single-nucleotide polymorphisms within the fibroblast growth factor 20 gene revealed association from a risk allele (rs12720208) in the 3′-UTR (Wang et al, 2008a). Functional assays showed that rs12720208 disrupts a binding site for miR-433 that leads to increased expression of fibroblast growth factor 20 and subsequent increases in α-synuclein expression that can cause PD (Wang et al, 2008a). Further, miR-7 that is expressed mainly in neurons binds to the α-synuclein mRNA 3′-UTR to repress protein expression, which protects cells against oxidative stress (Junn et al, 2009). Studies in the MPTP neurotoxin model of PD also suggest that decreased expression of miR-7 results in increased expression of α-synuclein (Junn et al, 2009).
MicroRNAs in Prion Diseases
Prion diseases are fatal, degenerative diseases, and include Creutzfeldt–Jakob disease in humans, bovine spongiform encephalopathy in cattle, and Scrapie in sheep and goats. Studies suggest that microRNA expression might provide a potential marker for prion diseases in animals and humans. For example, the expression of 15 microRNAs was dysregulated in mouse brains infected with mouse-adapted scrapie (Saba et al, 2008). Of these, miR-342-3p, miR-320, let-7b, miR-328, miR-128, miR-139-5p, and miR-146a showed >2.5-fold upregulation, whereas miR-338-3p and miR-337-3p showed >2.5-fold downregulation. Prediction analysis revealed many mRNA targets, including 119 mRNAs dysregulated in mouse scrapie (Saba et al, 2008). In addition, studies in a monkey model of Creutzfeldt–Jakob disease revealed a significant upregulation of miR-342-3p, consistent with upregulation of miR-342-3p in brain samples from humans with sporadic Creutzfeldt–Jakob disease (Montag et al, 2009).
Concluding Remarks
Cell biology and consequently, biomedical research, has undergone a paradigm shift in the short time since the discovery of small noncoding RNAs. As we have moved away from the notion that 95% of the genome is ‘junk DNA,' it is gratifying and daunting to discover that the genome is teeming with information, and gives rise to effectors both large and small that together form complex interactions which underlie life. Genetic variations in microRNAs and/or their mRNA targets which disrupt their interaction will likely emerge as the underlying cause of numerous human diseases and disorders. The important discovery of Piwi-interacting RNAs and endogenous siRNAs, which protect the genome from external threats and thus maintain the integrity of the genome, has cemented the idea that no area of cellular function will be left untouched by small RNAs. The implications for these tiny molecules in all aspects of cellular development and homeostasis present major challenges in terms of both technology development and bioinformatic analysis. That new small RNA families are still being discovered underscores the major challenges that lie ahead in deciphering the functions regulated by these effectors and in constructing interpretable data networks, which represent both individual and cooperative effects of small RNAs on cellular function, an undertaking of great significance.
Acknowledgments
The author acknowledges that many excellent original articles on microRNAs have been published over the last two decades. Owing to constraints, many of these central studies are cited in reviews, and exclusion of any particular article was not intentional. The author thanks her husband, friend and colleague, Dr Michael Roberts, for his constructive comments regarding this review, all of which improved the final article.
The author declares no conflict of interest.
References
- Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Barbato C, Ciotti MT, Serafino A, Calissano P, Cogoni C. Dicer expression and localization in post-mitotic neurons. Brain Res. 2007;1175:17–27. doi: 10.1016/j.brainres.2007.07.088. [DOI] [PubMed] [Google Scholar]
- Barbato C, Ruberti F, Cogoni C. Searching for MIND: microRNAs in neurodegenerative diseases. J Biomed Biotechnol. 2009;2009:871313. doi: 10.1155/2009/871313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–5241. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone FC, White RF, Spera PA, Ellison J, Currie RW, Wang X, Feuerstein GZ.1998Ischemic preconditioning and brain tolerance: temporal histological and functional outcomes, protein synthesis requirement, and interleukin-1 receptor antagonist and early gene expression Stroke 291937–1950.discussion 1950–1951 [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettens K, Brouwers N, Engelborghs S, Van Miegroet H, De Deyn PP, Theuns J, Sleegers K, Van Broeckhoven C. APP and BACE1 miRNA genetic variability has no major role in risk for Alzheimer disease. Hum Mutat. 2009;30:1207–1213. doi: 10.1002/humu.21027. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- Bicker S, Schratt G. microRNAs: tiny regulators of synapse function in development and disease. J Cell Mol Med. 2008;12:1466–1476. doi: 10.1111/j.1582-4934.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- Burian RM. On microRNA and the need for exploratory experimentation in post-genomic molecular biology. Hist Philos Life Sci. 2007;29:285–311. [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SP, Slack FJ. microRNA-mediated silencing inside P-bodies. RNA Biol. 2006;3:97–100. doi: 10.4161/rna.3.3.3499. [DOI] [PubMed] [Google Scholar]
- Chen PY, Meister G. microRNA-guided posttranscriptional gene regulation. Biol Chem. 2005;386:1205–1218. doi: 10.1515/BC.2005.139. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Christensen M, Schratt GM. microRNA involvement in developmental and functional aspects of the nervous system and in neurological diseases. Neurosci Lett. 2009;466:55–62. doi: 10.1016/j.neulet.2009.04.043. [DOI] [PubMed] [Google Scholar]
- Chua JH, Armugam A, Jeyaseelan K. MicroRNAs: biogenesis, function and applications. Curr Opin Mol Ther. 2009;11:189–199. [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hata A. Regulation of MicroRNA biogenesis: a miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral MicroRNAome. J Cereb Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Vemuganti R.2010Ischemic pre-conditioning alters cerebral microRNAs that are upstream to neuroprotective signaling pathways J Neurochemadvanced online publication 6 April 2010 [e-pub ahead to print] [DOI] [PMC free article] [PubMed]
- Dirnagl U, Becker K, Meisel A. Preconditioning and tolerance against cerebral ischaemia: from experimental strategies to clinical use. Lancet Neurol. 2009;8:398–412. doi: 10.1016/S1474-4422(09)70054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirnagl U, Simon RP, Hallenbeck JM. Ischemic tolerance and endogenous neuroprotection. Trends Neurosci. 2003;26:248–254. doi: 10.1016/S0166-2236(03)00071-7. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Forman JJ, Coller HA. The code within the code: microRNAs target coding regions. Cell Cycle. 2010;9:1533–41. doi: 10.4161/cc.9.8.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales ML, LaSalle JM. The role of MeCP2 in brain development and neurodevelopmental disorders. Curr Psychiatry Rep. 2010;12:127–134. doi: 10.1007/s11920-010-0097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang Q, Liu Y, Pan X. Cloning and identification of novel microRNAs from rat hippocampus. Acta Biochim Biophys Sin (Shanghai) 2007;39:708–714. doi: 10.1111/j.1745-7270.2007.00324.x. [DOI] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, De Strooper B. MicroRNA regulation of Alzheimer′s amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer′s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite KC, Adams VH, Hansen JC. Recent advances in MeCP2 structure and function. Biochem Cell Biol. 2009;87:219–227. doi: 10.1139/o08-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Res. 2008;36:D165–D169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Li MD. Nicotine modulates expression of miR-140*, which targets the 3′-untranslated region of dynamin 1 gene (Dnm1) Int J Neuropsychopharmacol. 2009;12:537–546. doi: 10.1017/S1461145708009528. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Standart N. How do microRNAs regulate gene expression. Sci STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington′s disease. Neurobiol Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci USA. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Konecna A, Heraud JE, Schoderboeck L, Raposo AA, Kiebler MA. What are the roles of microRNAs at the mammalian synapse. Neurosci Lett. 2009;466:63–68. doi: 10.1016/j.neulet.2009.06.050. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, Lao K, Kosik KS. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13:1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16:961–966. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei P, Li Y, Chen X, Yang S, Zhang J. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res. 2009;1284:191–201. doi: 10.1016/j.brainres.2009.05.074. [DOI] [PubMed] [Google Scholar]
- Li L, Xu J, Yang D, Tan X, Wang H. Computational approaches for microRNA studies: a review. Mamm Genome. 2010;21:1–12. doi: 10.1007/s00335-009-9241-2. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NK, Wang XF, Lu QB, Xu XM. Altered microRNA expression following traumatic spinal cord injury. Exp Neurol. 2009a;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XQ, Sheng R, Qin ZH. The neuroprotective mechanism of brain ischemic preconditioning. Acta Pharmacol Sin. 2009b;30:1071–1080. doi: 10.1038/aps.2009.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, De Simone G, Ferguson TB, Ford E, Furie K, Gillespie C, Go A, Greenlund K, Haase N, Hailpern S, Ho PM, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott MM, Meigs J, Mozaffarian D, Mussolino M, Nichol G, Roger V, Rosamond W, Sacco R, Sorlie P, Stafford R, Thom T, Wasserthiel-Smoller S, Wong ND, Wylie-Rosett J. Heart Disease and Stroke Statistics—2010 update. A report from the American Heart Association. Circulation. 2009;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusardi TA, Farr CD, Faulkner CL, Pignataro G, Yang T, Lan J, Simon RP, Saugstad JA. Ischemic preconditioning regulates expression of microRNAs and a predicted target, MeCP2, in mouse cortex. J Cereb Blood Flow Metab. 2010;30:744–756. doi: 10.1038/jcbfm.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: implications for Alzheimer Disease and other human CNS Disorders. Curr Genomics. 2009;10:154–168. doi: 10.2174/138920209788185252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF, Sonenberg N. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- Mishima T, Mizuguchi Y, Kawahigashi Y, Takizawa T, Takizawa T. RT-PCR-based analysis of microRNA (miR-1 and -124) expression in mouse CNS. Brain Res. 2007;1131:37–43. doi: 10.1016/j.brainres.2006.11.035. [DOI] [PubMed] [Google Scholar]
- Montag J, Hitt R, Opitz L, Schulz-Schaeffer WJ, Hunsmann G, Motzkus D. Upregulation of miRNA hsa-miR-342-3p in experimental and idiopathic prion disease. Mol Neurodegener. 2009;4:36. doi: 10.1186/1750-1326-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV. RNA-mediated transcriptional gene silencing in human cells. Curr Top Microbiol Immunol. 2008;320:211–224. doi: 10.1007/978-3-540-75157-1_10. [DOI] [PubMed] [Google Scholar]
- Nan X, Cross S, Bird A.1998Gene silencing by methyl-CpG-binding proteins Novartis Found Symp 2146–16.discussion 16–21, 46–50 [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L, Klausen M, Helboe L, Nielsen FC, Werge T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PLoS One. 2009;4:e7225. doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington′s disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CS, Tang SJ. Regulation of microRNA expression by induction of bidirectional synaptic plasticity. J Mol Neurosci. 2009;38:50–56. doi: 10.1007/s12031-008-9158-3. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms. Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Pontes O, Pikaard CS. siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Genet Dev. 2008;18:197–203. doi: 10.1016/j.gde.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Rovelet-Lecrux A. Recent insights into the molecular genetics of dementia. Trends Neurosci. 2009;32:451–461. doi: 10.1016/j.tins.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J Neurosci Res. 2009;87:1435–1448. doi: 10.1002/jnr.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redshaw N, Wheeler G, Hajihosseini MK, Dalmay T. microRNA-449 is a putative regulator of choroid plexus development and function. Brain Res. 2009;1250:20–26. doi: 10.1016/j.brainres.2008.11.020. [DOI] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–D177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr Opin Neurobiol. 1995;5:566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- Schratt G. Fine-tuning neural gene expression with microRNAs. Curr Opin Neurobiol. 2009;19:213–219. doi: 10.1016/j.conb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi P, Loukianiouk S, Bohne-Lang A, Kenzelmann M, Kuffer S, Maertens S, Eils R, Grone HJ, Gretz N, Brors B. Argonaute—a database for gene regulation by mammalian microRNAs. Nucleic Acids Res. 2006;34:D115–D118. doi: 10.1093/nar/gkj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hubel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR. Synaptic enrichment of microRNAs in adult mouse forebrain is related to structural features of their precursors. Biol Direct. 2008;3:44. doi: 10.1186/1745-6150-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G. microRNA regulation of synaptic plasticity. Neuromolecular Med. 2009;11:133–140. doi: 10.1007/s12017-009-8065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Steitz JA, Vasudevan S. miRNPs: versatile regulators of gene expression in vertebrate cells. Biochem Soc Trans. 2009;37:931–935. doi: 10.1042/BST0370931. [DOI] [PubMed] [Google Scholar]
- Stenzel-Poore MP, Stevens SL, King JS, Simon RP. Preconditioning reprograms the response to ischemic injury and primes the emergence of unique endogenous neuroprotective phenotypes: a speculative synthesis. Stroke. 2007;38:680–685. doi: 10.1161/01.STR.0000251444.56487.4c. [DOI] [PubMed] [Google Scholar]
- Stenzel-Poore MP, Stevens SL, Xiong Z, Lessov NS, Harrington CA, Mori M, Meller R, Rosenzweig HL, Tobar E, Shaw TE, Chu X, Simon RP. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362:1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, Jeyaseelan K. Expression profile of microRNAs in young stroke patients. PLoS One. 2009;4:e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Tsai NP, Lin YL, Wei LN. MicroRNA mir-346 targets the 5′-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem J. 2009;424:411–418. doi: 10.1042/BJ20090915. [DOI] [PubMed] [Google Scholar]
- van den Berg A, Mols J, Han J. RISC-target interaction: cleavage and translational suppression. Biochim Biophys Acta. 2008;1779:668–677. doi: 10.1016/j.bbagrm.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008a;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer′s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008b;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Baker MB, Moore JP, Searles CD. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun. 2010;393:643–648. doi: 10.1016/j.bbrc.2010.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg MS, Wood MJ. Short non-coding RNA biology and neurodegenerative disorders: novel disease targets and therapeutics. Hum Mol Genet. 2009;18:R27–R39. doi: 10.1093/hmg/ddp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Sayer JA. Naturally occurring antisense RNA: function and mechanisms of action. Curr Opin Nephrol Hypertens. 2009;18:343–349. doi: 10.1097/MNH.0b013e32832cb982. [DOI] [PubMed] [Google Scholar]
- Weyrich AS, Lindemann S, Tolley ND, Kraiss LW, Dixon DA, Mahoney TM, Prescott SP, McIntyre TM, Zimmerman GA. Change in protein phenotype without a nucleus: translational control in platelets. Semin Thromb Hemost. 2004;30:491–498. doi: 10.1055/s-2004-833484. [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Yelamanchili SV, Fox HS. Defining larger roles for ‘tiny' RNA molecules: role of miRNAs in neurodegeneration research. J Neuroimmune Pharmacol. 2009;5:63–69. doi: 10.1007/s11481-009-9172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Deng Z, Huang H, Hamblin M, Xie C, Zhang J, Chen YE. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010;38:17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y. Regulation of the mammalian nervous system by microRNAs. Mol Pharmacol. 2009;75:259–264. doi: 10.1124/mol.108.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]