Abstract
The assembly of a pre-B cell receptor (pre-BCR) composed of an Ig μ heavy chain (μH-chain), the surrogate light (SL) chain, and the Igα/β dimer is critical for late pro-B cells to advance to the pre-B cell stage. By using a transgenic mouse model, in which μH-chain synthesis is solely driven by a tetracycline-controlled transactivator, we show that de novo synthesis of μH-chain in transgenic pro-B cells not only induces differentiation but also proliferation. This positive effect of μH-chain synthesis on proliferation requires the presence of SL chain and costimulatory signals provided by stromal cells or IL-7. We conclude that pre-BCR signaling induces clonal expansion of early pre-B cells.
B lymphocytes mature from hematopoietic stem cells through a series of developmental stages that are characterized by sequential DNA rearrangements of Ig gene segments, first at the Ig heavy (H) chain and then at the Ig light (L) chain locus (1). Developing B cell precursors must pass several checkpoints, at which they are screened for the presence of functional Ig chains (2). An early checkpoint was identified at the transition from the pro-B to the pre-B cell stage through experiments with transgenic and gene-targeted mice. For example, B cell development is blocked at the pro-B cell stage in mutant mice unable to generate a productive IgH gene (3–5) or to assemble a pre-B cell receptor (pre-BCR) consisting of an Ig μH-chain, surrogate light (SL)-chain, and Igα/β (6–9). Therefore, only those pro-B cells that synthesize a μH-chain able to form a functional pre-BCR with the SL chain (10–13) will pass this early checkpoint and develop into pre-B cells. In addition, pre-BCR signals have been implicated in the transient down-regulation of the recombination activating genes RAG1 and RAG2 (14) and the redirection of the V(D)J-recombinase from the IgH to the IgL locus in early pre-B cells (15).
Further, it has been proposed that pre-BCR signals not only induce differentiation but also proliferation of pre-B cells (2, 16). This hypothesis is supported by the observations that the inability to form a membrane-bound pre-BCR blocks entry of pre-B cells into the large proliferating pre-B-II compartment (16, 17) and that pre-BCR-positive pre-B cells are in cell cycle (18). In addition, transgenic μH chain expression leads to an increase in numbers of B lineage cells in the bone marrow of RAG−/− mice (19, 20). However, the effect of pre-BCR signals on the proliferative capacity of pre-B cells remains controversial, and it has been suggested that the pre-BCR provides rather a signal for survival than for proliferation (21).
To determine the effect of de novo synthesis of a pre-BCR on the proliferation of pre-B cells, we developed an inducible gene expression system for a transgenic μH-chain, which is based on the tetracycline transactivator system developed by Gossen and Bujard (22). Here, we directly demonstrate that de novo synthesis of the pre-BCR induces proliferative expansion of μH-chain-positive pre-B cells.
Materials and Methods
Constructs.
The plasmid pRSP6 containing the rearranged VH-D-JH gene segment from the hybridoma SP6 (23) (GenBank accession no. X56936) and all exons of the constant region of the μH-chain gene was a kind gift of A. Traunecker (Basel, Switzerland). First, a 700-bp XbaI–EcoRI fragment containing the intronic IgH enhancer was deleted. A BclI restriction site 45 bp upstream of the start codon and the EcoRI site downstream of the cμ-exons were used to first isolate the promoter- and enhancerless μ-gene and clone it into the vector pUHD10-3 downstream of the minimal promoter with heptamerized upstream tet operator (tetO)-sequence (22). From the resulting vector tet-μ, a 10.9-kb XhoI fragment containing the tetO-sequence and the Ig sequence was released, and the DNA fragment was electroeluted from an agarose gel and used for injection into fertilized oocytes.
To generate the Ig-tetracycline-controllable transactivator (tTA) vector, an AatII/ClaI fragment containing the IgH-enhancer (700 bp) and a minimal promoter with a synthetic octamer motif were excised from pUCβ-globin (24) and cloned into pUHG15.1 (22) digested with AatII and EcoRI. An AatII–AseI fragment was used for injection of fertilized oocytes.
Mice.
The Ig-tTA and the tet-μ fragments were individually microinjected into fertilized oocytes of (C57BL/6 × DBA/2)F2 mice. Offspring were initially typed by Southern blot hybridization and routinely by PCR. Rag-2−/− mice (4), obtained from Fred Alt (Boston, MA), and λ5−/− mice (7) were bred in individually ventilated cages under pathogen-free conditions. Mice of 6–8 wk of age were used for the experiments. Tetracycline was administered in the drinking water (supplemented with 2% sucrose) at a concentration of 200 μg/ml.
Antibodies and Flow Cytometric Analysis.
The following rat mAbs were purchased from PharMingen: FITC- and allophycocyanin-conjugated RA-6B2 (CD45R, B220), biotinylated 7D4 (anti-mouse CD25, TAC), biotinylated 2B8 (anti-mouse c-kit), and PE-conjugated 1D3 (anti-mouse CD19). The rat monoclonal antibody SL156 (anti-pre-B cell receptor) (18) was conjugated with biotin using standard procedures. FITC-conjugated goat anti-mouse IgM (μH-chain specific) was obtained from Southern Biotechnology Associates (Birmingham, AL). The purified anti-IL-7 receptor antibody (A7R34) (25) and anti-c-kit antibody (ACK-2) (26) were kind gifts of S. Nishikawa (Kumamoto, Japan). The SP6 H-chain specific anti-idiotypic mAb 20.5 was a kind gift of P. Cazenave (Institute Pasteur, Paris).
Single-cell suspensions from bone marrow were prepared by flushing out cells from femurs with either ice-cold staining buffer (PBS containing 2% FCS and 0.1% NaN3) or culture medium. Cells were incubated with a combination of FITC-, PE-, biotin-, or allophycocyanin-conjugated antibodies in staining buffer, washed with staining buffer, incubated for 15 min with PE-streptavidin (Southern Biotechnology Associates) to reveal the biotin reagent, and finally washed with staining buffer. Cells were analyzed on an EPICS XL flow-cytometer (Coulter) by using the WINMDI software (http://facs.scripps.edu).
For the detection of cytoplasmic μH-chain, sorted cells or cells from cultures were fixed with 4% paraformaldehyde, permeabilized with 0.1% Tween-20 in PBS, and stained with FITC-conjugated goat anti-mouse μ antibodies.
Numbers of cultured viable cells were determined by flow cytometry. Cells were resuspended from the wells, mixed with a defined concentration of Flow-Count fluorospheres (Coulter) and 1 μg/ml propidium iodide (PI), and analyzed by flow cytometry. Viable cell counts were calculated from the number of fluorospheres and the number of PI-negative cells falling into the lymphoid gate according to forward- and side-scatter characteristics.
Sorting and Culturing of Pro-B Cells.
Pro-B cells from bone marrow were isolated by using anti-CD19- or anti-B220-coated magnetic beads following the instructions of the supplier (Miltenyi Biotec, Auburn, CA). Reanalysis revealed that this population contains >96% CD19+ and B220+ cells, respectively. RPMI medium 1640 supplemented with 50 μM β-mercaptoethanol, 1 mM glutamine, and 5% FCS was used for all cell culture experiments. Pro-B cells isolated from bone marrow were cultured on γ-irradiated ST2 cells in medium with or without IL-7 (27). Tetracycline hydrochloride was added to the culture medium at a concentration of 50 ng/ml.
Western Blot Analysis.
Cellular extracts of 106 cells were separated by SDS/PAGE under reducing conditions in a 12% polyacrylamide gel. Proteins were transferred to a nylon membrane, and the presence of μH-chain was revealed by a goat anti-μ antiserum followed by horseradish peroxidase-conjugated rabbit anti-goat IgG antibodies and enhanced chemiluminescence (Amersham, Freiburg, Germany).
Results
Generation of Tetracycline-Responsive μH-Chain Transgenic Mice.
To establish a tetracycline regulatory gene expression system for a transgenic μH-chain, we generated two transgenic mouse lines (Fig. 1). The first line contains a transgene that encodes a tetracycline-controllable transactivator (tTA) under the transcriptional control of an Ig-enhancer driven lymphocyte-specific expression cassette (24). Three of six Ig-tTA-transgenic founder lines showed tetracycline-controlled expression of a tetO-controlled luciferase gene in lymphoid organs (not shown). In the absence of tetracycline, line 1.5 showed the highest expression levels of tetO-controlled luciferase in the bone marrow and was used for further breedings. Luciferase expression in the absence of tetracycline was also detected in thymus, spleen, and lymph node and to some extends in skeletal muscle (not shown). In other organs as well as in the presence of tetracycline, luciferase expression was at background levels. To generate the second line (tet-μ), the known intronic IgH enhancer sequences were deleted from a productively rearranged μH-chain gene, and the 5′ Ig-VH-promoter region was replaced with a minimal promoter and the tetracycline operator sequence, tetO. One transgenic tet-μ founder line was obtained. Expression of the transgenic μH-chain derived from the TNP-specific hybridoma SP6 (23) was undetectable in tet-μ transgenic mice with SP6 H-chain-specific anti-idiotypic antibodies and with a sensitive reverse transcription–PCR in bone marrow and spleen (data not shown).
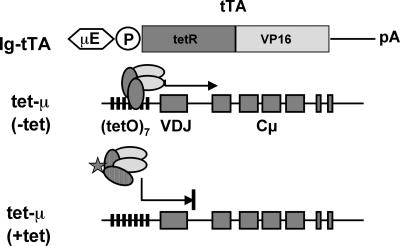
Figure 1.
Schematic representation of the tetracycline-controlled expression of a transgenic μH-chain gene. The tetracycline-controlled transactivator composed of the tetracycline repressor (tetR) and a viral transactivator (VP16) is driven by a minimal promoter containing a synthetic octamer motif and the 700-bp fragment of the intronic IgH enhancer (Ig-tTA construct). In the tet-μ construct, the known intronic IgH enhancer sequence was deleted from a productively rearranged μH-chain gene, and the 5′ Ig-VH-promoter region was replaced with a minimal promoter and the tetracycline resistance operon (tetO).
Expression of Transgene-Encoded μH Chain.
To analyze the expression of the tetO-controlled transgenic μH-chain in the absence of endogenous μH-chain expression, we crossed the transgenic Ig-tTA and tet-μ lines with mutant rag-2−/− mice (4). Rag-2−/− mice do not synthesize endogenous μH-chain because the V(D)J recombination machinery is inactivated. Therefore, B cell development is blocked at the pro-B cell stage in these mice. However, the constitutive expression of a transgenic μH-chain in rag-2−/− mice results in a rescue of the developmental block and allows differentiation of cells from the pro-B to the pre-B stage (19, 20). Single-transgenic rag-2−/− lines were finally intercrossed to obtain double-transgenic Ig-tTA/tet-μ (dTg) mice on a rag-2−/− genetic background. In the absence of tetracycline, the tTA binds to the tetO sequence, which should result in transcriptional activation of the μ-gene in a double-transgenic Ig-tTA/tet-μ mouse. In the presence of tetracycline, tTA fails to bind to tetO and the expression of the μ gene should be terminated (Fig. 1).
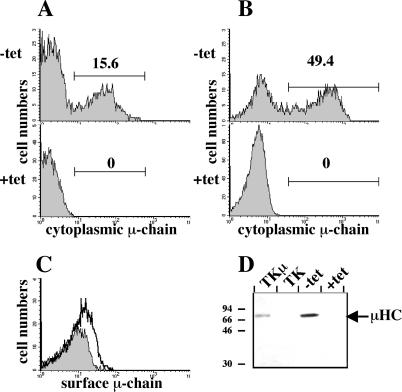
To determine whether in vivo expression of the transgenic μH-chain gene can be controlled by tetracycline, dTg rag-2−/− littermates received drinking water with or without tetracycline for 7 days. Because of the fast turnover of pro-B and pre-B cells in the bone marrow (28) and because of the fast pharmacokinetics of the tetracycline control system (29), we expected a complete down-regulation of the μH-chain after in vivo administration of tetracycline for 7 days. To analyze μH-chain expression in transgenic mice, total bone marrow cells were removed, fixed with formaldehyde, and stained with fluorescein-conjugated antibodies against μH-chain. Flow cytometric analysis revealed that 15% of freshly isolated bone marrow cells from dTg rag-2−/− mice that had not received tetracycline stained with anti-μH-chain antibodies (Fig. 2A). In contrast, μH-chain-positive cells were not detected in the bone marrow of dTg rag-2−/− mice that had received tetracycline in the drinking water (Fig. 2A). To determine whether the expression of the transgenic μH-chain can be induced in in vitro cultures, pro-B cells isolated from the bone marrow of dTg rag-2−/− mice that had received tetracycline for 7 days were cultured for 2 days on stroma cells with IL-7 in the presence or absence of tetracycline. In the absence of tetracycline, transgenic μH-chain was detected by cytoplasmic flow cytometry in about 50% of the cells (Fig. 2B). In contrast, μH-chain-positive cells could not be detected in cultures containing tetracycline (Fig. 2B). The heterogeneous expression of the tetracycline-controlled expression of the μH-chain gene after withdrawal of tetracycline seems to be a common feature of tetracycline-inducible cell lines and transgenic mice. Hypermethylation of the tetracycline responsive promoter or mosaic expression of the tTA gene (30, 31) might be responsible for the observation that μH-chain could not be detected in all transgenic pro-B cells after the removal of tetracycline. Similar results were obtained when we analyzed in vitro cultured CD19-positive bone marrow cells from dTg rag-2−/− mice for the expression of μH-chain on the cell surface. Surface μH-chain could only be detected on cells when cultured in the absence of tetracycline (Fig. 2C). The low expression levels of surface μH-chain are comparable with the low levels of surface μH-chain detected on pre-B cells isolated ex vivo (18, 32). The tetracycline-controlled transgenic μH-chain forms a pre-BCR on the cell surface, because cells can be stained with the monoclonal antibody SL156, which detects μH-chain only when bound to SL-chain (ref. 18, and data not shown). The de novo expression of the μ-chain induced differentiation of the cells in vitro, as c-kit was partially down-regulated and CD25 was up-regulated (not shown). Further, we detected by Western blot analysis a full-length μH-chain in cellular extracts of cultured B lymphoid cells from dTg rag-2−/− mice, but only when the culture medium did not contain tetracycline (Fig. 2D). Because μH-chain could not be detected in cells by flow cytometry and Western blot analysis in the presence of tetracycline, the repression of the transgenic μH-chain by tetracycline appears rather tight in vivo as well as in vitro. In support of this idea, we did not detect a specific signal in sorted bone marrow B lymphoid cells from mice that had received tetracycline in the drinking water with a sensitive μH-chain transgene-specific reverse transcription–PCR assay (not shown). The concentration of tetracycline required to completely repress transgenic, tetO-driven transcription in vitro was 5 ng/ml and is at least 100 times below the dose that shows any measurable toxic effect on cultured B cell precursors (data not shown). We conclude from these results that we have generated a mouse model that allows tight tetracycline-controlled in vivo and in vitro expression of a tetO-controlled μH-chain transgene in normal precursor B cells.
Figure 2.
Analysis of tetracycline-controlled expression of transgenic μH-chain in bone marrow cells of dTg rag-2−/− mice. (A) Flow cytometric analysis of cytoplasmic μH-chain in total bone marrow cells from dTg rag-2−/− mice. Bone marrow cells were isolated from mice that had received for at least 7 days drinking water without and with tetracycline. The percentages of cells falling within each gate are indicated. (B) Flow cytometric analysis of cytoplasmic μH-chain expression in in vitro cultured CD19-positive B cell precursors. CD19-positive cells were isolated from dTg rag-2−/− mice that had received tetracycline for 7 days and cultured for 48 h on stromal cells and IL-7 in the absence or presence of tetracycline. (C) Flow cytometric analyses of expression of μH-chain on the cell surface of in vitro cultured CD19-positive B cell precursors. The histograms obtained by flow cytometry of cells that were cultivated in the presence (shaded histograms) or absence (open histograms) of tetracycline are overlaid. (D) Western blot analysis of μH-chain production in in vitro cultured CD19-positive cells from dTg rag-2−/− mice. TK and TK.μ are Abelson murine leukemia virus-transformed pre-B cell lines that served as negative and positive controls for μH-chain expression, respectively. Molecular masses of standard proteins are indicated in kDa on the left and the position of μH-chain on the right of the blot.
Regulation of B Cell Differentiation by Tetracycline in Vivo.
To determine whether the induction of the tetO-controlled transgenic μH-chain rescues the block at the pro-B cell stage in rag-2−/− mice, we analyzed B cell developmental stages in the bone marrow of nontransgenic, single-transgenic, and dTg rag-2−/− mice by flow cytometry (Fig. 3). As expected, in nontransgenic (Fig. 3, Upper), single transgenic (not shown), and dTg rag-2−/− mice that had received tetracycline in the drinking water (Fig. 3, Middle), virtually all B220+ B lineage cells are pro-B cells, because they coexpress the pro-B cell markers c-kit (26, 33) and CD43 [not shown (34)], but not the pre-B cell-specific marker CD25 (17). In contrast, in the bone marrow of dTg rag-2−/− mice that had not received tetracycline (that is, μH-chain is produced), most of the B220+ cells had a pre-B cell phenotype, as c-kit and CD43 were down-regulated and CD25 was up-regulated (Fig. 3, Lower). In addition, the relative number of B220+ cells was increased about 2-fold in the bone marrow population of dTg rag-2−/− mice when compared with that of nontransgenic rag-2−/− mice or dTg rag-2−/− mice that had received tetracycline. Hence, early B cell development from the pro-B to the pre-B cell stage can be controlled in vivo by tetracycline in our transgenic mouse model.
Figure 3.
Flow cytometric analysis of bone marrow cells from nontransgenic and dTg (Ig-tTA/tet-μ) rag-2−/− mice. Bone marrow cells from 6-wk-old rag-2−/− and dTg rag-2−/− mice that had received drinking water with (200 μg/ml) and without tetracycline were stained with fluorochrome-conjugated antibodies indicated adjacent to the axes. Cells falling into the lymphoid gate were analyzed. The numbers indicate percentages of cells contained within each quadrant. Similar results were obtained in experiments with five rag-2−/− mice and three dTg rag-2−/− mice. The percentage of CD25+ cells among all B220+ cells in dTg rag-2−/− mice that did not receive tetracycline varied between 45 and 81% in four different mice analyzed.
De Novo Expression of Transgenic μH-Chain Induces Proliferation of Pre-B Cells.
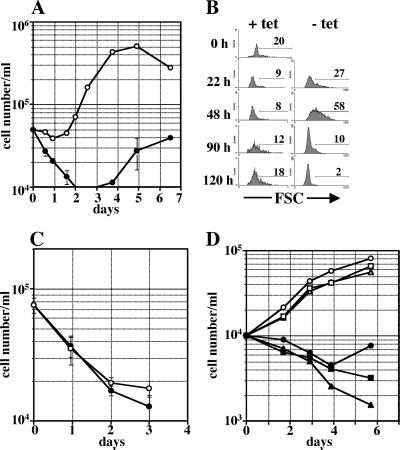
Because the expression of the transgenic μH-chain induces differentiation of pro-B cells into pre-B cells in vivo (Fig. 3), we asked whether de novo synthesis of μH-chain in pro-B cells could also induce proliferation. A rapid, complete withdrawal of tetracycline from the circulation of a mouse, which is essential to induce μH-chain expression, cannot be accomplished in vivo. Removal of tetracycline can, however, be accomplished more rapidly in short-term cultures of freshly isolated pro-B cells. Early B cell progenitors in the bone marrow are in close contact to stromal cells (35), and such stromal cell contact has been shown to be beneficial for survival, proliferation, and differentiation in in vitro culture systems (27, 36, 37). To examine the effect of de novo synthesis of the transgenic μH-chain on the proliferative capacity of B cell progenitors, we, therefore, used a physiologic in vitro system, in which pro-B cells were cultivated directly on stromal cells. CD19-positive pro-B cells were isolated from the bone marrow of dTg rag-2−/− mice that had received tetracycline for at least 7 days in the drinking water. In these pro-B cells, μH-chain was undetectable (see Fig. 2B). The isolated cells were cultivated on a layer of irradiated stromal cells in the presence or absence of tetracycline and numbers of viable cells were determined by flow cytometry over a period of 7 days. In the presence of tetracycline (i.e., μH-chain expression is repressed), numbers of B lymphoid cells gradually decreased in the individual cultures over 4 days, presumably because they were dying by apoptosis (Fig. 4A). However, after 4–5 days of culture, increasing numbers of viable CD19+, μ-negative cells were detected (not shown). The subpopulation growing under these culture conditions on stromal cells may represent very primitive B cell progenitors growing on stromal cells only, as described by Era et al. (37). In the absence of tetracycline, μH-chain expression was detected 2 h after initiating the cultures and reached its maximum after 12 h (not shown). In these cultures, B lymphoid cells expanded after a short lag period and numbers of viable cells increased at least 8-fold over 5 days in culture (Fig. 4A). Thus, de novo synthesis of a tetO-controlled, transgenic μH-chain in normal pro-B cells induces proliferation. As determined by forward light scatter analysis, the expansion of cells coincided with an increase in the number of large cells already 1 day after initiating the cultures (Fig. 4B). After 5 days of culture, most of the living cells were small in size. Thereafter, the numbers of living cells declined rapidly. These results are consistent with the proposed differentiation program of B cell development, in which large pre-BCR-positive pre-B cells undergo a certain number of cell divisions and thereafter spontaneously differentiate into small pre-B cells (2, 38). Proliferation of pro-B cells from nontransgenic rag-2−/− mice was not influenced by tetracycline when cultured with stroma cells and IL-7, indicating that the low concentration of tetracycline in the culture medium had no effect on pro-B cell viability or proliferation. The concentration of tetracycline (of 50 ng/ml) is at least 10 times below the dose that shows any measurable effect on pro-B cell proliferation on stromal cells in the presence of IL-7 (not shown).
Figure 4.
Effect of de novo synthesis of transgenic μH-chain on proliferation of in vitro cultured B cell precursors. CD19+ pro-B cells were isolated from various dTg mice that had received tetracycline (200 μg/ml) in the drinking water for at least 7 days and cultured as indicated in the absence (open symbols) and presence (filled symbols) of tetracycline. Cell numbers were determined by flow cytometry. (A) Pro-B cells from dTg rag-2−/− mice were plated on ST-2 stromal cells and cultivated in the presence (●) or absence (○) of tetracycline. Mean values and standard errors were calculated from triplicates. In five independent experiments an 8- to 14-fold increase in cell numbers was observed 5 days after initiating the cultures in the absence of tetracycline. (B) Forward light scatter characteristics of B cell precursors from dTg rag-2−/− mice cultured on stromal cells in the presence (+ tet) or absence (− tet) of tetracycline. Percentages of large cells are indicated. (C) CD19+ pro-B cells from dTg rag-2−/−/λ5−/− mice were plated on ST-2 stromal cells and cultured in the presence (●) or absence (○) of tetracycline over a period of 3 days. (D) CD19+ pro-B cells from dTg rag-2−/− mice were plated on stromal cells and cultivated with (filled symbols) or without (open symbols) tetracycline in the presence of blocking antibodies specific for c-kit (rectangles, 20 μg/ml) or the IL-7Rα-chain (triangles, 20 μg/ml).
Requirements for μH-Chain Induced Proliferation.
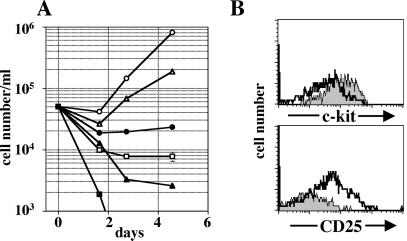
To determine whether the formation of a functional pre-BCR is critical for induction of proliferation of pre-B cells, dTg rag-2−/− mice were crossed with mice containing a targeted deletion of the λ5 gene that encodes one component of the surrogate light chain (7). Pro-B cells were isolated from the bone marrow of dTg rag-2−/−/λ5−/− mice that received tetracycline for 7 days, and the isolated cells were cultivated on a layer of irradiated stromal cells in the presence or absence of tetracycline. Numbers of cultured B lymphoid cells from dTg rag-2−/−/λ5−/− mice declined over 3 days, regardless whether tetracycline was present in the culture medium (Fig. 4C). Thus, the formation of a functional pre-BCR is critical for inducing proliferation of pre-B-cells after de novo synthesis of a μH-chain. To determine which stroma cell signals are critical for the proliferation of pre-B cells after de novo synthesis of the pre-BCR, we isolated pro-B cells from dTg rag-2−/− mice that had received tetracycline and cultured them on stroma cells with and without tetracycline in the presence of blocking monoclonal antibodies against either the receptor tyrosine kinase c-kit or the α-chain of the IL-7 receptor. Signals by both receptors are critical for cell cycle progression of pro-B-cells (27, 39). As shown in Fig. 4D, both antibodies had only a slight inhibitory effect on pre-BCR-induced proliferation of pre-B cells. However, the same concentrations of antibodies inhibited the stromal cell/IL-7-induced proliferation of pro-B cells from nontransgenic rag-2−/− mice by 70% in case of the anti-c-kit antibody and completely in the case of the anti-IL-7R antibody (not shown; ref. 40). The late outgrowth of a small population of CD19+ cells in the presence of tetracycline (see Fig. 4 A and D, ●) was inhibited by either anti-c-kit or anti-IL-7 antibodies (Fig. 4D), indicating that this population of cells might represent early pro-B cells. These results show that the positive effect of de novo synthesis of the transgenic μH-chain on pre-B cell proliferation does not require signals provided by the receptor tyrosine kinase c-kit or the IL-7Rα-chain. Because the induction of proliferation after de novo expression of a μH-chain depends on the capacity to form a pre-BCR, it is possible that a ligand for the pre-BCR on stromal cells is required. We, therefore, used a culture system without stromal cells in flat-bottom culture plates. Under these conditions, cell death was observed in the absence of any other growth factors, regardless of whether tetracycline was present or absent from the medium (Fig. 5A). The early and rapid cell death of ex vivo-isolated dTg pro-B cells could be inhibited by IL-7 in the culture medium. In the presence of tetracycline and high concentrations of IL-7, the number of viable cells remained relatively constant for 5 days in culture, whereas, in the absence of tetracycline, the numbers of the precursor B cells increased significantly (Fig. 5A). Furthermore, differentiation is induced in the absence of tetracycline, as the expression of c-kit is down-regulated and CD25 expression is up-regulated (Fig. 5B). It has recently been proposed that the pre-BCR-signal modulates the responsiveness of pre-B cells to IL-7 (41). We, therefore, cultivated ex vivo isolated dTg pro-B cells in the presence of 1,000-fold lower concentrations of IL-7 (25 pg/ml) in the presence or absence of tetracycline. Under these culture conditions, pro-B cells rapidly died in the presence of tetracycline. In contrast, numbers of pre-B cells increased significantly when tetracycline was omitted from the culture medium (Fig. 5B). Thus, the presence of a putative ligand for the pre-BCR on stroma cells is not necessary in our system to induce proliferation and differentiation after de novo synthesis of the pre-BCR.
Figure 5.
Effect of de novo synthesis of transgenic μH-chain on proliferation of in vitro-cultured B cell precursors in the absence of stroma cells. (A) Pro-B cells from dTg rag-2−/− mice were plated in cell culture medium and cultivated with (filled symbols) or without (open symbols) tetracycline in the absence (squares) or presence of 25 ng/ml (circles) or 25 pg/ml (triangles) IL-7. Cell numbers were determined by flow cytometry. (B) Flow cytometric analyses of expression of c-kit and CD25 on the cell surface of in vitro cultured CD19-positive B cell precursors. The histograms obtained by flow cytometry of cells that were cultivated in the presence (shaded histograms) or absence (open histograms) of tetracycline are overlaid.
Discussion
We describe here a transgenic mouse model, in which the expression of an Ig μ-heavy-chain gene is solely driven by a tetracycline-controlled transactivator. By using this transgenic mouse model, we clearly show that the de novo synthesis of μH-chain in transgenic pro-B cells not only induces differentiation but also proliferation of these cells. We further found that proliferation induced by de novo synthesis of μH-chain in our experiments depends on the presence of the so-called surrogate light chain that, together with the μH-chain, forms a pre-B cell receptor. Therefore, our results directly demonstrate a function of the pre-B cell receptor in inducing the proliferative expansion of pre-B cells producing a functional μH-chain, i.e., one that pairs with SL-chain. It is conceivable that prematurely expressed Igκ-transgenes that have been shown to rescue B cell development in λ5-deficient mice (42, 43) can substitute for the SL-chain receptor in inducing proliferative expansion of pre-B cells producing a functional μH-chain. We further found that the number of small pre-B cells increased in our in vitro culture system after induction of μH-chain. These findings are in good accordance with the earlier proposed four to six cell divisions of μH-chain-positive, large pre-B cells, and the differentiation of these cells into small resting pre-B cells (38). The molecular events implicated in the transition of large cycling pre-B cells into the pool of small, resting pre-B cells are completely unclear. On possible mechanism might be that the down-regulation of SL-chain expression during the proliferative expansion phase terminates the signal for proliferation.
Based on the findings that high concentrations of IL-7 alone promote the in vitro growth of pro-B and pre-BI cells for a limited period (27) and that bone marrow B lymphopoiesis in IL-7−/− mice is blocked at the pro-B cell stage, it was proposed that IL-7 induces proliferative signals in pre-BCR-positive pre-B cells (44). Based on our findings in this manuscript, we favor the idea that IL-7 or other stromal cell-derived factors rather enable than induce proliferative signals in pre-BCR-positive pre-B cells. In support of this idea, proliferation of freshly isolated pro-B cells could be observed on μH-chain induction when either stroma cells or IL-7 were present in our in vitro culture system. Further, low concentrations of IL-7, which might better resemble the in vivo situation, were sufficient to support proliferation of cells in our in vitro culture system, but only when tetracycline was absent (i.e., μH-chain expression was on) from the culture medium. These results confirm and extend recent findings, suggesting that the signals induced by the pre-BCR induces molecular changes in the pre-B cell to become more sensitive for costimulatory signals provided by IL-7 or other factors derived from stromal cells (37, 41). IL-7-independent signals from the stromal cell microenvironment may include stem cell factor (39), SDF-1 (45), CD44 (46) VLA-4 (47), TSLP (48), or a combination of these factors.
Another possible explanation for the positive effect of the pre-BCR on proliferation could be that stroma cells or IL-7 provide the stimulus for proliferation and the pre-BCR signal induces anti-apoptotic signals. This seems unlikely, because proliferation of single pre-BCR-positive pre-B cells from bone marrow of normal mice occurs in plain culture medium, presumably because pre-BCR-positive cells have received the signals from the stroma cell already in vivo (49). The cells that proliferate under these culture conditions might carry a productive V(D)J rearrangement and, thus, initiated entry into cell cycle already in vivo. Their capacity to proliferate might, therefore, be independent from further growth promoting and anti-apoptotic signals.
The fact that stroma cells are required for pre-BCR induced proliferation of pre-B cells in our culture system supports the idea that the interaction between a putative stroma cell ligand and a pre-BCR might be a prerequisite to initiate pre-BCR-mediated signals in pre-B cells (50). Our finding that low concentrations of IL-7 induce proliferation and differentiation of pro-B cells after de novo induction of the pre-BCR in the absence of stroma cells indicates that such an interaction is not required for proliferation. Although there is currently no experimental evidence for a ligand for the pre-BCR, our findings do not rule out that such a ligand exists in vivo on adjacent pre-B cells, the plasma membrane of the same cell, or even on stromal cells.
We show here that a stringent inducible regulation of a transgene can be achieved with the tetracycline regulatory system (22) in B lymphoid cells in vivo and that induction of μH-chain synthesis induces differentiation of pro-B cells and promotes proliferation of pre-B cells. Our inducible expression system will facilitate the analysis of pre-BCR-induced downstream signaling events in normal B-lymphoid precursors and the characterization of molecular events that control allelic exclusion at the IgH-chain locus.
Acknowledgments
We thank Dr. Bujard for the pUHD10–3 and the pUHD15–1 plasmids, Urs Müller for his help in generating transgenic mice, and Drs. Ingrid Haas, Antonius Rolink, and Matthias Wabl for critical reading of our manuscript. This work was supported in part by the Deutsche Forschungsgemeinschaft through SFB466 (to T.H.W. and H.-M. J.), and by the Deutsche Forschungsgemeinschaft through SFB465 (to J.H. and T.W.). The Basel Institute was founded and is supported by Hoffmann La-Roche.
Abbreviations
- H
heavy chain
- L
light chain
- pre-BCR
pre-B cell receptor
- SL
surrogate light
- RAG
recombinant activating gene
- tetO
tet-operator
- tTA
tetracycline-controllable transactivator
- dTg
double transgenic
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041492098.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041492098
References
- 1.Alt F W, Yancoupoulos G D, Blackwell T K, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melchers F, Haasner D, Grawunder U, Kalberer C, Karasuyama H, Winkler T, Rolink A. Annu Rev Immunol. 1994;12:209–225. doi: 10.1146/annurev.iy.12.040194.001233. [DOI] [PubMed] [Google Scholar]
- 3.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 4.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Stall A M, Alt F W. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Trounstine M, Alt F W, Young F, Kurahara C, Loring J F, Huszar D. Int Immunol. 1993;5:647–656. doi: 10.1093/intimm/5.6.647. [DOI] [PubMed] [Google Scholar]
- 6.Kitamura D, Roes J, Kuhn R, Rajewsky K. Nature (London) 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 7.Kitamura D, Kudo A, Schaal S, Muller W, Melchers F, Rajewsky K. Cell. 1992;69:823–831. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 8.Gong S, Nussenzweig M C. Science. 1996;272:411–414. doi: 10.1126/science.272.5260.411. [DOI] [PubMed] [Google Scholar]
- 9.Kline G H, Hartwell L, Beck-Engeser G B, Keyna U, Zaharevitz S, Klinman N R, Jäck H M. J Immunol. 1998;161:1608–1618. [PubMed] [Google Scholar]
- 10.Pillai S, Baltimore D. Nature (London) 1987;329:172–174. doi: 10.1038/329172a0. [DOI] [PubMed] [Google Scholar]
- 11.Karasuyama H, Kudo A, Melchers F. J Exp Med. 1990;172:969–972. doi: 10.1084/jem.172.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsubata T, Reth M. J Exp Med. 1990;172:973–976. doi: 10.1084/jem.172.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimoto N, Kubagawa H, Ohno T, Gartland G L, Stankovic A K, Cooper M D. Proc Natl Acad Sci USA. 1991;88:6284–6288. doi: 10.1073/pnas.88.14.6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grawunder U, Leu T M J, Schatz D G, Werner A, Rolink A G, Melchers F, Winkler T H. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 15.Constantinescu A, Schlissel M S. J Exp Med. 1997;185:609–620. doi: 10.1084/jem.185.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karasuyama H, Rolink A, Shinkai Y, Young F, Alt F W, Melchers F. Cell. 1994;77:133–143. doi: 10.1016/0092-8674(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 17.Rolink A, Grawunder U, Winkler T H, Karasuyama H, Melchers F. Intern Immunol. 1994;6:1257–1264. doi: 10.1093/intimm/6.8.1257. [DOI] [PubMed] [Google Scholar]
- 18.Winkler T H, Rolink A, Melchers F, Karasuyama H. Eur J Immunol. 1995;25:446–450. doi: 10.1002/eji.1830250221. [DOI] [PubMed] [Google Scholar]
- 19.Spanopoulou E, Roman C A, Corcoran L M, Schlissel M S, Silver D P, Nemazee D, Nussenzweig M C, Shinton S A, Hardy R R, Baltimore D. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 20.Young F, Ardman B, Shinkai Y, Lansford R, Blackwell T K, Mendelsohn M, Rolink A, Melchers F, Alt F W. Genes Dev. 1994;8:1043–1057. doi: 10.1101/gad.8.9.1043. [DOI] [PubMed] [Google Scholar]
- 21.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 22.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusconi S, Kohler G. Nature (London) 1985;314:330–338. doi: 10.1038/314330a0. [DOI] [PubMed] [Google Scholar]
- 24.Annweiler A, Muller U, Wirth T. Nucleic Acids Res. 1992;20:1503–1509. doi: 10.1093/nar/20.7.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudo T, Nishikawa S, Ohno N, Akiyama N, Tamakoshi M, Yoshida H, Nishikawa S. Proc Natl Acad Sci USA. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogawa M, Matsuzaki Y, Nishikawa S, Hayashi S I, Kunisada T, Sudo T, Kina T, Nakauchi H, Nishikawa S I. J Exp Med. 1991;174:63–71. doi: 10.1084/jem.174.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolink A, Kudo A, Karasuyama H, Kikuchi Y, Melchers F. EMBO J. 1991;10:327–336. doi: 10.1002/j.1460-2075.1991.tb07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park Y H, Osmond D G. J Exp Med. 1987;165:444–458. doi: 10.1084/jem.165.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furth P A, St-Onge L, Boger H, Gruss P, Gossen M, Kistner A, Bujard H, Hennighausen L. Proc Natl Acad Sci USA. 1994;91:9302–9306. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harkin D P, Bean J M, Miklos D, Song Y H, Truong V B, Englert C, Christians F C, Ellisen L W, Maheswaran S, Oliner J D, Haber D A. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 31.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, Bujard H. Proc Natl Acad Sci USA. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lassoued K, C A, N, Billips L, Kubagawa H, Monteiro R C, Le Bien T W, Cooper M D. Cell. 1993;73:73–86. doi: 10.1016/0092-8674(93)90161-i. [DOI] [PubMed] [Google Scholar]
- 33.Rolink A, Streb M, Nishikawa S I, Melchers F. Eur J Immunol. 1991;21:2609–2612. doi: 10.1002/eji.1830211044. [DOI] [PubMed] [Google Scholar]
- 34.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobsen K, Osmond D G. Eur J Immunol. 1990;20:2395–2404. doi: 10.1002/eji.1830201106. [DOI] [PubMed] [Google Scholar]
- 36.Kincade P W, Lee G, Pietrangeli C E, Hayashi S, Gimble J M. Annu Rev Immunol. 1989;7:111–143. doi: 10.1146/annurev.iy.07.040189.000551. [DOI] [PubMed] [Google Scholar]
- 37.Era T, Ogama D L, Nishikawa S I, Okamoto M, Honjo T, Akaji K, Miyasaki J I, Yamamura K. EMBO J. 1991;10:337–342. doi: 10.1002/j.1460-2075.1991.tb07954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Decker D J, Boyle N E, Koziol J A, Klinman N R. J Immunol. 1991;146:350–361. [PubMed] [Google Scholar]
- 39.Yasunaga M, Wang F, Kunisada T, Nishikawa S. J Exp Med. 1995;182:315–323. doi: 10.1084/jem.182.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winkler T H, Melchers F, Rolink A G. Blood. 1995;85:2045–2051. [PubMed] [Google Scholar]
- 41.Marshall A J, Fleming H E, Wu G E, Paige C J. J Immunol. 1998;161:6038–6045. [PubMed] [Google Scholar]
- 42.Papavasiliou F, Jankovic M, Nussenzweig M C. J Exp Med. 1996;184:2025–2030. doi: 10.1084/jem.184.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelanda R, Schaal S, Torres R M, Rajewsky K. Immunity. 1996;5:229–239. doi: 10.1016/s1074-7613(00)80318-0. [DOI] [PubMed] [Google Scholar]
- 44.von Freeden-Jeffry U, Vieira P, Lucian L A, McNeil T, Burdach S E, Murray R. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagasawa T, Nakajima T, Tachibana K, Iizasa H, Bleul C C, Yoshie O, Matsushima K, Yoshida N, Springer T A, Kishimoto T. Proc Natl Acad Sci USA. 1996;93:14726–14729. doi: 10.1073/pnas.93.25.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyake K, Medina K L, Obo S, Hamaoka T, Kincade O W. J Exp Med. 1990;171:477–488. doi: 10.1084/jem.171.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyake K, Weissman I L, Greenberger J S, Kincade P W. J Exp Med. 1991;173:599–608. doi: 10.1084/jem.173.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin S D, Koelling R M, Friend S L, Isaksen D E, Ziegler S F, Perlmutter R M, Farr A G. J Immunol. 1999;162:677–683. [PubMed] [Google Scholar]
- 49.Rolink A G, Winkler T, Melchers F, Andersson J. J Exp Med. 2000;191:23–32. doi: 10.1084/jem.191.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melchers F, Karasuyama H, Haasner D, Bauer S, Kudo A, Sakaguchi N, Jameson B, Rolink A. Immunol Today. 1993;14:60–68. doi: 10.1016/0167-5699(93)90060-X. [DOI] [PubMed] [Google Scholar]