Abstract
Background
Genome wide association (GWA) studies identified single nucleotide polymorphisms (SNPs) that are associated with increased hepatic fat or elevated liver enzymes, presumably reflecting nonalcoholic fatty liver disease (NAFLD).
Aim
To investigate whether these SNPs are associated with histological severity in a large cohort of NAFLD patients.
Methods
1117 (894 adults/223 children) individuals enrolled in NASH-Clinical Research Network and National Institutes of Health Clinical Center studies with histologically-confirmed NAFLD were genotyped for six SNPs that are associated with hepatic fat or liver enzymes in GWA studies.
Results
In adults, 3 SNPs on chromosome 22 showed associations with histological parameters of nonalcoholic steatohepatitis (NASH). After adjustment for age, gender, diabetes and alcohol consumption, the minor allele of rs738409C/G, a nonsynonymous coding SNP in the PNPLA3 (adiponutrin) gene encoding an I148M change, was associated with steatosis (p=0.03), portal inflammation (p=2.5×10−4), lobular inflammation (p=0.005), Mallory-Denk bodies (p=0.015), NAFLD activity score (NAS, p=0.004) and fibrosis (p=7.7×10−6). Two other SNPs in the SAMM50-PNPLA3 cluster demonstrated similar associations. Three SNPs on chromosome 10 in the CPN1-ERLIN1-CHUK region were independently associated with fibrosis (p=0.010). In children, no SNP was associated with histological severity. However, the rs738409 G allele was associated with younger age at the time of biopsy in multivariate analysis (p=0.045).
Conclusions
In this large cohort of histologically-proven NAFLD, we confirm the association of the rs738409G allele with steatosis and describe its association with histological severity. In pediatric patients, the high-risk rs738409G allele is associated with an earlier presentation of disease. We also describe a hitherto unknown association between SNPs at a chromosome 10 locus and the severity of NASH fibrosis.
Keywords: Non-Alcoholic Fatty Liver Disease, PNPLA3, Adiponutrin, rs738409, CHUK
Introduction
Nonalcoholic fatty liver disease (NAFLD) has become the most common cause of chronic liver disease and has an estimated prevalence of 20%-30% in the general population and 67–75% in the obese population (1, 2). This disease ranges from simple steatosis to nonalcoholic steatohepatitis (NASH) and has been closely associated with obesity, insulin resistance, diabetes, metabolic syndrome, and hyperlipidemia (3, 4). The precise mechanism responsible for the development of the NASH phenotype is yet to be elucidated (5). Metabolic derangements associated with NAFLD such as obesity and diabetes are known to have a strong underlying genetic component. Moreover, familial clustering of steatosis, NASH and cryptogenic cirrhosis has been reported, suggesting that genetic factors may contribute to the development of NAFLD (6–8). An important initial step in the identification of these genetic factors is through the use of genome-wide association (GWA) studies. These studies evaluate the genotypic-phenotypic association in large population-based cohorts and have identified susceptibility loci in numerous diseases (9).
Recently, GWA studies have identified several single nucleotide polymorphisms (SNPs) that are associated with increased hepatic fat content (10) or with elevated liver enzymes (11). Of these, the PNPLA3 locus on chromosome 22, and especially the non-synonymous coding SNP rs738409 C/G (I148M), emerged as an important genetic variant associated with the presence of NAFLD. These findings were confirmed by candidate-gene studies assessing the associations with this SNP in several cohorts of different ethnicity (12–15).
These previous studies have focused on the genetic variability associated with the presence of NAFLD, mainly diagnosed using non-invasive markers. However, whether these genetic variants also determine disease severity is currently unknown. In the present study, we investigated the association of well-replicated SNPs identified from published literature with histological parameters of disease severity in a large cohort of patients with NAFLD.
Methods
Study Population
The patient population consisted of participants in one of three National Institutes of Health (NIH) sponsored NASH-Clinical Research Network (NASH-CRN) multicenter studies and a cohort of individuals with NASH at the NIH Clinical Center. The NASH-CRN studies include: 1) the NAFLD natural history database; 2) adult individuals enrolled in the Pioglitazone versus Vitamin E versus Placebo for the Treatment of Nondiabetic Patients with Nonalcoholic Steatohepatitis (PIVENS) trial (16), and 3) the Treatment of NAFLD in Children (TONIC)(17). The NIH cohort consists of patients enrolled in two pilot treatment studies of NASH: 1) A pilot study of Pioglitazone treatment for nonalcoholic steatohepatitis (18), and 2) A pilot study of Metformin for the treatment of nonalcoholic steatohepatitis (19). The design and enrollment criteria for these studies have been previously reported.
Individuals included in the analysis had histological evidence of NAFLD or NASH on a liver biopsy obtained prior to enrollment. All liver biopsy specimens from the NASH-CRN studies were scored by the pathology committee of the network in a blinded manner; biopsies from the NIH pilot studies were evaluated by a single pathologist (DEK). Biopsies were scored according to the NASH-CRN scoring system (20, 21) (Supplementary table 1). All studies were approved by the institutional review boards and all patients gave written informed consent for participation and for analysis of genetic material.
Population control samples are from a cohort of 336 community-dwelling Caucasian men aged ≥50 years who were enrolled in a study of bone mineral density in Pittsburgh, Pennsylvania (22). Participants were recruited by population-based mailings to age-eligible men. Samples were selected regardless of the bone mineral density or health status of participants. Written informed consent was obtained from all participants and the protocol was approved by University of Pittsburgh Institutional Review Board.
SNP selection and Genotyping
Six SNPs that were previously shown to be associated with liver fat (on non-invasive tests) or elevated liver enzymes (10, 11)(Table 2) were chosen for genotyping. DNA was obtained from peripheral mononuclear cells or Epstein-Barr virus-immortalized B cell lines.
Table 2.
Allele frequency of SNPs in adult Caucasians with NAFLD and control populations
| SNP | Chromosome and location |
Nearest Gene |
Alleles | HapMap– Caucasian (CEU) Frequency |
Control Caucasian cohort frequency |
NAFLD adult Caucasian cohort Frequency |
p-value1 |
|---|---|---|---|---|---|---|---|
| rs11597390 | 10:101861435 | CPN1 | G/A | 0.675/0.325 | 0.666/0.334 | 0.482 | |
| rs11597086 | 10:101953705 | CHUK | A/C | 0.575/0.425 | 0.584/0.416 | 0.611/0.389 | 0.034 |
| rs11591741 | 10:101976501 | CHUK | G/C | 0.575/0.425 | 0.584/0.416 | 0.607/0.393 | 0.072 |
| rs738409 | 22:44324727 | PNPLA3 | C/G | 0.767/0.233 | 0.772/0.228 | 0.495/0.505 | 9×10−143* |
| rs2281135 | 22:44332570 | PNPLA3 | C/T | 0.792/0.208 | 0.835/0.165 | 0.632/0.368 | 6×10−98* |
| rs2143571 | 22:44391686 | SAMM50 | G/A | 0.808/0.192 | 0.831/0.169 | 0.680/0.320 | 2×10−54* |
2-sided χ2 test for comparison between NAFLD adult Caucasians and control Caucasians
rs11597390 could not be genotyped in controls. 2-sided χ2 test is reported for comparison between NAFLD adult Caucasians and Caucasians from HapMap
Statistically significant (corrected α = 0.0083, Bonferroni procedure)
The SNPs were genotyped using the Sequenom MassARRAY iPLEX Gold platform (Sequenom, Inc; San Diego, CA), with PCR primers purchased from Invitrogen (Carlsbad, CA). Genotyping concordance was 99.9% as determined by inclusion of 301 replicate NAFLD patient samples and 100% using 96 replicate control population samples. The average genotyping call rate across all SNP assays was 97.9% and all call rates were >94%.
Measurements and data
Body mass index (BMI) was calculated as weight [kg]/Height [m]2. In pediatric patients, BMI was adjusted to age, according to the World Health Organization (WHO) growth charts for the appropriate age and gender (23) and the Z-score was used for analysis. The frequency of alcoholic beverage consumption was assessed using the AUDIT questionnaire. Race and ethnicity were self-reported.
Statistical analysis
The univariate association of SNP genotypes with histological ordinal variables was assessed using the Jonckheere-Terpstra test for ordered alternatives. To determine the appropriate genetic model to be used for multivariable analysis, a modification of the method suggested by Minelli et al (24) was used. The outcome was binarized and the univariate odds ratios (OR) were calculated for comparison between homozygotes of the minor allele and homozygotes of the major allele (denoted ORGG) and heterozygotes compared to homozygotes of the major allele (denoted ORGg). The ratio between log ORGg and log ORGG, λ, captures the genetic mode of inheritance, where λ values of 0, 0.5 and 1 are consistent with a recessive, additive (co-dominant) and dominant models, respectively. We arbitrarily defined λ<0.25 as recessive, 0.25≤λ<0.75 as additive and 0.75≤λ as dominant. Bootstrap analysis (500 samples with no replacement, 1000 repetitions) was used to assess the dispersion of λ values, using the median λbootstrap and the percentage of λbootstrap values that were classified similarly to the actual λ.
Ordinal logistic regression was used for multivariable analyses. Logit link function was selected for uniformly distributed dependent variables, while for normally distributed variables, a Probit link function was used. Binary variables were assessed using binary logistic regression, reporting odds ratios and 95% confidence intervals (CI). Since ordinal regression does not provide meaningful odds ratios, a secondary analysis was performed, in which dependent variables were binarized and binary logistic regression was applied to obtain odds ratios. The genetic heredity model suggested by the λ value was used in the ordinal and binary logistic regressions.
Liver enzymes were log-transformed to achieve a normal distribution and their association with SNPs was analyzed using one-way ANOVA for univariate analysis and linear regression for multivariate one. Age, gender, BMI, alcohol consumption and the presence of diabetes mellitus type 2 were selected a priori as covariates in multivariable analyses. A two-sided significance level of α<0.05 was used for all significant tests.
When more than one SNP was tested for association with an attribute, the Bonferroni correction for multiple comparisons was used. To correct for testing for multiple histological dependent parameters, the false discovery rate (FDR) was calculated using the Benjamini-Hochberg procedure (25). The FDR significance level was set at q<0.05. Correction was not used in univariate analyses, as these were only used to select SNPs for multivariable analyses. Statistical analyses were performed using SPSS v.13.0 and Microsoft Excel.
In order to observe the association of genetic variability with the full spectrum of histological disease, we limited the primary analysis to adult patients (aged 18 or more at the time of biopsy). Pediatric cases were used in a secondary analysis, to extend the results obtained from the adult population.
Results
SNP genotypes were available from 894 adult patients with NAFLD (Table 1), the majority of whom (766 patients, 85.7%) were Caucasians. A definite histological diagnosis of NASH was obtained in 59.4% of patients. SNPs that are associated with NAFLD are expected to be enriched in this cohort relative to the general population. We compared the allele frequencies in the adult Caucasian patients to that of the Caucasian control cohort and to the reported frequency in 120 healthy persons of northern European descent in the HapMap project (19). Highly significant enrichment of the minor allele was found for 3 SNPs in the PNPLA3 gene or its vicinity on chromosome 22, rs738409, rs2281135 and rs2143571 (Table 2). The minor allele was less frequent in 3 SNPs on chromosome 10, but only one of those, rs11597086 located near the CHUK gene, achieved borderline significance. The allelic frequencies in the Caucasian control population were similar to those reported in HapMap Caucasians.
Table 1.
Patient characteristics
| Adult Cohort | Pediatric Cohort | p-value (for comparison with adults) |
|
|---|---|---|---|
| N | 894 | 223 | |
| Male gender | 337 (37.7%) | 162 (72.6%) | p<0.001a |
| Data Source | |||
| NASH-CRN database | 642 (72%) | 91 (41%) | |
| PIVENS | 213 (24%) | 0 (0%) | |
| NIH trials | 39 (4%) | 0 (0%) | |
| TONIC | 0 (0%) | 132 (59%) | |
| Race (%) | <0.001a | ||
| Caucasian | 82.1 | 67.3 | |
| African American | 2.3 | 0.9 | |
| Asian | 5.4 | 2.7 | |
| American Indian | 3.2 | 18.8 | |
| Other | 7 | 9.4 | |
| > 1 race | 3.9 | 0.9 | |
| Hispanic ethnicity | 97 (10.9%) | 144 (64%) | <0.001a |
| Age | 47.9±11.6 (18–76) | 12.4 (6–17) | |
| BMI | 34.4±6.4 (20–59.5) | +3.3 SD (1.5–7.5)b | |
| ALT (median, range) | 58 (10–538) | 82 (16–328) | <0.001c |
| AST | 42 (10–480) | 49 (14–317) | <0.001c |
| GGT | 45 (5–793) | 33 (5 – 208) | 0.003c |
| Alcoholic drinks <= 2–4/month | 96.1% | 98.6% | 0.089e |
| DM Type 1 | 4 (0.5%) | 0 | 0.586e |
| DM Type 2 | 244 (28.5%) | 9 (4%) | <0.001a |
| NAS (median, range) | 4 (0–8) | 4 (1 – 8) | 0.08d |
| Steatosis grade | <0.001d | ||
| 0 (< 5%) | 6.2% | 2.2% | |
| 1 (5–33%) | 39.2% | 30% | |
| 2 (33–66%) | 21.8% | 28.7% | |
| 3 (> 66%) | 22.9% | 39% | |
| Fibrosis stage | <0.001a | ||
| 0 (no fibrosis) | 22.4% | 28.3% | |
| 1 (zone 3 or periportal only) | 26.5% | 40.8% | |
| 2 (zone 3 and periportal) | 18.1% | 15.2% | |
| 3 (bridging) | 20.8% | 13.5% | |
| 4 (cirrhosis) | 10.1% | 1.8% | |
| NASH diagnosis | <0.001a | ||
| No NASH | 22% | 24.7% | |
| Indeterminate | 19% | 43.9% | |
| Definite NASH | 59% | 31.4% |
2-sided χ2 test
BMI for children reported in standard deviations from age- and gender-fitted norms
Student’s t-test
Mann-Whitney U test
Fisher’s exact test
The 3 SNPs on the chromosome 22 locus were highly correlated with each other (Spearman’s rho 0.599–0.775, p<0.001 for all pair-wise comparisons). Since rs738409 has the highest level of significance, is the only non-synonymous coding SNP amongst the three, and is potentially a functionally relevant mutation (26), it was chosen as the representative SNP for that locus for further analyses.
Association with histological parameters of disease activity
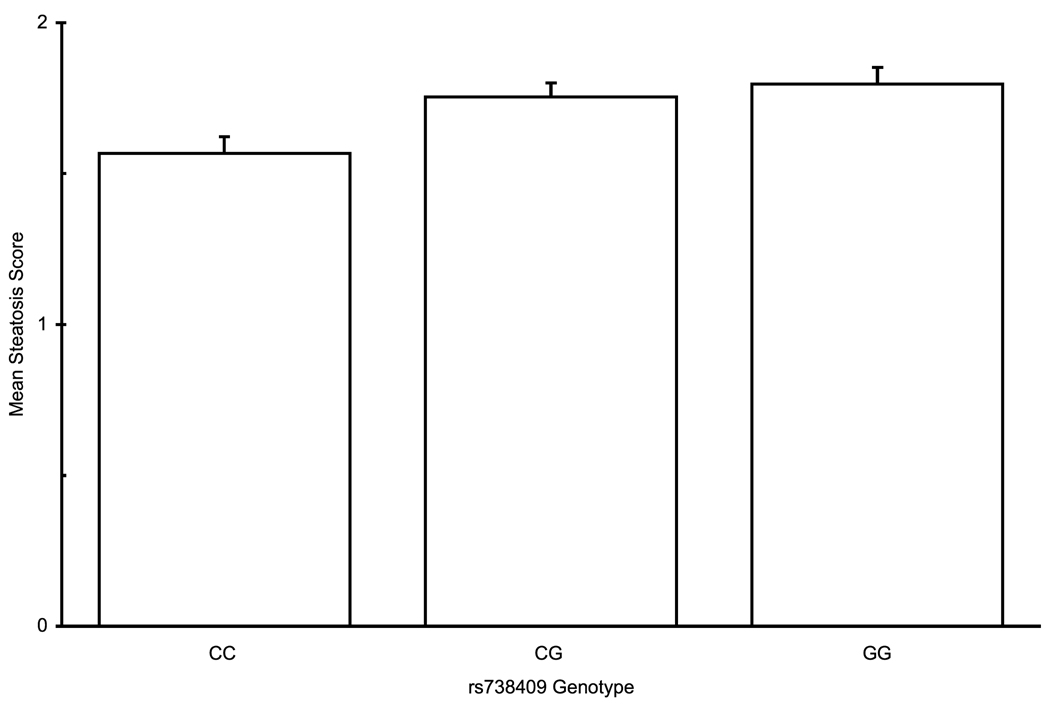
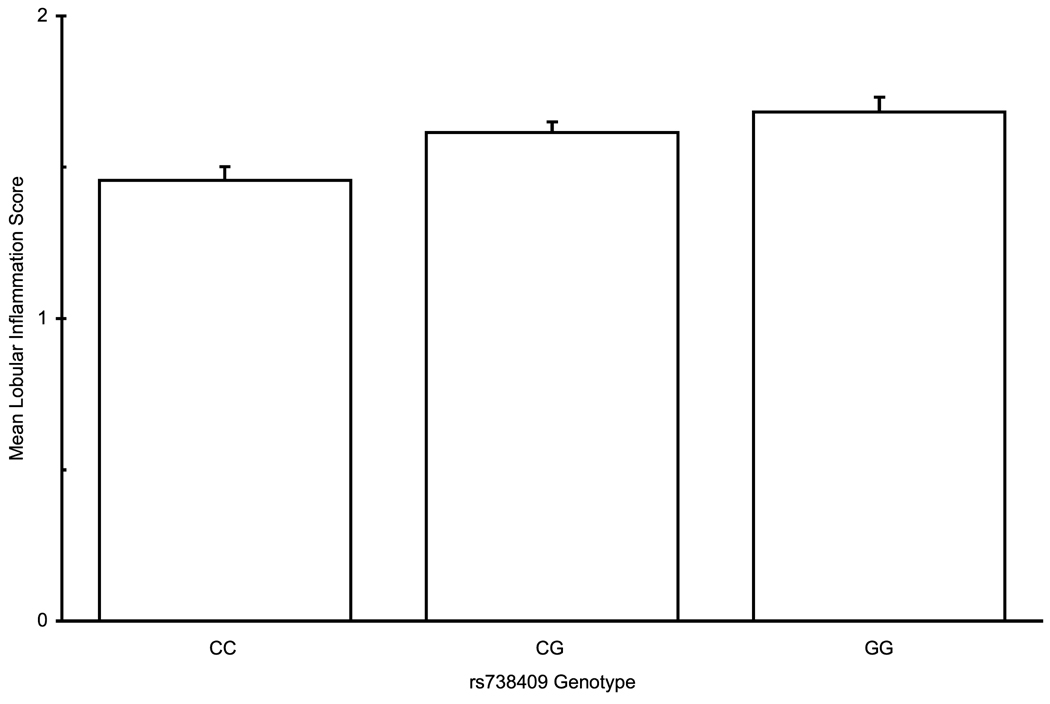
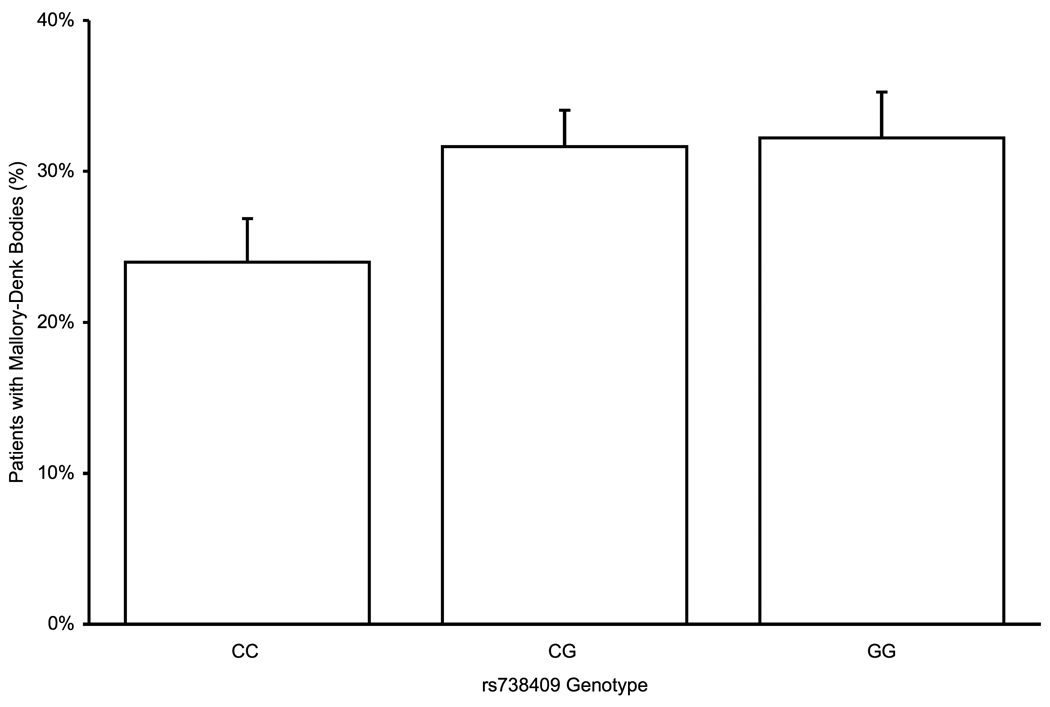
Histological parameters of NAFLD are assessed along several axes – steatosis, inflammation, cellular injury, and fibrosis. On univariate analysis, rs738409 was significantly associated with increased steatosis, portal inflammation, and lobular inflammation, and showed a near-significant trend for association with the presence of Mallory-Denk bodies (Table 3). For all of these parameters, genotypes containing the minor allele rs738409 G allele were associated with more severe disease, in a dominant pattern, whereas with portal inflammation, the association is better fitted by an additive model (Figure 1a–c). After adjustment for age, gender, BMI, diabetes type 2 and alcohol consumption, all associations remained significant. No association was found with hepatocellular ballooning. Since the results of ordinal regression cannot be easily converted to odds ratios, we used multivariate binary logistic regression to estimate the magnitude of effect. Patients with at least one minor allele of rs738409 were more likely to have a steatosis score ≥ 2 (OR 1.46, 95% CI 1.07–2.01). Similarly, the OR for lobular inflammation was 1.84 (1.33–2.55) and for Mallory-Denk bodies it was 1.55 (1.07–2.26). For portal inflammation, the OR for the presence of each minor allele (additive model) was 1.57 (1.24–1.99). These OR are of the same order of magnitude as those of other binary variables, such as gender or the presence of diabetes (Table 3).
Table 3.
Univariate and multivariable regression and odds ratio for association of rs738409 genotype with histological parameters of disease severity in adults with NAFLD
| Predictor | Steatosis | Portal Inflammation | Lobular Inflammation | Mallory-Denk Bodies | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate p-value |
Multivariate p-valueb (FDR q– valueh) |
OR (95% CI)d | Univariate p-value |
Multivariate p-valueb (FDR q– value) |
OR (95% CI)e | Univariate p-value |
Multivariate p-valueb (FDR q– value) |
OR (95% CI)f | Univariate p-valueg |
Multivariate p-valueg (FDR q– value) |
OR (95% CI)g | |
| rs738409 | 0.005a | 0.012 (0.033) |
1.46 (1.07– 2.01) |
0.009a | 2.5×10−4 (0.017) |
1.57(1.24– 1.99) |
4×10−4a | 0.002 (0.025) | 1.84 (1.33– 2.55) |
0.058 | 0.015 (0.042) | 1.6 (1.46– 3.07) |
| Age | 1.1×10−5b | 5.5×10−5 | 0.98 (0.97– 0.99) |
4.5×10−7b | 8.1×10−6 | 1.03 (1.02– 1.05) |
0.998b | 0.098 | 0.99 (0.98– 1.01) |
1.3×10−9 | 1.4×10−4 | 1.03 (1.01– 1.05) |
| Female Gender |
0.564c | 0.075 | 1.20 (0.89– 1.62) |
0.008c | 0.754 | 1.01 (0.69– 1.48) |
0.001c | 4.8×10−5 | 1.60 (1.18– 2.17) |
1.7×10−8 | 7.3×10−5 | 2.12 (1.46– 3.07) |
| BMI | 0.756b | 0.937 | 0.99 (0.97– 1.01) |
5×10−5b | 4×10−5 | 1.04 (1.01– 1.06) |
0.103b | 0.020 | 0.98 (0.96– 1.00) |
0.382 | 0.952 | 1.00 (0.98– 1.03) |
| Diabetes Type 2 |
0.075c | 0.266 | 0.83 (0.60– 1.13) |
9.7×10−5c | 0.010 | 1.72 (1.19– 2.48) |
0.649c | 0.795 | 0.97 (0.70– 1.33) |
7.2×10−7 | 1.5×10−4 | 1.95 (1.38– 2.75) |
| Alcohol Consumption |
0.171a | 0.043 | 0.91 (0.44– 1.87) |
6.9×10−5a | 0.088 | 0.53 (0.18– 1.55) |
0.694a | 0.336 | 1.52 (0.73– 3.18) |
1.8×10−4 | 0.06 | 1.380 (0.606– 3.143) |
Jonkheere-Terpestra test
Ordinal regression
Mann-Whitney U
Multivariable binary logistic regression for steatosis ≥ 33% vs. < 33%. rs738409 entered using dominant model (i.e. genotype CG or GG vs. CC)
Multivartiable binary logistic regression for more than mild portal inflammation vs. none or mild. rs738409 entered using additive model
Multivariable binary logistic regression for lobular inflammation ≥ 2 foci vs. < 2. rs738409 entered using dominant model
Multivariable binary logistic regression. rs738409 entered using dominant model
False discovery rate evaluated for association with SNP only. q<0.05 is significant.
Figure 1.
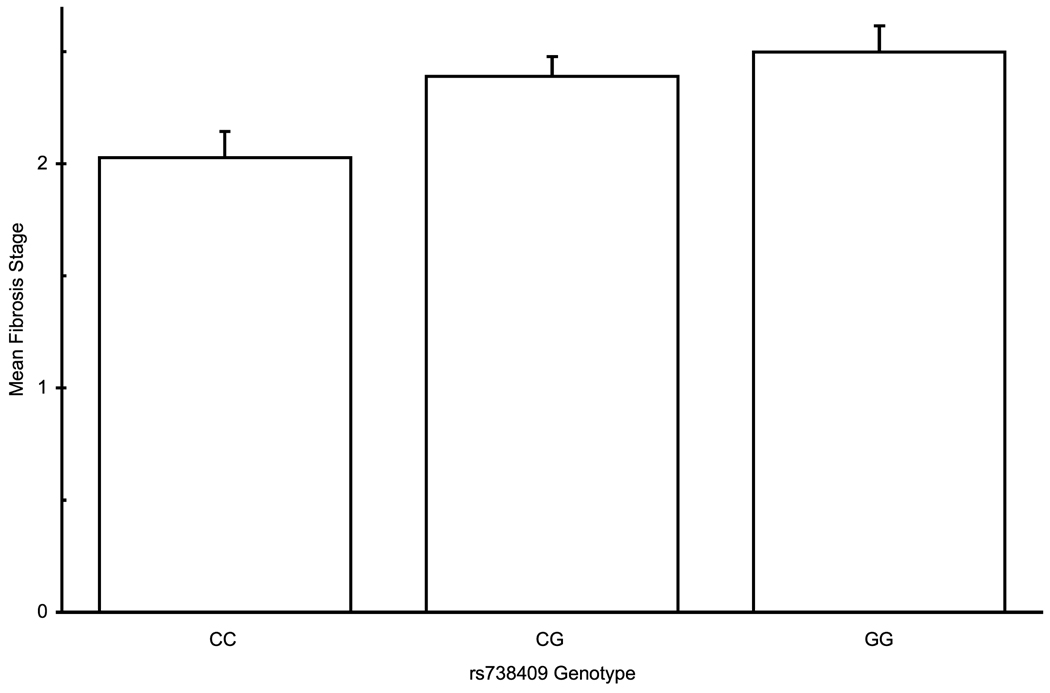
PNPLA3 rs738409 genotype association with histolgical attributes: (a) Association with steatosis in a dominant pattern (λ=0.79, median λbootstrap=0.80, 55% of λbootstrap values classified as dominant) (b) Lobular inflammation, dominant pattern (λ=0.99, median λbootstrap=0.99, 90% classified as dominant) (c) Presence of Mallory-Denk bodies , dominant pattern (λ=0.93, median λbootstrap=0.91, 71% classified as dominant) (d) Fibrosis scores, additive hereditary pattern (λ=0.43, median λbootstrap=0.42, 57% classified as additive). Error bars denote standard error of the mean.
The association of rs738409 with hepatic steatosis could theoretically explain the association with inflammation and Mallory-Denk bodies. However, even when the degree of steatosis was included in the multivariable model, the association with inflammation and Mallory-Denk bodies remained significant (Table 4).
Table 4.
The association of rs738409 genotype with histological parameters of NAFLD before and after inclusion of steatosis in the model
| Dependent Parameter |
Multivariatea | Multivariate with steatosisb |
|---|---|---|
| Portal inflammation | 2.5×10−4 | 1.2×10−4 |
| Lobular inflammation | 0.005 | 0.008 |
| Mallory-Denk bodiesc | 0.015 | 0.017 |
| Ballooning | 0.970 | 0.864 |
| Fibrosis | 7.7×10−6 | 2.7×10−6 |
Ordinal regression adjusted for age, sex, BMI, alcohol consumption and diabetes type 2.
Ordinal regression adjusted for age, sex, BMI, alcohol consumption, diabetes type 2 and steatosis score.
Binary logistic regression (univariate and adjusted multivariable)
The NAFLD activity score (NAS) is defined as the sum of the scores for lobular inflammation, ballooning and steatosis and is proposed as a measure of overall activity of the disease(21). As expected from the analyses of individual components, carriers of the rs738409 G allele had higher NAS on univariate (p=0.009) and multivariable (p=0.004) analyses in a dominant pattern (λ = 0.991). Homozygotes or heterozygotes for rs738409 G had a mean NAS score of 4.52±1.69, compared to 4.11±1.72 in homozygotes for rs738409 C, and were more likely to have NAS > 3 with an OR of 1.56 (CI 1.12–2.17).
The two other SNPs on chromosome 22 in the SAMM50-PNPLA3 gene region (rs2281135 and rs2143571) demonstrated similar associations as those seen with rs738409. No other SNP was associated with any of the histological parameters of activity.
Association with fibrosis
The rs738409 SNP was associated with higher fibrosis scores (Table 5) in an additive pattern (λ=0.43), with an average fibrosis score that was 0.47 stage higher for patients with genotype GG compared to CC (mean score 2.50 vs. 2.03, Figure 1d). This association was highly significant even after controlling for steatosis, inflammation, and the presence of Mallory-Denk bodies, suggesting that the association of rs738409 with fibrosis may be independent of its association with other parameters of disease severity. The adjusted OR for having at least bridging fibrosis was 1.50 (CI 1.19–1.89) for each G allele present.
Table 5.
Univariate or multivariable regression for the association of SNPs with NASH fibrosis score in adults.
| Univariate p-value | Multivariate p-valueb (FDR q– valued) |
|
|---|---|---|
| rs738409 | 0.005a | 1.9×10−5 (0.008) |
| rs11591741 | 0.001a | 0.010 |
| Age | 1.8×10−13b | 9.2×10−10 |
| Gender | 2.3×10−4c | 0.998 |
| BMI | 0.009b | 0.062 |
| Diabetes Type 2 | 1.1×10−15c | 1.2×10−10 |
| Alcohol consumption | 1.9×10−13a | 0.003 |
Both SNPs are applied as an additive inheritance model
Jonkheere-Terpestra test
Ordinal regression
Mann-Whitney U
False discovery rate evaluated for association with rs738409 only. q<0.05 is significant.
Three SNPs (rs11591741, rs11597086 and rs11597390), all on chromosome 10 in the CPN1-ERLIN1-CHUK gene region and known to be in linkage disequilibrium with each other, were also found to be associated with fibrosis. Of these three SNPs, rs11591741 had the most significant association (minor allele C associated with increased fibrosis) and was selected as the representative SNP for further analysis. Age, gender, BMI, diabetes and alcohol consumption were also associated with fibrosis. When both rs738409 and rs11591741 were included in the multivariable analysis, both retained significance (Table 5).
Effect of race on genetic association
As only a small number of patients were non-Caucasians (~18%), we were unable to control for race in multivariable analyses. When limiting the analyses to Caucasian adults only, all of the associations remained statistically significant. The number of non-Caucasian patients was too small to allow for statistical significance analysis. However, the direction and magnitude of the effect of rs738409 genotype was similar to that seen in Caucasians for steatosis, inflammation and NAS (Supplementary table 2). There was less clear relationship between rs738409 and fibrosis in non-Caucasians, probably because of the small number.
Association with spectrum of NAFLD
For every patient, the pathologists assigned a diagnosis of definite steatohepatitis, borderline steatohepatitis or not steatohepatitis, based on accepted criteria (27). We compared the frequency of rs738409 G in Caucasian patients from 3 populations – patients with a definite diagnosis of steatohepatitis (n=438), patients with steatosis only (taken from the group classified as not steatohepatitis and defined as liver fat, minimal inflammation, no cellular injury and no fibrosis, n=82, Supplementary Table 3) and 336 population controls. The allelic frequency of rs738409 G did not differ between the steatohepatitis (49.2%) and simple steatosis (51.8%, p=0.54) groups, but was markedly higher in both groups than normal controls (22.8%, p=2×10−13 and 4×10−26 for the comparison with steatosis and steatohepatitis, respectively). On univariate and multivariable genotypic analysis, rs738409 did not differentiate steatohepatitis from steatosis alone, even after controlling for potential confounders.
Association with liver enzymes
On univariate analysis, the minor allele of rs738409 was associated with increased serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST)(p=0.027, p=0.023, respectively). On multivariable linear regression, rs738409 genotype maintained significance with both enzymes (ALT, regression coefficient 0.027, p=0.03; AST, regression coefficient 0.024, p=0.033; additive model). Similar associations were seen with the other two SNPs in the PNPLA3 vicinity.
The minor allele of rs11591741, a SNP in the CHUK gene of chromosome 10, was associated with decreased ALT on univariate analysis (p=0.025). This association remained significant on multivariable linear regression, adjusted for age, BMI, diabetes, gender and alcohol consumption (regression coefficient −0.028, p=0.038, additive model). No significant association was observed with AST on univariate or multivariable analysis. Similar findings were seen for rs11597086, a closely linked SNP to rs11591741. There was no association between the third chromosome 10 SNP of rs11597390 and aminotransferases.
Results in pediatric patients
Liver histology was available for 223 pediatric patients (Table 1). The patients in this cohort were more likely to be males and Hispanic in comparison to the adult cohort, were less likely to be diabetic, and had higher aminotransferases but lower GGT levels. Histologically, the pediatric cohort had more steatosis, a non-significant trend towards a decreased NAS, and milder fibrosis, with portal fibrosis (1c) predominating, as previously described (28). Additionally, these patients were less likely than the adult patients to have a confirmed diagnosis of NASH and were more likely to have a borderline diagnosis.
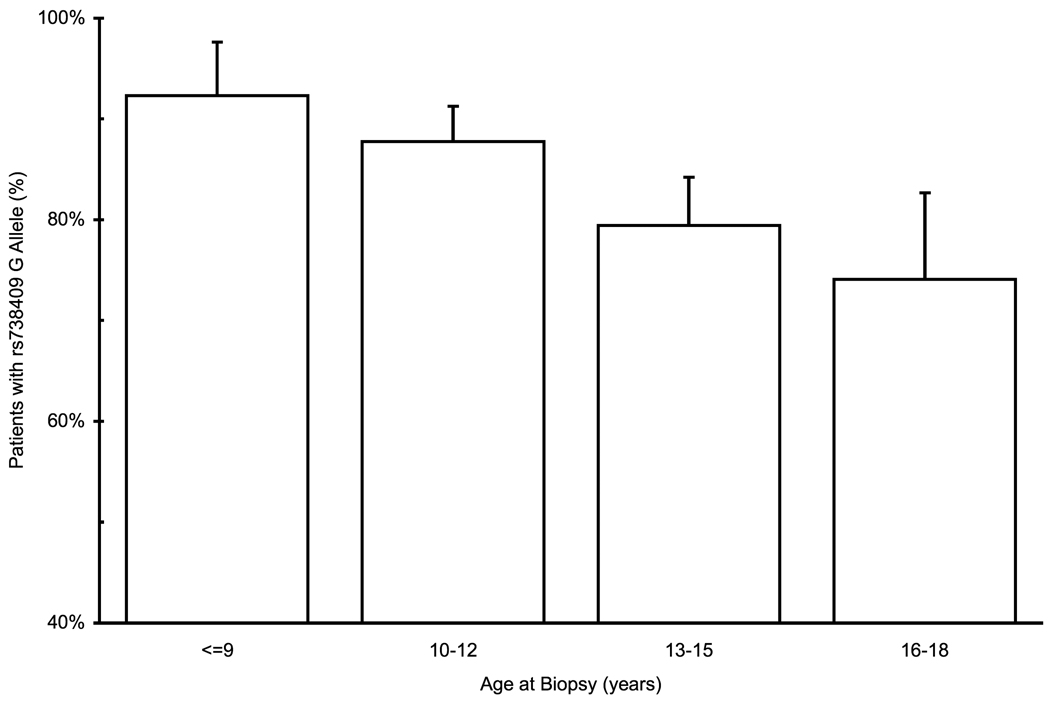
No association was found for any SNP with histologic parameters in the pediatric patients. However, patients carrying the rs738409 G allele, tended to be younger by 11 months at the time of biopsy, in a dominant pattern (Figure 2a) and the association with the minor allele was monotonous with age (Figure 2b). This association reached borderline significance in univariate analysis (p=0.054, Student’s t-test) and was significant on multivariable analysis, (p=0.045, linear regression adjusted for gender and BMI Z-score). Hispanic ethnicity was also associated with younger age at time of biopsy in univariate and multivariable analyses. Hispanics were more likely to carry the rs738409 G allele (91% vs. 70% of non-Hispanics, p=9×10−5, Fisher’s exact test) and thus, when both Hispanic ethnicity and rs738409 genotype were entered as parameters in the regression, both became non-significant predictors. However, even when analyzing Hispanics and non-Hispanics separately, the G allele was associated with a younger age at biopsy in both groups, although the difference was not statistically significant, probably due to a smaller number of patients.
Figure 2.
Association of rs738409 genotype with age at time of the biopsy in pediatric patients. (a) Average age at biopsy according to rs738409 genotype. (b) Frequency of rs738409 G allele according to age at time of biopsy. Error bars denote standard error of the mean.
Discussion
In this study, using a large cohort of patients with NAFLD and well-defined histological information, we confirm the association of the minor alleles of rs738409 and other SNPs in the PNPLA3 gene region with liver steatosis and liver enzymes. More importantly, we show a strong association of PNPLA3 with histological parameters of disease activity (inflammation and Mallory-Denk bodies) and with fibrosis severity, associations that are independent of the effect on steatosis. Thus, the presence of the minor allele of rs738409 is a strong predictor of NAFLD histological severity.
Interestingly, the rs738409 G allele was present in the same frequency in patients with NASH or patients with steatosis alone, despite its marked enrichment in both groups compared to population controls, and despite its association with disease severity. This is probably related to the lack of association between rs738409 and hepatocyte ballooning, a histological feature that heavily favors a diagnosis of NASH by the pathologist (27). Another explanation may result from a potential selection bias. In order to be included, patients had to have been referred to a hepatologist and have an indication for a liver biopsy. Thus, it is possible that the patients with steatosis only are not fully representative of simple steatosis and have very mild features of NASH (perhaps obscured by sampling error).
One study to date has investigated the association of rs738409 with histological parameters of NAFLD (29) by comparing 172 patients with NAFLD (103 of whom had liver biopsies) to 94 normal controls. In that study, the minor allele was associated with histological steatosis and overall severity, similar to our findings. Our larger sample size enabled us to study in greater detail the associations with specific components of histological severity and to demonstrate their independence of steatosis grade. In contrast to our findings, Sookoian et al (29) observed a significant difference in rs738409 genotypes between patients with NASH and steatosis. The two studies differ in the population prevalence of the minor allele, in the definition of NASH and in the recruitment strategy of patients. Further studies are necessary to resolve this discordant result.
NAFLD is becoming increasingly prevalent in pediatric patients (30). We did not identify an association between the PNPLA3 SNPs and severity in the pediatric cohort, probably because of a milder disease at an early stage of the NASH natural course, as evident by the smaller proportion of pediatric patients with a definite NASH diagnosis. However, the presence of the rs738409 G allele was associated with a younger age at the time of liver biopsy, suggesting a younger age of disease presentation.
There are important racial and ethnic differences in the prevalence of fatty liver disease and the predisposition to NASH; patients of Hispanic ethnicity are more likely to have NASH and NAFLD, while African-Americans seem to be relatively protected (31). The racial distribution in our adult cohort did not allow us to determine whether the effects of genetic variation explain the ethnic-racial differences or are independent of them.
PNPLA3 encodes for adiponutrin, a membrane-bound protein expressed primarily in adipose tissue (32). The expression level of adiponutrin mRNA is tightly regulated by food intake (33, 34) through changes in insulin levels (35, 36). The function of adiponutrin is not fully understood. It has close homology with adipose triglyceride lipase (37) and seems to have both triglyceride lipase and acyl-CoA-independent transacylase activity (38). rs738409C->G is a nonsynonymous coding SNP, creating an I148M change in the adiponutrin protein. Recently, this mutation was suggested to impair triglyceride hydrolysis in hepatocytes, a potential explanation for increased triglyceride accumulation (26). Interestingly, despite the association with liver steatosis, this SNP does not seem to be associated with insulin resistance (10, 12, 13) or with obesity (39, 40). It has been reported to be associated with lower cholesterol and LDL-cholesterol levels (14), but whether this is related to the liver disease is unclear. It is possible that another genetic variant, strongly linked to rs738409, may be responsible for the difference in phenotypes.
We have also found an association of NASH fibrosis with several SNPs on chromosome 10, in the vicinity of the CHUK1 gene, encoding IKKα which is involved in the NFκB signaling and CPN1 which encodes carboxypeptidase N, a metalloproteinase that regulates the activity of kinins and anaphylotoxins. Despite the association with increased fibrosis, the minor alleles at this locus are associated with lower ALT in our cohort as well as in other studies (11). This apparently paradoxical association is explained by the non-monotonous relationship between ALT and fibrosis (increasing fibrosis stage associated initially with higher ALT and then with lower ALT) in patients with NAFLD. These SNPs are not associated with any other histological aspect of NAFLD, and thus, it is unclear whether they are associated with NASH fibrosis specifically, or with fibrosis in other liver diseases as well. Further studies are needed to confirm their role in NAFLD and other liver diseases.
Our findings of association with histological parameters reach conventional, but not GWAS-level significance. Although this is often advocated in genetic studies, we believe it may not be applicable in this case for several reasons. First, we have performed a candidate-gene analysis with only 6 SNPs, as compared to hundreds of thousands in GWA studies. Second, as we tested associations with several phenotypes, we corrected for multiple comparisons using the FDR procedure (25). Since the histological attributes we tested are not independent of each other, the use of the Bonferroni correction would have been too conservative and would greatly reduce the power to detect an association. The FDR procedure is much more suitable for this situation (41). Third, the separate and independent association of rs738409 with different histological aspects of NAFLD, all in the same direction of effect, strongly supports a true association. Finally, the histological scoring scheme for NAFLD is semi-quantitative and has a limited dynamic range. This factor markedly reduces the ability to achieve high significance values that can be observed with discrete traits such as liver enzymes or liver fat quantification by MRS.
In summary, we confirm the association of the rs738409 G allele in the PNPLA3 (adiponutrin) gene with steatosis in the largest cohort of histologically-proven NAFLD reported to date. Furthermore, this is the first study to describe an association of this SNP with histological parameters of disease severity, including inflammation, Mallory-Denk bodies and fibrosis. In pediatric patients, the high-risk allele seems to be associated with earlier presentation of disease. Our findings suggest that the rs738409 G allele may predispose patients to fat accumulation in the liver, but that other factors, environmental or hereditary, may be required for the development of inflammation, cellular injury and fibrosis. However, once patients develop NASH, the rs738409 G allele predisposes them to worse injury. In addition, we report an association of SNPs in the PNPLA3 locus and in the CPN1-CHUK locus on chromosome 10 with severity of NASH fibrosis. The mechanism underlying these associations requires further investigation.
Supplementary Material
Acknowledgments
Financial support:
The Nonalcoholic Steatohepatitis Clinical Research Network (NASH CRN) is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (grants U01DK061718, U01DK061728, U01DK061731, U01DK061732, U01DK061734, U01DK061737, U01DK061738, U01DK061730, U01DK061713), and the National Institute of Child Health and Human Development (NICHD). Several clinical centers use support from General Clinical Research Centers or Clinical and Translational Science Awards in conduct of NASH CRN Studies (grants UL1RR024989, M01RR000750, M01RR00188, UL1RR02413101, M01RR000827, UL1RR02501401, M01RR000065, M01RR020359). This work was supported by the intramural programs of NIDDK and the National Cancer Institute (NCI).
We are grateful for the technical assistance of Ronda Sapp and Yoon Park, and the statistical advice of Dr. Xiongce Zhao and Prof. Yoav Benjamini.
Abbreviations
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- GWA
genome wide association
- SNP
single nucleotide polymorphism
- NIH
National Institutes of Health
- CRN
clinical research network
- BMI
body-mass index
- WHO
World Health Organization
- OR
odds ratio
- CI
confidence intervals
- FDR
false detection rate
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- GGT
gamma-glutamyl transferase
- DM
diabetes mellitus
- NAS
NAFLD activity score
- MRS
magnetic resonance spectroscopy
References
- 1.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 2.Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 3.Adams LA, Lymp JF, St Sauver J, Sanderson SO, Lindor KD, Feldstein A, Angulo P. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Neuschwander-Tetri BA, Caldwell SH. Nonalcoholic steatohepatitis: summary of an AASLD Single Topic Conference. Hepatology. 2003;37:1202–1219. doi: 10.1053/jhep.2003.50193. [DOI] [PubMed] [Google Scholar]
- 5.Sanyal AJ. Mechanisms of Disease: pathogenesis of nonalcoholic fatty liver disease. Nat Clin Pract Gastroenterol Hepatol. 2005;2:46–53. doi: 10.1038/ncpgasthep0084. [DOI] [PubMed] [Google Scholar]
- 6.Browning JD, Kumar KS, Saboorian MH, Thiele DL. Ethnic differences in the prevalence of cryptogenic cirrhosis. Am J Gastroenterol. 2004;99:292–298. doi: 10.1111/j.1572-0241.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 7.Struben VM, Hespenheide EE, Caldwell SH. Nonalcoholic steatohepatitis and cryptogenic cirrhosis within kindreds. Am J Med. 2000;108:9–13. doi: 10.1016/s0002-9343(99)00315-0. [DOI] [PubMed] [Google Scholar]
- 8.Willner IR, Waters B, Patil SR, Reuben A, Morelli J, Riely CA. Ninety patients with nonalcoholic steatohepatitis: insulin resistance, familial tendency, and severity of disease. Am J Gastroenterol. 2001;96:2957–2961. doi: 10.1111/j.1572-0241.2001.04667.x. [DOI] [PubMed] [Google Scholar]
- 9.LA H, HA J, JP M, TA M. A Catalog of Published Genome-Wide Association Studies. 2009 Available at: www.genome.gov/gwastudies. In;
- 10.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, Zhang W, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kantartzis K, Peter A, Machicao F, Machann J, Wagner S, Konigsrainer I, Konigsrainer A, et al. Dissociation between Fatty Liver and Insulin Resistance in Humans carrying a Variant of the Patatin-like Phospholipase 3 Gene. Diabetes. 2009 doi: 10.2337/db09-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotronen A, Johansson LE, Johansson LM, Roos C, Westerbacka J, Hamsten A, Bergholm R, et al. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52:1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- 14.Kollerits B, Coassin S, Kiechl S, Hunt SC, Paulweber B, Willeit J, Brandstatter A, et al. A common variant in the adiponutrin gene influences liver enzyme levels. J Med Genet. 2009 doi: 10.1136/jmg.2009.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romeo S, Sentinelli F, Dash S, Yeo GS, Savage DB, Leonetti F, Capoccia D, et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int J Obes (Lond) 2010;34:190–194. doi: 10.1038/ijo.2009.216. [DOI] [PubMed] [Google Scholar]
- 16.Chalasani NP, Sanyal AJ, Kowdley KV, Robuck PR, Hoofnagle J, Kleiner DE, Unalp A, et al. Pioglitazone versus vitamin E versus placebo for the treatment of non-diabetic patients with non-alcoholic steatohepatitis: PIVENS trial design. Contemp Clin Trials. 2009;30:88–96. doi: 10.1016/j.cct.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavine JE, Schwimmer JB, Molleston JP, Scheimann AO, Murray KF, Abrams SH, Rosenthal P, et al. Treatment of nonalcoholic fatty liver disease in children: TONIC trial design. Contemp Clin Trials. 2009 doi: 10.1016/j.cct.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Promrat K, Lutchman G, Uwaifo GI, Freedman RJ, Soza A, Heller T, Doo E, et al. A pilot study of pioglitazone treatment for nonalcoholic steatohepatitis. Hepatology. 2004;39:188–196. doi: 10.1002/hep.20012. [DOI] [PubMed] [Google Scholar]
- 19.Loomba R, Lutchman G, Kleiner DE, Ricks M, Feld JJ, Borg BB, Modi A, et al. Clinical trial: Pilot study of metformin for the treatment of nonalcoholic steatohepatitis. Aliment Pharmacol Ther. 2008 doi: 10.1111/j.1365-2036.2008.03869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunt EM, Kleiner DE, Wilson LA, Unalp A, Behling CE, Lavine JE, Neuschwander-Tetri BA. Portal chronic inflammation in nonalcoholic fatty liver disease (NAFLD): a histologic marker of advanced NAFLD-Clinicopathologic correlations from the nonalcoholic steatohepatitis clinical research network. Hepatology. 2009;49:809–820. doi: 10.1002/hep.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 22.Glynn NW, Meilahn EN, Charron M, Anderson SJ, Kuller LH, Cauley JA. Determinants of bone mineral density in older men. J Bone Miner Res. 1995;10:1769–1777. doi: 10.1002/jbmr.5650101121. [DOI] [PubMed] [Google Scholar]
- 23.Geneva: World Health Organization; The WHO child growth standards BMI-for-age. 2006
- 24.Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The choice of a genetic model in the meta-analysis of molecular association studies. Int J Epidemiol. 2005;34:1319–1328. doi: 10.1093/ije/dyi169. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 26.He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, Cohen JC, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh MM, Brunt EM. Pathology of nonalcoholic fatty liver disease. Am J Clin Pathol. 2007;128:837–847. doi: 10.1309/RTPM1PY6YGBL2G2R. [DOI] [PubMed] [Google Scholar]
- 28.Schwimmer JB, Behling C, Newbury R, Deutsch R, Nievergelt C, Schork NJ, Lavine JE. Histopathology of pediatric nonalcoholic fatty liver disease. Hepatology. 2005;42:641–649. doi: 10.1002/hep.20842. [DOI] [PubMed] [Google Scholar]
- 29.Sookoian S, Castano GO, Burgueno AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loomba R, Sirlin CB, Schwimmer JB, Lavine JE. Advances in pediatric nonalcoholic fatty liver disease. Hepatology. 2009;50:1282–1293. doi: 10.1002/hep.23119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohanty SR, Troy TN, Huo D, O'Brien BL, Jensen DM, Hart J. Influence of ethnicity on histological differences in non-alcoholic fatty liver disease. J Hepatol. 2009;50:797–804. doi: 10.1016/j.jhep.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 32.Baulande S, Lasnier F, Lucas M, Pairault J. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem. 2001;276:33336–33344. doi: 10.1074/jbc.M105193200. [DOI] [PubMed] [Google Scholar]
- 33.Liu YM, Moldes M, Bastard JP, Bruckert E, Viguerie N, Hainque B, Basdevant A, et al. Adiponutrin: A new gene regulated by energy balance in human adipose tissue. J Clin Endocrinol Metab. 2004;89:2684–2689. doi: 10.1210/jc.2003-031978. [DOI] [PubMed] [Google Scholar]
- 34.Polson DA, Thompson MP. Macronutrient composition of the diet differentially affects leptin and adiponutrin mRNA expression in response to meal feeding. J Nutr Biochem. 2004;15:242–246. doi: 10.1016/j.jnutbio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Faraj M, Beauregard G, Loizon E, Moldes M, Clement K, Tahiri Y, Cianflone K, et al. Insulin regulation of gene expression and concentrations of white adipose tissue-derived proteins in vivo in healthy men: relation to adiponutrin. J Endocrinol. 2006;191:427–435. doi: 10.1677/joe.1.06659. [DOI] [PubMed] [Google Scholar]
- 36.Moldes M, Beauregard G, Faraj M, Peretti N, Ducluzeau PH, Laville M, Rabasa-Lhoret R, et al. Adiponutrin gene is regulated by insulin and glucose in human adipose tissue. Eur J Endocrinol. 2006;155:461–468. doi: 10.1530/eje.1.02229. [DOI] [PubMed] [Google Scholar]
- 37.Kershaw EE, Hamm JK, Verhagen LA, Peroni O, Katic M, Flier JS. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 2006;55:148–157. [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 39.Johansson LE, Johansson LM, Danielsson P, Norgren S, Johansson S, Marcus C, Ridderstrale M. Genetic variance in the adiponutrin gene family and childhood obesity. PLoS One. 2009;4:e5327. doi: 10.1371/journal.pone.0005327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johansson LE, Lindblad U, Larsson CA, Rastam L, Ridderstrale M. Polymorphisms in the adiponutrin gene are associated with increased insulin secretion and obesity. Eur J Endocrinol. 2008;159:577–583. doi: 10.1530/EJE-08-0426. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.