Abstract
A new methodology using hydrogen/deuterium amide exchange (HDX) to determine the binding affinity of protein-peptide interactions is reported. The method, based on our previously established approach, PLIMSTEX (Protein Ligand Interaction by Mass Spectrometry, Titration and H/D Exchange) [J. Am. Chem. Soc. 2003, 125, 5252–5253], makes use of a dilution strategy (dPLIMSTEX) for HDX, using the mass of the peptide ligand as readout. We employed dPLIMSTEX to study the interaction of calcium-saturated calmodulin with the opioid peptide β-endorphin as a model system; the affinity results are in good agreement with those from traditional PLIMSTEX and with literature values obtained by using other methods. We show that the dPLIMSTEX method is feasible to quantify an antigen-antibody interaction involving a 3-nitrotyrosine modified peptide in complex with a monoclonal anti-nitrotyrosine antibody. A dissociation constant in the low nanomolar range, was determined, and a binding stoichiometry of antibody/peptide of 1:2 was confirmed. In addition, we determined that the epitope in the binding interface contains a minimum of five amino acids. The dPLIMSTEX approach is a sensitive and powerful tool for the quantitative determination of peptide affinities with antibodies, complementary to conventional immuno-analytical techniques.
Introduction
Protein-peptide interactions are important in basic biophysics and in research aimed at development of therapeutic agents. Antibody-antigen binding affinity and specificity are of special interest; they are essential aspects in immune function investigation, assay development, biomarker discovery, and rational drug or vaccine design, [1, 2]. Affinity determination of antibody-peptide interactions is key to characterize recognition specificities of antibodies, to delineate the antigenic determinant (epitope mapping) [3, 4], and to probe the antibody-binding ability of synthetic peptides derived from the amino acid sequence of the antigen protein when the epitope is linear or continuous [2, 5]. Binding affinity has been typically assessed by using enzyme-linked immunosorbent assay (ELISA), surface plasmon resonance (SPR) including Biacore, and isothermal calorimetry [1].
The advantages of a mass-spectrometry-based approach for measuring binding constants of protein-ligand systems include high accuracy, high sensitivity, and straightforward data analysis [6, 7]. Many MS methods employ hydrogen/deuterium amide exchange (HDX), which also has been widely used to determine protein conformations, binding interfaces, and dynamics [8–10]. SUPREX (Stability of Unpurified Proteins from Rates of H/D Exchange) makes use of the extent of protein HDX in various concentrations of denaturants to determine the affinities of protein-ligand complexes [11–19]. SPROX (Stability of Proteins from Rates of Oxidation) takes a similar approach, adopting hydrogen peroxide-mediated methionine oxidation instead of HDX as the readout [20]. PLIMSTEX (Protein Ligand Interaction by Mass Spectrometry, Titration and H/D Exchange), previously developed in the Gross laboratory, tracks changes in the extent of protein HDX at various ligand/protein ratios to give affinity, stoichiometry and conformational changes that occur upon ligand binding [21–26]. Thus far, there are no reports of extending these MS-based methods to antibody-antigen binding.
Nevertheless, HDX is effective for epitope mapping to probe antibody-antigen interactions [2, 27–31]. HDX takes advantage of the drastically slower kinetics of HDX rates in regions of an antigen shielded by binding with an antibody [2, 29–31]. In many cases, HDX results are combined with affinity measurements from SPR to obtain more certain epitope delineation [28, 30, 31].
Here we report a novel adaption of PLIMSTEX by incorporating a dilution strategy, dPLIMSTEX, for affinity determination of protein-peptide systems. One design goal of the dilution strategy is to minimize protein consumption in the measurement. In addition, unlike traditional PLIMSTEX that monitors the protein mass as HDX occurs, dPLIMSTEX uses HDX of the ligand peptide as readout. Given that the mass of the peptide is more readily measured than that of the antibody, the approach should add precision and accuracy to the determination. Moreover, with dPLIMSTEX, the problems of low ionization efficiency and relatively small and hard-to-measure changes in mass of an antibody are avoided. The approach also provides information on the minimum number of amino acids constituting the epitope at the binding interface.
Experimental Procedures
Materials
Porcine calmodulin (MW 16790) was purchased from Ocean Biologics Co. (Edmonds, WA). The monoclonal mouse anti-3-nitrotyrosine antibody (SC-Ab, 39B6) was obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA), and was raised against a 3-(2-(4-hydroxy-3-nitrophenyl) acetamido) propionyl-bovine serum albumin immunogen conjugate. β-Endorphin, deuterium oxide (D2O), formic acid (FA), hydrochloric acid (HCl), calcium chloride (CaCl2), potassium chloride (KCl), 3-(N-morpholino)propanesulfonic acid (MOPS) hemisodium salt, N-(2-hydroxyethyl)-1-piperazineethanesulfonic (HEPES) acid, HEPES sodium salt were obtained from Sigma-Adrich (St. Louis, MO). All reagents and solvents were of analytical grade or highest available purity.
Peptide Synthesis
The antigenic peptide, prostacyclin synthase (PCS) (79–91), containing 3-nitrotyrosine [DPHS83Y(NO2)DAVVWEPR; PCS2a], was synthesized on a semi-automated peptide synthesizer (EPS-221, Intavis, Langenfeld, Germany) by solid-phase peptide synthesis (SPPS) with Fmoc/t-butyl protection chemistry. The peptide was purified by reversed phase-high performance liquid chromatography (RP-HPLC), and its purity confirmed by electrospray ionization mass spectrometry (ESI-MS).
HDX Protocol
Solutions of calmodulin saturated with calcium (CaM·4Ca2+) and β endorphin were prepared in aqueous buffer containing 200 mM KCl, 50 mM MOPS (pH 7.3) and 10 mM CaCl2. Solutions of anti-3-nitrotyrosine antibodies and PCS2a were prepared in a buffer containing 150 mM KCl and 10 mM HEPES (pH 7.4). Protein-peptide mixture solutions were incubated for 2 h at room temperature to establish equilibrium before starting HDX. To initiate the HDX, a solution of protein-peptide complex or peptide alone was mixed with an equal volume of D2O buffer (with the same salt composition as in the aqueous solution) at room temperature. After 65 sec, the exchange was quenched by adding ice-cold HCl solution (0.5 M) to afford a final pH of ~2.5, and the sample was quickly frozen in liquid nitrogen until desalting and analysis. For the β-endorphin:CaM system, both traditional PLIMSTEX and the new dPLIMSTEX were performed to permit comparison; a C18 guard column was used for desalting, as described previously [21, 22, 25]. For the antibody-peptide system, C18 ziptips (Millipore, Billerica, MA) were used to desalt the antigenic peptide. After the frozen sample was removed from the liquid nitrogen, introduction to the ziptip was initiated when half the volume of the sample was melted and the other half remained frozen. The ziptip procedures were performed with ice-cold solvents within 30–50 sec to minimize back-exchange. The desalted peptide solution was immediately injected into the mass spectrometer through a steel T-union (Cobert Associates, St. Louis, MO) that was connected with an isocratic LC flow of 50% acetonitrile containing 0.1% formic acid at 40 μL/min. The T-union and the capillaries for the LC and mass spectrometer connections were placed in an ice bath. HDX of the free peptide using the ziptip desalting method was performed to determine the back-exchange extent of this method. The HDX shift was compared to the theoretical number of exchangeable amide protons on the peptide backbone, allowing us to deduce that the back-exchange was below 20%. Free peptide HDX defined the asymptote (D0, the mass of the free peptide after HDX) in the data modeling; its determination acted as a standard experiment that was performed daily to check the experimental settings and to assist with trouble-shooting when needed.
The adoption of C18 ziptip greatly facilitated peptide desalting and concentrating in the case of antigen-antibody complexes, given that the C18 column used for traditional PLIMSTEX would easily get clogged by injection of large antibody molecules. We found that C18 guard columns quickly lost their efficiency for peptide-binding and desalting after 1–2 injections of an antibody-peptide mixture. To overcome this problem, antibody immobilization has been often effectively employed which enables the separation of antibody by precipitation [27–29]; however immobilization approaches cannot be used in dPLIMSTEX as a solution method. The use of C18 ziptips (200 Å pore size) obviated the problem of a guard column and provided efficient and reproducible desalting. Furthermore, the ziptips were used only once and disposed, preventing sample carryover. C4 ziptips (300 Å pore size) were also evaluated, but with them, no or poor mass spectral signal was seen for the peptide when its concentration was comparable to that of the protein, probably because the ziptip had insufficient capacity to bind the peptide in the presence of antibody.
Mass Spectrometry
For β-endorphin:CaM system, ESI mass spectra were acquired in the positive-ion mode on a Micromass Q-TOF Ultima (Waters, Manchester, UK) with a Waters nanoAcquity UPLC, or on a LCQ Deca XP plus (Thermo, San Jose, CA) with a Waters CapLC system (Manchester, UK). For the antibody-peptide system, an LTQ Orbitrap XL (Thermo, San Jose, CA) was used for ESI acquisition in the positive-ion mode. For the measurements on the LTQ Orbitrap or LCQ Deca XP plus, the spray voltage was 4.0 kV, and the capillary temperature was 250 °C. The other instrument parameters were optimized based on the tuning for the most abundant charge-state peak from the peptide of interest, and the zoom scan mode was utilized. On the Q-TOF instrument, the capillary voltage was 3.2 kV, and the source and desolvation temperatures were 80 and 180 °C, respectively. The cone and desolvation gas flows were 40 and 400 L/h, respectively.
Data Processing
The mass centroids of isotopic pattern envelops from peptide mass spectra were calculated, and the data were sorted into sets with each containing data from experiments having the same macromolecule concentration, MT. A modeling program implemented with Mathcad V14_M020 professional (MathSoft, Inc., Cambridge, MA) was designed to fit all sets of data with different MT and to extract Ka (affinity constant) and ΔD1 (difference in deuterium uptake between bound and unbound state of the peptide). More details are below.
Modeling Procedure
The process to quantify the affinity of antibody peptide binding began with a plot of the peptide HDX mass versus the ratio of total ligand concentration [LigT] to the total macromolecule (protein) concentration [MT], referred to as a dPLIMSTEX curve. The objective of the process was to obtain the best-fit model curve to the dPLIMSTEX data; from the best-fit curve were extracted the binding constants and the changes in HDX that occurred upon binding. For the general case of 1:n protein:ligand binding model, where n is the number of binding sites for the same ligand, these unknown parameters included the overall binding constants (βi, i = 1 to n, which is the product of all the stepwise macroscopic binding constants Kj, j = 1 to i), and ligand deuterium shifts upon binding (ΔDi, i = 1 to n, which is the difference between the average deuterated mass of the bound ligand in the ith complex and that of the apo-ligand form). Program components used for calculation of the solution equilibrium remained the same from previous implementations of PLIMSTEX [23]: D0 is the HDX mass of the free peptide; [Lig] and [M] represent free ligand concentration and free macromolecule concentration in the solution phase equilibrium, respectively.
The most challenging task in the modeling procedure was to establish the relationship between the free solution ligand concentration [Lig] and the total ligand concentration [LigT]. As we reported before [23], [Lig] can be inferred from [LigT] by numerically solving the ordinary differential equation formed by the analytical derivative of the solution ligand concentration with respect to the total ligand concentration as shown in eq 1 (β0 = 1).
| (1) |
In the case of 1:1 binding systems, eq 1 can be simplified to give eq 2.
| (2) |
The differential equation 1 or 2 was solved by integration from the initial state of zero [Lig] at zero [LigT] by using the “Rkadapt” function in Mathcad software. The integration produced a lookup table for the free ligand concentration as a function of the total ligand concentration; the table had a constant step size of 1000 intervals over the ligand range in the experiment. Each experimental total ligand concentration value had a matching value in the lookup table.
In addition to the solution-phase equilibrium as modeled above, the model must also include the mass shift signal function. Because dPLIMSTEX uses the peptide mass as readout for HDX, this function gave the relationship for the average HDX mass of the ligand peptide, D, that was expressed by the deuterium shifts of the ligand molecules from each species, as shown in eq (3).
| (3) |
If [M] was expressed as a function of βi, [Lig], and the total macromolecule concentration [MT] using binding polynomial, eq 4 can be derived from eq 3.
| (4) |
For a 1:1 binding system (i.e. n = 1), βi became the affinity binding constant Ka. Correspondingly, eq 4 can be converted to eq 5.
| (5) |
Dilution data were sorted into sets with each containing data from experiments having the same macromolecule concentration [MT]. In each trial of the nonlinear least squares search (NLLS), a theoretical curve based on the trial parameters was computed for each of the macromolecule concentrations, the squares of the residuals between the corresponding data and theoretical curves for all the experiment ligand-macromolecule concentration ratios were summed, and the square root of the sum was taken for the value to be minimized.
Once a best-fit PLIMSTEX curve was obtained from the process, the solution values of the unknown parameters (βi and ΔDi) and the deviation between the experimental data and the fitted curve (RMS) were reported. In most circumstances, multiple independent trials under the same experimental conditions were performed for each data point. A bootstrap resampling method [32] was used to evaluate the precision of the solution parameters. At each titration point, the data were resampled by randomly selecting with replacement the same number of times as the number of trials available at that point. This was done for all the titration points to form a “new” data set. Rather than refitting each of a large number of the new sample sets, each new set of solution parameters was estimated to first order by using the residuals produced by the new samples. Second-order statistics of the collection of new solution parameters were computed as described previously [23].
Results and Discussion
Dilution Strategy for dPLIMSTEX
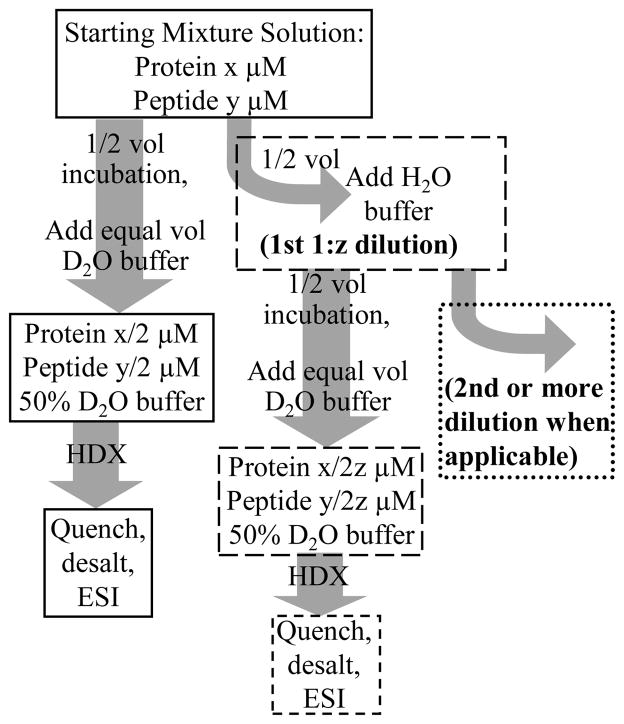
The efficiency of dPLIMSTEX stems from a procedure whereby we used a fraction of the sample for measurement. Instead of disposing of the unused portion, that fraction was diluted and used again. In the typical workflow of the dilution steps, a half volume of the starting protein-peptide solution was equilibrated directly to form the complex, and the other half volume was diluted by aqueous buffer before incubation (outlined in Scheme 1). The latter solution was further divided and diluted by the same procedure. The dilution steps can be continued until the concentration of the analyte becomes so low that it is no longer detectable by the mass spectrometer.
Scheme 1.
Typical work flow for the dilution strategy used in dPLIMSTEX. The terms x and y are the initial concentrations (μM) of protein and peptide, respectively, and z is the dilution factor.
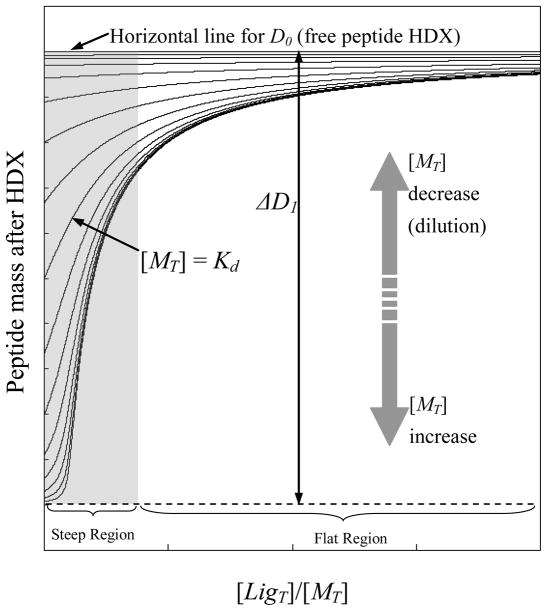
As an illustration of the expected results from a dPLIMSTEX approach, we used the modeling program to generate a series of theoretical curves based on a 1:1 binding system with a specific Ka and ΔD1 (Figure 1). The upper horizontal line represents the value of D0, the free peptide mass after HDX determined without any macromolecule present; this value is an average of replicate measurements. The curves below correspond to dilution series of different [MT] values, where [MT] refers to the total concentration of the macromolecule, with a dilution factor of two (i.e., each curve represents a 2-fold dilution from the curve immediately below it). When the [LigT]/[MT] ratio is low, generally the dPLIMSTEX curves exhibit relatively large slopes and curvatures, as shown in the shadowed area (Figure 1) denoted as the “steep region”. The curves become flat, and their slopes smaller in the “flat region”, and they eventually approach the upper horizontal line when the [LigT]/[MT] ratio reaches infinity. Data distributed in the “steep region” are more sensitive to changes of binding affinity and dilution factor, thus carrying more information for determining fitting parameters (Ka and ΔD1). The data modeling outcome, Ka and ΔD1, depend on the shape of the fitting curves and the mass difference between data points; thus, it is important to measure all the data points under the same experimental conditions so that the inevitable back-exchange will be consistent for all the experiments.
Figure 1.
A series of theoretical dPLIMSTEX curves generated from the modeling program implemented in Mathcad. The curves were calculated based on a 1:1 binding system with Ka = 1 μM and ΔD1 = 5 for illustration.
Application of dPLIMSTEX to a model system
We chose calmodulin and its binding to β-endorphin as a model system to evaluate dPLIMSTEX and to compare it with traditional PLIMSTEX. Calmodulin exhibits high-affinity, calcium-dependent binding to several peptide hormones and neurotransmitters [33–36]. β-Endorphin, an opioid peptide containing 32 amino acids, binds calmodulin with a 1:1 stoichiometry and a dissociation constant of 1.9 – 3.8 μM (25°C, pH 7.3), as determined by fluorescence of dansylcalmodulin [33], and of 2.5 – 4.6 μM (4°C, pH 7.0) as determined by equilibrium dialysis [35].
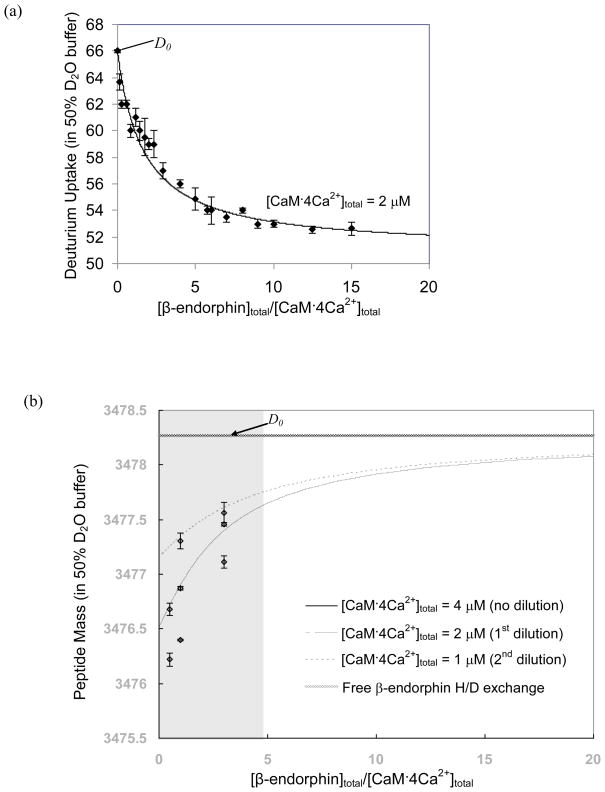
Conventional PLIMSTEX yields a titration curve (Figure 2a) that gives a binding constant (Ka) for CaM·4Ca2+ and β-endorphin of (2.2 ± 0.1)×105 M−1 (Kd = 4.5 ± 0.2 μM), which agrees well with the literature values [33, 35]. The ΔD1 value is 15.0; given that the D2O content is 50% for the HDX, there are approximately 30 backbone amide protons of CaM·4Ca2+ that become protected when binding to β-endorphin. This number is similar to that determined by PLIMSTEX for the interaction between CaM·4Ca2+ and melittin [21], suggesting that similar conformational changes occur when calmodulin binds to each ligand [37].
Figure 2.
Traditional PLIMSTEX (a) and dPLIMSTEX (b) fitting curves for CaM 4Ca2+: β-endorphin. The error bars shown for the data points represent the standard deviation from duplicate independent experiments. The D0 point in (a), is a single point for the apo state of the protein and also a “fit parameter”, whereas for (b), the D0 mass is for the unbound peptide, as such, D0 is depicted as a line representing the asymptote for the other curves in (b).
The dPLIMSTEX fitting result, shown in Figure 2b, affords a binding constant Ka of (2.05 ± 0.09)×105 M−1 (Kd = 4.9 ± 0.2 μM), which is in good agreement with both the result from traditional PLIMSTEX and those from the literature [33, 35]. Although the curve fitting seems to involve only a few data points, an important and additional data point is D0 (the mass of the unbound β-endorphin submitted to HDX in the same media as for the complexes). Furthermore, this point is an average value from repeated measurements of the peptide (free β-endorphin) mass after HDX. This is the asymptote for the curves when [β-endorphin]total/[CaM·4Ca2+]total becomes infinite. The best-fitting curves separate well from each other because the experimental [CaM·4Ca2+]total values are close to Kd. It should be emphasized that these three curves do not represent separate fits. Rather, the curves arise in the same model from the same fit parameter values (Ka, ΔD1). The curves are mutually dependent and simultaneously generated by the modeling procedure from fitting all the eight data points in Figure 2b (at three different [β-endorphin]total/[CaM·4Ca2+]total ratios) and the D0, which is the asymptote. The deviation between the best-fitted curves and the experimental data RMS is 0.061. Thus, any conclusions that dPLIMSTEX uses a sparse data set should be reconsidered.
Unlike the ΔD1 value determined by PLIMSTEX, which is a measure of the number of sites protected in CaM by the binding, the ΔD1 from dPLIMSTEX reflects the number of protected amides on the peptide and complements that from PLIMSTEX. The ΔD1 obtained by dPLIMSTEX is 3.96 ± 0.09. Correcting for the concentration of D2O (50%) in the exchange media, we conclude that at least eight backbone amides from β-endorphin are protected upon binding to CaM·4Ca2+. Comparing the sequence of β-endorphin to that of other calmodulin-binding peptides or proteins, Malencik et al. [33] suggested that the active region of β-endorphin, containing about eight amino acids, is located between a hydrophobic region and a basic region. Using a series of amino- and carboxy-terminal deletion peptides, Giedroc et al. [38] identified the residues 14–25 as the domain of β-endorphin responsible for the inhibition of calmodulin function.
The dilution approach employed in dPLIMSTEX allows one to minimize the quantity of macromolecule used in the determination for a certain system compared to that used in traditional PLIMSTEX. For example, in the dPLIMSTEX experiment shown in Figure 2b, eight data points at three different [β-endorphin]total/[CaM·4Ca2+]total ratios were used, consuming approximately 200 picomoles of protein for duplicate independent measurements. The experiment shown in Figure 2a, however, consumed ~840 picomoles of protein for duplicate determinations of 21 titration points. This latter quantity could be reduced to 480 picomoles by using 12 titration points, which has been used for 1:1 binding in traditional PLIMSTEX [21, 22, 25]. Note that these quantities are not absolute for a dPLIMSTEX or traditional PLIMSTEX method; given that the quantities are highly dependent on the affinity of the target interaction system (the ideal condition to work with in dPLIMSTEX is when the macromolecule concentrations are comparable to Kd value, as shown in Figure 1). Furthermore, all the eight data points for dPLIMSTEX are distributed in the “steep region” of the fitting curves, where the data are more informative for the unknown fitting parameters (Ka and ΔD1) than those in the “flat region”. Furthermore, given that the peptide is reporter for dPLIMSTEX, only it, and not the protein, is consumed in setting up and optimizing the mass spectrometric measurements.
Application to peptide-antibody immune complexes
An application of dPLIMSTEX to peptide-antibody immune complexes, explored here for the first time, aims to characterize the recognition specificity of a monoclonal antibody against the 3-nitrotyrosine peptide. A Tyr-nitrated peptide of PCS was used as a model system for this application. In previous work, it was shown that prostacyclin synthase (PCS) in bovine aortic microsomes undergoes specific nitration upon treatment with peroxynitrite at Tyr-430, as identified by high resolving power mass spectrometry [39]. To investigate the recognition specificities of 3-nitro-tyrosine antibodies, a commercially available monoclonal antibody, SC-Ab (39B6), was used. The interaction of the antibody with PCS2a from PCS (79–91) in which Tyr-83 was nitrated [39–41] was measured by dPLIMSTEX.
Typically, high affinity antibody (IgG type) antigen interactions have Ka values in the range of 108–1010 M−1 (dissociation constant Kd in the nanomolar (nM) range) [1]. This suggests ideal experiments to be performed at nM concentrations to obtain sensitive data modeling. Considering the detection limit of current mass spectrometers operating in the ESI mode, we chose the lowest concentration of antibody and antigenic peptide that afford suitable mass spectra for accurate centroid calculation for all data series. For the SC-Ab/PCS2a interaction, the lowest concentrations used here were 41 and 20.5 nM, respectively, for the SC-Ab and the PCS2a peptide.
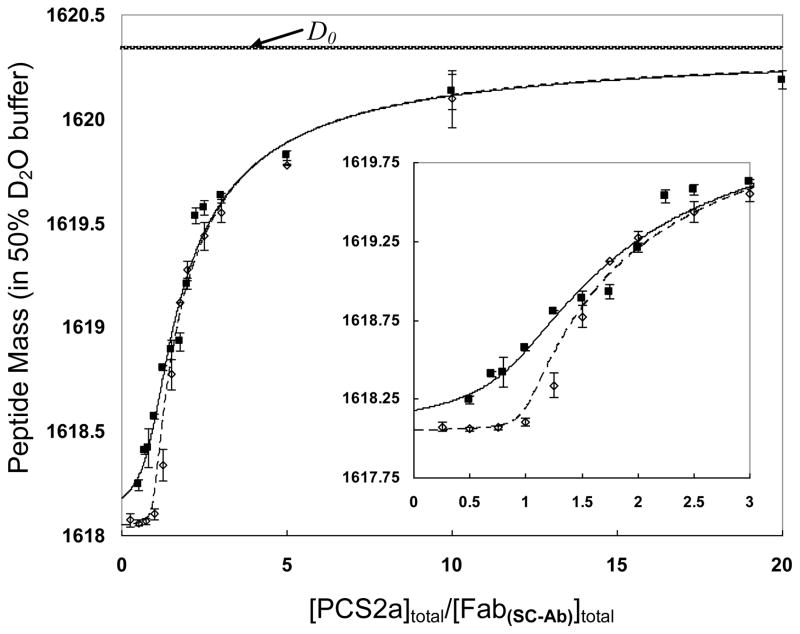
Fitting the data for SC-Ab/PCS2a interaction, shown as Figure 3, afforded a binding constant Ka of (3.4 ± 0.3)×108 M−1 (Kd = 3.0 ± 0.3 nM). The two fitted curves, representing a 16-fold dilution from the lower dashed curve to the upper solid curve, are well-separated only in the “steep region” because the lowest macromolecule concentration is still one magnitude higher than the Kd value. Hence, we collected more data points in the “steep region” to define better the curve shape and to afford higher modeling sensitivity. A two-fold dilution (e.g., as we made in the case of CaM·4Ca2+:β-endorphin) is not appropriate here because it made the dilution series data almost overlapping (data not shown) and lacked efficiency to provide a good fit. RMS for the modeling is 0.094, representing the small deviation between best fit and the experimental data, thereby substantiating the reliability of the fitting results.
Figure 3.
dPLIMSTEX fitting curves for SC-Ab:PCS2a system. The concentration of antibody Fab regions (shown as [Fab(SC-Ab)]) is used in place of the antibody concentration in the modeling. [Fab(SC-Ab)]total for the dashed line (----) is 660 nM, whereas [Fab(SC-Ab)]total for the solid line (____) is 41 nM. The error bars shown for the data points represent the standard deviation from duplicate independent experiments. The D0 mass is for the unbound peptide, and is depicted as a line representing the asymptote for the other curves.
In addition, the binding stoichiometry was also assessed by the dPLIMSTEX approach. IgG antibodies frequently bind two antigens, each Fab region interacting with one antigen molecule, but 1:1 antibody/antigen binding stoichiometry was observed in other cases [2]. The dashed fitted curve in Figure 3, obtained when the macromolecule concentration was more than two orders of magnitude larger than Kd, is almost a “sharp-break” curve that reveals the binding stoichiometry. This break takes place when [PCS2a]total/[Fab(SC-Ab)]total = 1. Note that we used the Fab concentration, and not the antibody concentration, in the analysis and in the plot. Given that there are two Fab regions per antibody, the SC-Ab:PCS2a is a 1:2 binding system, as is normally the case. This outcome also indicates that the two binding events per antibody are independent.
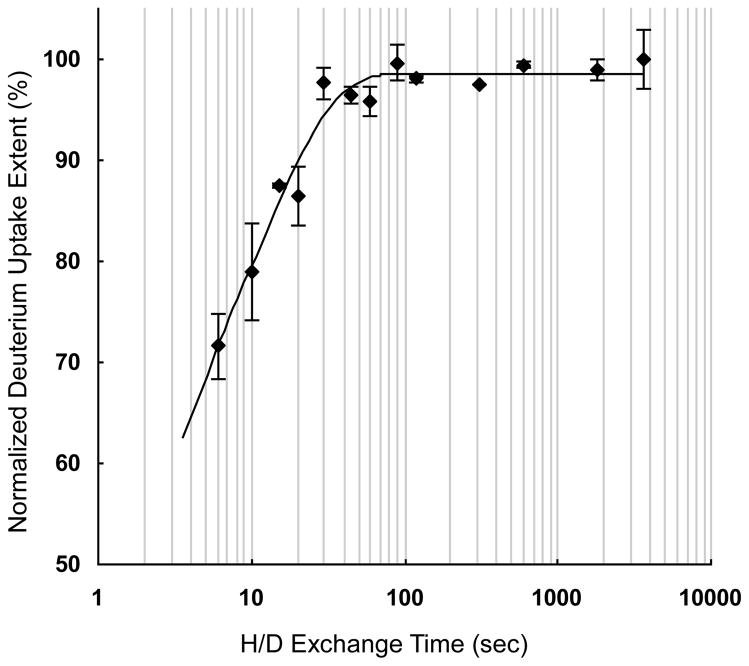
The ΔD1 from the modeling of SC-Ab:PCS2a is 2.3 ± 0.1, indicating that at least ~5 backbone amide protons (correcting for the % D2O in the exchange medium) of the antigenic peptides are sequestered from HDX upon binding and suggesting that at least five amino acids constitute the epitope of the antigen peptide. When the HDX kinetics curve of PCS2a was treated by two-group modeling (Figure 4), we found an average chemical exchange rate constant kex of free PCS2a at 0.69 sec−1 for 58% exchangeable amide protons and 0.077 sec−1 for the remaining 42% exchangeable amide protons. Compared to the association rate kon of the SC-Ab:PCS2a complex [42], kex ≫ kon[M]. Therefore, the backbone amide hydrogens in the peptide ligand are likely to undergo correlated exchange owing to the relatively slow association rate (i.e., HDX occurs during the time following dissociation of the complex and before the peptide can form a new complex [43]). This can be considered as an inevitable “leak” or loss of measured protected amide sites in the ΔD1. Consequently, the actual number of amide protons that becomes sequestered upon complex formation is larger than that calculated from ΔD1. The influence of correlated exchange and binding dynamics on the determination of Ka and ΔD1 by dPLIMSTEX and the offset of these parameters will be discussed in detail in future work.
Figure 4.
The kinetic curve for H/D exchange of free PCS2a peptide. All the data of deuterium uptake was normalized to the highest average uptake value. The error bars shown for the data points represent the standard deviation from duplicate independent experiments.
CONCLUSIONS
Compared to traditional PLIMSTEX, dPLIMSTEX has several advantages for determining the affinity of an antibody-peptide complex. The first is that dPLIMSTEX monitors the peptide mass shift after HDX, which is less subject to error because the ΔD1 value is relatively large fraction of the number of amides in the peptide. Second, the peptide has high ionization efficiency, whereas, at least several micrograms [44] are required for each analysis of antibodies to afford good mass spectra.
dPLIMSTEX appears to be a sensitive quantitative method to study protein-peptide interactions. It determines the binding affinities and binding stoichiometry, and estimates the extent of the interaction region of the peptide ligand. The dilution series approach used in dPLIMSTEX offers the opportunity to use a minimum amount of macromolecule, especially for cases involving large proteins. The application of dPLIMSTEX to antibody-antigen complexes offers the capability to characterize the specificity and recognition motif of antibodies in solution. More work using a series of antigenic peptides is in progress.
Acknowledgments
The work was supported by the National Centers for Research Resources of the NIH under Grant No. 2P41RR000954, the Deutscher Akademischer Austausch Dienst (DAAD, Bonn, Germany; PP 502/09) and the Deutsche Forschungsgemeinschaft (DFG, Bonn, Germany; FOR-753). The authors also thank Dr. Justin B. Sperry for useful discussions and Ms. Manohari Silva for advice on incorporating ziptips and the T-union for the measurements.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Delves P, Martin S, Burton D, Roitt I. Roitt’s Essential Immunology. 11. Wiley: 2006. pp. 86–110. [Google Scholar]
- 2.Hager-Braun C, Tomer KB. Determination of protein-derived epitopes by mass spectrometry. Expert Rev Proteomics. 2005;2:745–756. doi: 10.1586/14789450.2.5.745. [DOI] [PubMed] [Google Scholar]
- 3.Suckau D, Kohl J, Karwath G, Schneider K, Casaretto M, Bittersuermann D, Przybylski M. Molecular Epitope Identification by Limited Proteolysis of an Immolbilized Antigen-Antibody Complex and Mass-Spectrometric Peptide-Mapping. Proc Natl Acad Sci U S A. 1990;87:9848–9852. doi: 10.1073/pnas.87.24.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefanescu R, Iacob RE, Damoc EN, Marquardt A, Amstalden E, Manea M, Perdivara I, Maftei M, Paraschiv G, Przybylski M. Mass spectrometric approaches for elucidation of antigen-anti body recognition structures in molecular immunology. Eur J Mass Spectrom. 2007;13:69–75. doi: 10.1255/ejms.849. [DOI] [PubMed] [Google Scholar]
- 5.Reineke U, Kramer A, Schneider-Mergener J. Antigen sequence- and library-based mapping of linear and discontinuous protein-protein-interaction sites by spot synthesis. Combinatorial Chemistry in Biology. 1999;243:23–36. doi: 10.1007/978-3-642-60142-2_2. [DOI] [PubMed] [Google Scholar]
- 6.Schermann SM, Simmons DA, Konermann L. Mass spectrometry-based approaches to protein-ligand interactions. Expert Rev Proteomics. 2005;2:475–485. doi: 10.1586/14789450.2.4.475. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald MC, West GM. Painting Proteins with Covalent Labels: What’s In the Picture? J Am Soc Mass Spectrom. 2009;20:1193–1206. doi: 10.1016/j.jasms.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Busenlehner LS, Armstrong RN. Insights into enzyme structure and dynamics elucidated by amide H/D exchange mass spectrometry. Arch Biochem Biophys. 2005;433:34–46. doi: 10.1016/j.abb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annual Review of Biophysics and Biomolecular Structure. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 10.Suchanova B, Tuma R. Folding and assembly of large macromolecular complexes monitored by hydrogen-deuterium exchange and mass spectrometry. Microbial Cell Factories. 2008:7. doi: 10.1186/1475-2859-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powell KD, Fitzgerald MC. Measurements of protein stability by H/D exchange and matrix-assisted laser desorption ionization mass spectrometry using picomoles of material. Anal Chem. 2001;73:3300–3304. doi: 10.1021/ac0100805. [DOI] [PubMed] [Google Scholar]
- 12.Powell KD, Ghaemmaghami S, Wang MZ, Ma LY, Oas TG, Fitzgerald MC. A general mass spectrometry-based assay for the quantitation of protein-ligand binding interactions in solution. J Am Chem Soc. 2002;124:10256–10257. doi: 10.1021/ja026574g. [DOI] [PubMed] [Google Scholar]
- 13.Powell KD, Wales TE, Fitzgerald MC. Thermodynamic stability measurements on multimeric proteins using a new H/D exchange- and matrix-assisted laser desorption/ionization (MALDI) mass spectrometry-based method. Protein Sci. 2002;11:841–851. doi: 10.1110/ps.3820102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell KD, Wang MZ, Silinski P, Ma LY, Wales TE, Dai SY, Warner AH, Yang XY, Fitzgerald MC. The accuracy and precision of a new H/D exchange- and mass spectrometry-based technique for measuring the thermodynamic stability of proteins. Anal Chim Acta. 2003;496:225–232. [Google Scholar]
- 15.Roulhac PL, Powell KD, Dhungana S, Weaver KD, Mietzner TA, Crumbliss AL, Fitzgerald MC. SUPREX (stability of unpurified proteins from rates of H/D exchange) analysis of the thermodynamics of synergistic anion binding by ferric-binding protein (FbpA), a bacterial transferrin. Biochemistry. 2004;43:15767–15774. doi: 10.1021/bi0481848. [DOI] [PubMed] [Google Scholar]
- 16.Wang MZ, Shetty JT, Howard BA, Campa MJ, Patz EF, Fitzgerald MC. Thermodynamic analysis of cyclosporin A binding to cyclophilin A in a lung tumor tissue lysate. Anal Chem. 2004;76:4343–4348. doi: 10.1021/ac049536j. [DOI] [PubMed] [Google Scholar]
- 17.Dai SY, Gardner MW, Fitzgerald MC. Protocol for the thermodynamic analysis of some proteins using an H/D exchange- and mass spectrometry based technique. Anal Chem. 2005;77:693–697. doi: 10.1021/ac048967z. [DOI] [PubMed] [Google Scholar]
- 18.Dai SY, Fitzgerald MC. Accuracy of SUPREX (stability of unpurified proteins from rates of H/D exchange) and MALDI mass spectrometry-derived protein unfolding free energies determined under non-EX2 exchange conditions. J Am Soc Mass Spectrom. 2006;17:1535–1542. doi: 10.1016/j.jasms.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 19.Tang L, Hopper ED, Tong Y, Sadowsky JD, Peterson KJ, Gellman SH, Fitzgerald MC. H/D exchange- and mass spectrometry-based strategy for the thermodynamic analysis of protein-ligand binding. Anal Chem. 2007;79:5869–5877. doi: 10.1021/ac0700777. [DOI] [PubMed] [Google Scholar]
- 20.West GM, Tang L, Fitzgerald MC. Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass-spectrometry based strategy. Anal Chem. 2008;80:4175–4185. doi: 10.1021/ac702610a. [DOI] [PubMed] [Google Scholar]
- 21.Zhu MM, Rempel DL, Du ZH, Gross ML. Quantification of protein-ligand interactions by mass spectrometry, titration, and H/D exchange: PLIMSTEX. J Am Chem Soc. 2003;125:5252–5253. doi: 10.1021/ja029460d. [DOI] [PubMed] [Google Scholar]
- 22.Zhu MM, Rempel DL, Zhao J, Giblin DE, Gross ML. Probing Ca2+-induced conformational changes in porcine calmodulin by H/D exchange and ESI-MS: Effect of cations and ionic strength. Biochemistry. 2003;42:15388–15397. doi: 10.1021/bi035188o. [DOI] [PubMed] [Google Scholar]
- 23.Zhu MM, Rempel DL, Gross ML. Modeling data from titration, amide H/D exchange, and mass spectrometry to obtain protein-ligand binding constants. J Am Soc Mass Spectrom. 2004;15:388–397. doi: 10.1016/j.jasms.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Zhu MM, Chitta R, Gross ML. PLIMSTEX: a novel mass spectrometric method for the quantification of protein-ligand interactions in solution. Elsevier Science Bv; 2005. [Google Scholar]
- 25.Sperry JB, Shi XG, Rempel DL, Nishimura Y, Akashi S, Gross ML. A mass spectrometric approach to the study of DNA-binding proteins: Interaction of human TRF2 with telomeric DNA. Biochemistry. 2008;47:1797–1807. doi: 10.1021/bi702037p. [DOI] [PubMed] [Google Scholar]
- 26.Zhu MM, Chitta R, Gross ML. PLIMSTEX: a novel mass spectrometric method for the quantification of protein-ligand interactions in solution. Int J Mass Spectrom. 2005;240:213–220. [Google Scholar]
- 27.Yamada N, Suzuki E-I, Hirayama K. Identification of the interface of a large protein-protein complex using H/D exchange and Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 2002;16:293–299. doi: 10.1002/rcm.579. [DOI] [PubMed] [Google Scholar]
- 28.Baerga-Ortiz A, Hughes CA, Mandell JG, Komives EA. Epitope mapping of a monoclonal antibody against human thrombin by H/D-exchange mass spectrometry reveals selection of a diverse sequence in a highly conserved protein. Protein Sci. 2002;11:1300–1308. doi: 10.1110/ps.4670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coales SJ, Tuske SJ, Tomasso JC, Hamuro Y. Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography and mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:639–647. doi: 10.1002/rcm.3921. [DOI] [PubMed] [Google Scholar]
- 30.Lu J, Witcher DR, White MA, Wang X, Huang L, Rathnachalam R, Beals JM, Kuhstoss S. IL-1beta Epitope Mapping Using Site-Directed Mutagenesis and Hydrogen-Deuterium Exchange Mass Spectrometry Analysis. Biochemistry. 2005;44:11106–11114. doi: 10.1021/bi0505464. [DOI] [PubMed] [Google Scholar]
- 31.Obungu VH, Gelfanova V, Rathnachalam R, Bailey A, Sloan-Lancaster J, Huang L. Determination of the Mechanism of Action of Anti-FasL Antibody by Epitope Mapping and Homology Modeling. Biochemistry. 2009;48:7251–7260. doi: 10.1021/bi900296g. [DOI] [PubMed] [Google Scholar]
- 32.Caceci MS. Estimating Error Limits in Parametric Curve Fitting. Anal Chem. 1989;61:2324–2327. [Google Scholar]
- 33.Malencik DA, Anderson SR. Binding of Simple Peptides, Hormones, and Neurotransmitters by Calmodulin. Biochemistry. 1982;21:3480–3486. doi: 10.1021/bi00257a035. [DOI] [PubMed] [Google Scholar]
- 34.Malencik DA, Anderson SR. Binding of hormones and neuropeptides by calmodulin. Biochemistry. 1983;22:1995–2001. doi: 10.1021/bi00277a040. [DOI] [PubMed] [Google Scholar]
- 35.Sellinger-Barnette M, Weiss B. Interaction of Beta-Endorphin and Other Opioid-Peptides with Calmodulin. Mol Pharmacol. 1982;21:86–91. [PubMed] [Google Scholar]
- 36.Yoshino H, Wakita M, Izumi Y. Calcium-dependent changes in structure of calmodulin with substance P. J Biol Chem. 1993;268:12123–12128. [PubMed] [Google Scholar]
- 37.Kataoka M, Head JF, Seaton BA, Engelman DM. Melittin binding causes a large calcium-dependent conformational change in calmodulin. Proc Natl Acad Sci U S A. 1989;86:6944–6948. doi: 10.1073/pnas.86.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giedroc DP, Ling N, Puett D. Identification of Beta-Endorphin Residues 14–25 as a Region Involved in the Inhibition of Calmodulin-Stimulated Phosphodiesterase Activity. Biochemistry. 1983;22:5584–5591. doi: 10.1021/bi00293a020. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt P, Youhnovski N, Daiber A, Balan A, Arsic M, Bachschmid M, Przybylski M, Ullrich V. Specific nitration at tyrosine 430 revealed by high resolution mass spectrometry as basis for redox regulation of bovine prostacyclin synthase. J Biol Chem. 2003;278:12813–12819. doi: 10.1074/jbc.M208080200. [DOI] [PubMed] [Google Scholar]
- 40.Chiang CW, Yeh HC, Wang LH, Chan NL. Crystal structure of the human prostacyclin synthase. J Mol Biol. 2006;364:266–274. doi: 10.1016/j.jmb.2006.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulrich M, Petre A, Youhnovski N, Promm F, Schirle M, Schumm M, Pero RS, Doyle A, Checkel J, Kita H, Thiyagarajan N, Acharya KR, Schmid-Grendelmeier P, Simon HU, Schwarz H, Tsutsui M, Shimokawa H, Bellon G, Lee JJ, Przybylski M, Doring G. Post-translational tyrosine nitration of eosinophil granule toxins mediated by eosinophil peroxidase. J Biol Chem. 2008;283:28629–28640. doi: 10.1074/jbc.M801196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.This data will be published elsewhere.
- 43.Jorgensen TJD, Gardsvoll H, Dano K, Roepstorff P, Ploug M. Dynamics of urokinase receptor interaction with peptide antagonists studied by amide hydrogen exchange and mass spectrometry. Biochemistry. 2004;43:15044–15057. doi: 10.1021/bi048706j. [DOI] [PubMed] [Google Scholar]
- 44.Gadgil HS, Bondarenko PV, Pipes GD, Dillon TM, Banks D, Abel J, Kleemann GR, Treuheit MJ. Identification of cysteinylation of a free cysteine in the Fab region of a recombinant monoclonal IgG1 antibody using Lys-C limited proteolysis coupled with LC/MS analysis. Anal Biochem. 2006;355:165–174. doi: 10.1016/j.ab.2006.05.037. [DOI] [PubMed] [Google Scholar]