Abstract
The respiratory chain of Vibrio cholerae contains three bd-type quinol oxygen reductases as well as one cbb3 oxygen reductase. The cbb3 oxygen reductase has been previously isolated and characterized, however the natural mobile electron donor(s) which shuttles electrons between the bc1 complex and the cbb3 oxygen reductase is not known. The most likely candidates are the diheme cytochrome c4 and mono-heme cytochrome c5, which have been previously shown to be present in the periplasm of aerobically grown cultures of V. cholerae. Both cytochromes c4 and c5 from V. cholerae have been cloned and expressed heterologously in E. coli. It is shown that reduced cytochrome c4 is a substrate for the purified cbb3 oxygen reductase and can support steady state oxygen reductase activity of at least 300 e−1/s. In contrast, reduced cytochrome c5 is not a good substrate for the cbb3 oxygen reductase. Surprisingly, the dependence of the oxygen reductase activity on the concentration of cytochrome c4 does not exhibit saturation. Global spectroscopic analysis of the time course of the oxidation of cytochrome c4 indicates that the apparent lack of saturation is due to the strong dependence of KM and Vmax on the concentration of oxidized cytochrome c4. Whether this is an artifact of the in vitro assay or has physiological significance remains unknown. Cyclic voltammetry was used to determine that the midpoint potentials of the two hemes in cytochrome c4 are 240 mV and 340 mV (vs SHE), similar to the electrochemical properties of other c4-type cytochromes. Genomic analysis shows a strong correlation between the presence of a c4-type cytochrome and a cbb3 oxygen reductase within the β- and γ- proteobacterial clades, suggesting that cytochrome c4 is the likely natural electron donor to the cbb3 oxygen reductases within these organisms. These would include the β-proteobacteria Neisseria meningitidis and Neisseria gonnorhoeae, in which the cbb3 oxygen reductases are the only terminal oxidases in their respiratory chains, and the γ- proteobacterium Pseudomonas stutzeri.
Keywords: respiration, cytochrome c4, cytochrome c5, Vibrio cholera
The genome of Vibrio cholerae encodes four respiratory oxygen reductases (1). There are three bd-type oxygen reductases which use ubiquinol as the natural electron donor (2), and one cbb3-type heme-copper oxygen reductase (3), which is presumed to use a cytochrome c as its natural electron donor. The motivation of the current work is to identify the natural electron donor for the cbb3-type heme-copper oxygen reductase.
The cbb3-type oxygen reductases are members of the heme-copper superfamily which includes enzymes that perform oxygen reductase chemistry or nitric oxide reduction chemistry (4–7). The vast majority of currently identified oxygen reductases within the heme-copper superfamily are classified as being in one of three families, the A-, B- and C-families (6, 7). The cbb3-type oxygen reductases (7–11) are members of the C-family and represent over 20% of the sequences of heme-copper oxygen reductases from the currently known genome sequences. In the past decade, cbb3-type oxygen reductases have been purified and characterized from several bacterial species, including Rhodobacter sphaeroides (5, 12–17), Paracoccus denitrificans (18), Rhodobacter capsulatus (19–23), Vibrio cholerae (3, 24), Bradyrhizobium japonicum (25–31), Rhodothermus marinus (32), Pseudomonas stutzeri (33–37), Helicobacter pylori (38, 39), and Sulfurihydrogenibium azorense (10). It has been demonstrated by mass spectrometry that the cbb3-type oxygen reductases have the signature histidine-tyrosine cross linked co-factor within their active site, but that the tyrosine comes from a different transmembrane span than for the A-family and B-family heme-copper oxygen reductases (15, 24).
The physiological role of the cbb3-type oxygen reductases is generally defined by a high affinity for O2, allowing these enzymes to function at lower oxygen concentrations. Therefore these enzymes are often expressed under microaerophilic growth conditions (9). These enzymes are of particular interest because they are present in a number of pathogenic species and are proposed to be important for virulence by facilitating growth under conditions of low oxygen (40). The cbb3-type enzymes are the only oxygen reductases in Helicobacter pylori (41), Neisseria gonorrhoeae (42) and Neisseria meningitidis (43) and are, therefore, potential drug targets for these significant human pathogens.
To understand the physiological roles of the cbb3-type oxygen reductases, it is important to know the natural electron donor(s). In most cases, the natural electron donor is not known. One exception is the cbb3-type oxygen reductase from H. pylori, where the natural electron donor has been shown to be cytochrome c553 (39). When the electron donors are not known, the purified enzymes are usually assayed using the artificial electron donor TMPD (reduced by ascorbate) as the electron donor. In a few instances, reduced horse heart cytochrome c can function as an electron donor (25), though often these assays are also performed in the presence of TMPD.
The proteomes of V. cholerae strains have previously been analyzed for the presence of c-type cytochrome binding domains by searching for the canonical sequence CXXCH (44). A total of 48 CXXCH motifs were identified in 31 proteins, of which 14 have N-terminal signal sequences, indicating these are cytochrome c's and are components of the bacterial envelope. Under aerobic growth conditions, six major c-type cytochromes are visualized by SDS-PAGE analysis using a stain for covalently bound heme (44). Two of these six cytochromes c have been identified as the CcoO and CcoP subunits of the cbb3-type oxygen reductase (44). A third heme-containing band was tentatively identified as the YecK subunit of biotin sulfoxide reductase (44). It is likely that one of the heme-staining bands corresponds to the PetC gene, encoding the cytochrome c1 component of the bc1 complex, but this remains to be shown. The best candidates for the remaining two heme-staining bands in aerobically grown V. cholerae are the cycA and cycB gene products, corresponding to cytochrome c4 and cytochrome c5, respectively. Each of these cytochrome c's is predicted to have a cleaved N-terminal signal sequence and to be located within the periplasm. The predicted molecular weights of the mature cytochromes c4 and c5 (19.8 kDa and 11.5 kDa) also make these reasonable candidates for two of the heme-staining bands from aerobically grown V. cholerae (44). The V. cholerae genome also encodes two additional c-type cytochromes of unknown function that are predicted to be periplasmic (44). Their predicted molecular weights (9.1 kDa and 8.5 kDa) do not match the major heme-staining proteins reported for aerobically grown V. cholerae (44). It is likely, therefore, that cytochrome c4 and/or cytochrome c5, might be an electron donor to the cbb3-type oxygen reductase and responsible for shuttling electrons between the bc1 complex and the heme-copper oxygen reductase. For this reason, the genes encoding the di-heme cytochrome c4 and mono-heme cytochrome c5 were each cloned, expressed heterologously in E. coli and the proteins purified.
It is shown that cytochrome c5 does not function as an electron donor to the purified cbb3-type oxygen reductase from V. cholerae, whereas cytochrome c4 can support oxygen reductase activity at a rate of at least 300 e−1/s at 25°C.
Materials and Methods
Construction of expression plasmids
A heterologous E. coli expression system (45) was used to obtain recombinant cytochromes c4 and c5 from V. cholerae. Following the procedure successfully employed previously for the expression of cytochrome c552 from Thermus thermophilus (46), the DNA fragments encoding the predicted sequence of the mature cytochromes c4 and c5 were fused to a DNA fragment encoding the signal peptide (MKISIYATLAALSLALPAGA) of cytochrome c550 from Thiobacillus versutus (47). The genes of the mature cytochrome c's were obtained by PCR from genomic DNA. The oligonucleotides used were as follows: forward primer 5'-CAAGGCGCCCAGGCCCAAGGTAGTATCGAAG -3' and reverse primer 5'-AAGGATCCCTAGTGTAGGCCACCTAC -3' for cycA; forward primer 5'-CAAGGCGCCCAGGCTCTAACTGAAGCCGATA-3' and reverse primer 5'-AAGGATCCTTACAGGCCTGCGATCATA-3' for cycB. The primers were designed for introducing a 5' NarI site and a 3' BamHI site in order to create a full-length chimeric T. versutus / V. cholerae cycA or cycB gene and cloned into pET17b.
In addition, the 24th residue of cytochrome c5 was changed from Leu, to Gln to enhance the recognition by the E. coli signal peptidase to cleave the new cutting site. Both cytochromes c4 and c5 are predicted to have cleaved signal peptides and to be soluble, periplasmic proteins. The predicted cleavage sites are after Ala36 and Ala23 for cytochromes c4 and c5, respectively. This hybrid construct was then cloned into the commercial expression vector pET17b.
Cell growth and enzyme expression and purification
Conditions of cell growth and enzyme expression were similar to those previous reported (46). The recombinant expression plasmids were utilized in a strain that also contained plasmid pEC86, which expresses maturation genes for cytochrome c in E. coli (ccmABCDEFGH)(27). The two plasmids were co-transformed into E. coli BL21(DE3) competent cells which were then streaked on a Luria-Bertani (LB) agar plate containing 50 μg/ml ampicillin and 30 μg/ml chloramphenicol (27, 46). The cells were grown in 1 L of culture medium in a 2.8 L Fernbach flask to A600 = 0.8–1, and then expression of the cytochrome c was induced with 1 mM IPTG. After 7 hours, the cells were harvested by centrifugation at 7,000 × g for 15 min. Generally, about 10 g of cell paste was collected from each liter of growth media. The cell pellet was a reddish-brown color, indicating the overexpression of cytochrome c.
Protein purification
Each of the recombinant cytochrome c's was purified by column chromatography, using procedures similar to those used for the purification of recombinant cytochrome c552 from T. thermophilus (46). Briefly, the collected cell pellet was homogenized in 25 mM Tris-HCl (pH 7.5), 8 mM MgSO4, DNase I, 0.1% Triton X-100 and protease inhibitor cocktail. The suspended cell mixture was passed through a microfluidizer three times at a pressure of 20,000 psi. The cell debris was spun down at 8000 rpm for 30 min at 4□°C. The supernatant was loaded onto a 5×20 cm CM-52 cellulose column (Whatman) and eluted with a gradient of 0 to 1 M NaCl in 25 mM Tris-HCl (pH 7.5) buffer. Fractions which were reddish-brown were collected and concentrated using concentrators (Amicon) with YM-10 membranes. The concentrated protein was loaded onto a 2.6×100 cm gel filtration column (Sephacryl S-100 High Resolution; GE Healthcare) equilibrated with 100 mM potassium phosphate buffer (pH 7.0) at flow rate 0.4 ml/min. The protein was collected and dialyzed overnight against 25 mM Tris-HCl and then loaded onto the buffer-equilibrated 1.6×20 cm DEAE-5PW column (Toso-HaaS). The column was eluted using a gradient of 0 to 1 M NaCl in 25 mM Tris-HCl (pH 7.5) buffer. The eluted protein was dialyzed and concentrated again as described above and then flash-frozen in liquid nitrogen and stored at −80 °C. The final yield of the purified cytochromes c4 and c5 were about 15 mg and 5 mg per liter of cell culture, respectively.
SDS-PAGE analysis
The purified c-type cytochromes were analyzed using SDS-PAGE. Protein was visualized using Coomassie Blue and heme staining (48) was used to identify proteins containing covalently attached heme c. Pre-cast 15% SDS-PAGE gels from ISC BioExpress were used. After electrophoreses, the gels were then incubated in 15 mL of 6.3 mM 3,3`,5,5`-tetramethylbenzidine (TMBZ from Sigma) and 35 mL of 0.25 M sodium acetate, pH 5.0, for 1 h. The gels were then stained for heme by adding H2O2 to a final concentration of 30 mM.
Spectrophotometric Measurements
Spectra of the isolated cytochromes were acquired with a Shimadzu UV−vis-2101PC spectrophotometer. The enzyme sample (3–17 μM) was examined in 25 mM Tris-HCl buffer at pH 7.5. The enzymes were oxidized with 2 μL of 1 mM Fe(CN)6 and reduced with a small amount of solid dithionite, both obtained from Sigma. Spectra were scanned from 375 nm to 800 nm. The concentrations of the cytochrome c's were estimated from the spectra of the reduced cytochromes using extinction coefficients of 40 mM−1cm−1 at 553 nm for the di-heme cytochrome c4 (49, 50) and 20 mM−1cm−1 at 555 nm for the mono-heme cytochrome c5 (51).
Cyclic voltammetry
Reduction potentials were measured using protein film voltammetry (52, 53). The cytochrome c4 was applied directly to a freshly polished pyrolytic graphite-edge (PGE) electrode surface and then placed into solution in a thermostated all-glass cell encased in a Faraday cage. Since oxygen does not interfere with measurements in the range of 100 mV–500 mV (vs. SHE), measurements were performed aerobically. Analogue-scan cyclic voltammetry was performed using a Bioanalytical Systems (West Lafayette, IN) CV-27 voltammograph with an in-house amplifier, and results were recorded via an in-house program. Data were analyzed using Fourier transformation and Origin 7.5 (OriginLab Corp., Northampton, MA). Solution pH value was controlled by using 10 mM HEPES buffer containing 2 M NaCl at pH 7.0.
Purification of the cbb3-type oxidase from V. cholerae
The cbb3-type oxidase from V. cholerae was purified as previously described (3).
Steady state kinetics using an oxygen electrode
Cytochrome c oxidase activity was measured polarographically at 25 °C using a YSI model 53 oxygen meter. The standard reaction mixture contained 1.8 mL of 50 mM sodium phosphate (pH 6.5), 50mM NaCl, 0.05% dodecylmaltoside, 10 mM sodium ascorbate and the cytochrome c to be tested. The cytochrome c4 and cytochrome c5 concentrations were varied in the range of 1 μM to 100 μM. The O2 consumption reaction was initiated by the addition of the oxidase to a final concentration of 50 nM. The dependence of activity on ionic strength was measured by varying the concentration of NaCl from 10 mM to 150 mM using 20 μM of cytochrome c4 reduced by 10mM sodium ascorbate in 10 mM sodium phosphate (pH 6.5) buffer, 0.05% dodecylmaltoside.
Steady state kinetics by stopped flow spectrophotometry
To prepare the pre-reduced cytochrome c4, the sample was reduced with sodium dithionite and the excess dithionite was removed by gel filtration. The steady state kinetics between the cbb3 oxidase from V. cholerae and the pre-reduced cytochrome c4 was monitored at 25 °C with Applied Photophysics SX-17MV stopped-flow spectrometer as follow. One syringe of the stopped-flow apparatus was filled with 400 nM of the cbb3 oxygen reductase in 50 mM sodium phosphate (pH 6.5), 150 mM NaCl, 0.05 % dodecylmaltoside, while the pre-reduced cytochrome c4 in the same buffer was loaded into the other syringe. The concentration of cytochrome c4 was varied from ~ 1 μM to 60 μM for different measurements. After mixing, the reaction was followed spectrometrically from 400 nm −700 nm for 50 s −1000 s using a photodiode array. Using a least square fitting method (54), the spectrum at each time point was deconvoluted into the component spectra of reduced and oxidized cytochrome c4, which had been collected separately by mixing cytochrome c4 with either sodium dithionite or potassium ferricyanide. The concentrations of the reduced and oxidized cytochrome c4 at each time point were then used to calculate the reaction rate at each point. By using different starting concentrations of cytochrome c4 in different measurements, the reaction rates at various combinations of reduced and oxidized cytochrome c4 concentrations were obtained. The reaction rates were plotted against reduced cytochrome c4 concentrations at a fixed oxidized cytochrome c4 concentration, and the KM and Vmax for each plot were obtained by non-linear least square fitting to the standard Michaelis-Menten kinetics model. All data processing and analyses were performed with Mathematica.
Results
Expression and purification of recombinant cytochrome c4 and cytochrome c5
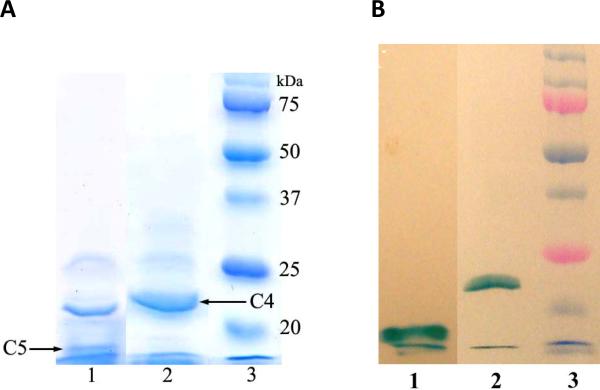
Transformants were found to express the cytochrome c's upon induction (46) and the recombinant proteins were isolated as described in the Methods section. The SDS-PAGE analysis of the recombinant cytochromes is shown in Figure 1. Staining with Coomassie Blue (Figure 1A), the purified cytochrome c4 has a single band near 21~22kDa which contains two covalently bound heme (Figure 1B). The expected molecular weight of the mature cytochrome c4 is 19,791 (187 amino acid residues) with 1,233 (two heme c moiety). Cytochrome c5 has a single heme-staining component running near the expected molecular weight of 11,480, but there is a major protein contaminant that does not contain heme. Since cytochrome c5 does not function as a reductant for the cbb3-type oxidase (see below), no further effort was made to improve the purification.
Figure 1.
SDS-PAGE of the isolated recombinant cytochromes c4 and c5. (A) 15% SDS gel stained with Coomassie blue. (B) Identical gel stained for heme. The lanes from left to right contain the cytochrome c5 (lane 1), cytochrome c4 (lane 2) and molecular weight standards (lane 3).
UV-vis spectroscopy
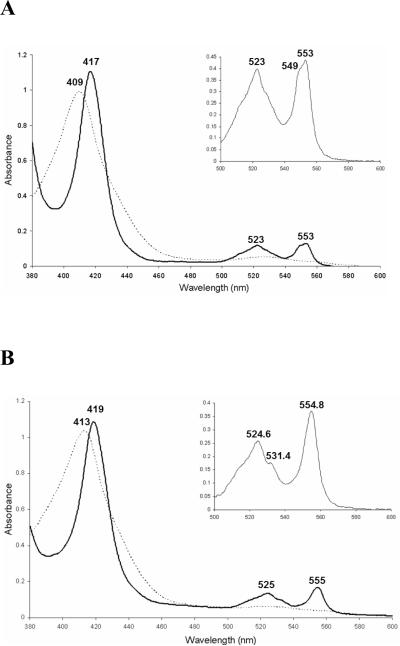
Figure 2 shows the spectra of the reduced and oxidized forms of cytochromes c4 and c5. The spectra of each are consistent with previously published reports of homologues from other organisms (49–51). In the reduced form, the absorption spectrum of cytochrome c4 has three maxima at 553nm, 523nm and 417nm, corresponding to the α-, β- and γ-(Soret) bands, respectively (Figure 2A). The maxima in the oxidized form of cytochrome c4 are at 528nm and 409 nm. The spectra show two diagnostic features shared by all cytochrome c4's: (1) A split α-band with maxima at 553nm and 549nm. (2) A low ratio of the amplitude of the α-band compared to the β-band, .
Figure 2.
Absorption spectra of fully reduced (solid line) and fully oxidased (broken line) V. cholerae (A) cytochrome c4 (3μM) and (B) cytochrome c5 (7μM). The insets show the α- and β-bands of the fully reduced cytochrome c4 (10μM) and cytochrome c5 (17μM). All the absorption spectra were detected under the condition of 25 mM Tris-HCl (pH=7.5), 25 °C.
Reduced cytochrome c5 has maxima at 554.8nm, 524.6nm and 419nm, corresponding to the α-, β- and γ -(Soret) bands, respectively (Figure 2B). The α/β ratio . The spectrum of the oxidized cytochrome c5 has two peaks at 526nm and 413nm. These features are consistent with previous reports of cytochrome c5's (51).
Electrochemistry
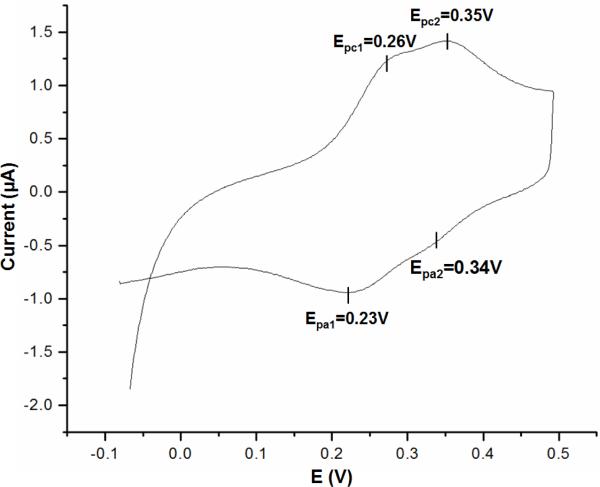
Cytochrome c4 was found to be amenable to direct electrochemical characterization by adhering the protein to a carbon electrode. The midpoint potentials of the two heme components of the recombinant cytochrome c4 were measured by cyclic voltammetry (Figure 3). Four well defined redox peaks are observed and the values of the two heme groups were determined to be approximately 240 mV and 340 mV (vs SHE).
Figure 3.
Cyclic voltammetry of the recombinant cytochrome c4 which was applied to a freshly polished pyrolytic PGE electrode. The solution contained 10 mM HEPES buffer and 2 M NaCl at pH 7.0. Values obtained (vs. SHE) were Epc1 = 0.26 V, Epc2 = 0.35 V, Epa1 = 0.23 V, Epa2 = 0.34 V and E1/2 = (Epc + Epa)/2 = 0.24 V and 0.34 V.
Rates of oxygen reduction by the V. cholerae cbb3-type oxidase using cytochromes c4 and c5
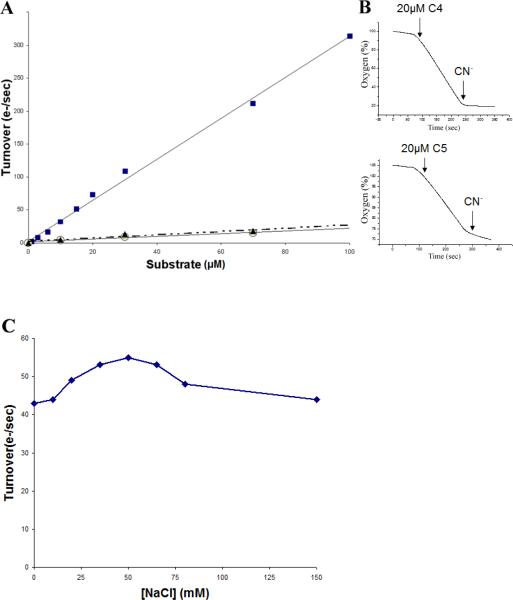
Figure 4A compares the rates of oxygen reduction, measured with an oxygen electrode, by the V. cholerae cbb3-type oxidase using ascorbate plus either i) horse heart cytochrome c, ii) recombinant cytochrome c4 or iii) recombinant cytochrome c5. These assays were performed in the absence of the mediator TMPD, which can itself function as a substrate for the cbb3-type oxidase. The data (Figure 4A) show that neither the horse heart cytochrome c nor cytochrome c5 is effective as a substrate, whereas cytochrome c4 is capable of supporting turnover at rates over 300 e−1/s at 25°C. This turnover is comparable to what is observed with 0.5 mM TMPD in the presence of ascorbate to maintain the TMPD reduced (not shown). The turnover using recombinant cytochrome c5 is more than a factor of 10 less (7%) than that observed with cytochrome c4, and horse heart cytochrome c is also a poor substrate (8%). Although the purity of recombinamt cytochrome c5 was not high, it is unlikely that the contaminants are inhibitory. The oxidase activity with cytochrome c4 is rapidly stopped upon the addition of 25 μM cyanide (Figure 4B), showing that the oxidase activity is catalyzed by the cbb3-type oxygen reductase. The data in Figure 4A do not indicate saturation even at concentrations of cytochrome c4 as high as 100 μM. Activity was compared using 50 nM of the oxidase and 20μM cytochrome c4, varying the concentration of NaCl from 1 mM to 150 mM (Figure 4C). These data indicate that the apparent binding of cytochrome c4 to the oxidase is not sensitive to ionic strength and that the lack of saturation of activity as a function of the concentration of cytochrome c4 is not due to weak binding due to the selected ionic strength. It was determined, however, that under the conditions used for the assays with the oxygen electrode, the ascorbate does not maintain the cytochrome c4 fully reduced.
Figure 4.
Steady state oxidase activity of the cbb3 oxygen reductase from V. cholerae. (A) Dependence on the concentration of cytochrome c4 (solid squares), cytochrome c5 (open circles) and horse heart cytochrome c (solid triangles). The reaction mixture contained 50 mM sodium phosphate (pH 6.5), 50mM NaCl, 0.05% dodecylmaltoside, 10 mM sodium ascorbate, and the indicated amount of the cytochrome c, in a total volume of 1.8 ml. The O2 consumption reaction was initiated by the addition of 50 nM of the oxidase. (B) Sensitivity of the oxidase activity to the addition of 25 μM cyanide. (C) Dependence of the oxidase activity on the concentration NaCl, measured with50 nM oxidase and 20 μM cytochrome c4, in the presence of 10mM sodium ascorbate, 10 mM sodium phosphate (pH 6.5) buffer and 0.05% dodecylmaltoside.
Stopped flow spectrophotometry
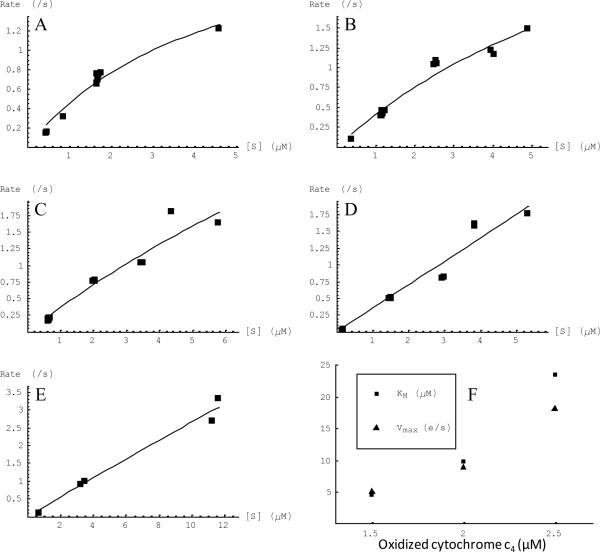
To better understand the lack of saturation in the steady state kinetics measurements using the oxygen electrode, steady state activity was also measured by stopped flow spectrophotometry, mixing pre-reduced cytochrome c4 with a solution containing the cbb3-type oxidase. The oxidation of cytochrome c4 was monitored by recording the change of the absorption spectrum from 400 nm to 700 nm using a photodiode array. The initial concentration of cytochrome c4 was varied from 1 to 60 μM and spectra were obtained as a function of time for each initial condition. The spectra were deconvoluted to quantify the concentration of oxidized and reduced cytochrome c4 in solution as a function of time. From this large matrix of data, the rate of oxidation of cytochrome c4 could be determined for a wide range of concentrations of both the reduced and oxidized forms of cytochrome c4. These data were plotted to show how the rate of oxidation varies as a function of reduced cytochrome c4 in the presence of a specific concentration of oxidized cytochrome c4 (Figure 5A–E). If the oxidized cytochrome c4 is a competitive inhibitor for the reduced cytochrome c4, one would expect that the data would show a constant Vmax and increasing KM as the concentration of oxidized cytochrome c4 increases. However, this is not the case, and both Vmax and KM increase as the concentration of oxidized cytochrome c4 increases (Figure 5F). One complication is that the spectral deconvolution with the least square method is not satisfactory in the case of cytochrome c4 because cytochrome c4 contains two hemes which have slightly different spectra. The assumption that either both are oxidized or both are reduced is not realistic. Attempts were not made to analyze the data with more complex models. Despite this, the current analysis indicates the following significant points.
-
1)
Oxidized cytochrome c4 has a strong effect on the activity between the cbb3-type oxidase and reduced cytochrome c4 The apparent KM increases in the presence of relatively small amounts of oxidized cytochrome c4 (Figure 5F). If the concentration of oxidized cytochrome c4 is greater than 2.5 μM, the KM of reduced cytochrome c4 is too large to obtain a reasonably accurate fitting to the Michaelis-Menten model.
-
2)
During the manipulation of the pre-reduced cytochrome c4 to load the stopped flow syringe, a significant amount of autoxidation occurs. Hence, none of the data represent a starting condition in which cytochrome c4 is actually fully reduced.
-
3)
Because of the spectroscopic complications of having two different heme components in cytochrome c4, data fits are not sufficient to extrapolate to obtain true values for either Vmax or KM. Phenomenologically, as the concentration of oxidized cytochrome c4 increases, both the apparent Vmax and KM increase. The inhibitory influence of oxidized cytochrome c4 and the increasing concentration of oxidized cytochrome c4 that accumulates as the reaction proceeds, explains the lack of saturation under conditions used for the oxygen electrode assays (Figure 4A).
Figure 5.
The rates of oxidation of reduced cytochrome c4 by the cbb3 oxygen reductase from V. cholerae at different oxidized cytochrome c4 concentrations determined by stopped-flow spectroscopy and least square fitting. The substrate [S] refers to reduced (not total) cytochrome c4. The oxidized cytochrome c4 concentrations were (A)1.5 μM; (B) 2.0 μM, (C) 2.5 μM; (D) 3.0 μM and (E) 7.5 μM. The curves show the non-linear least square fitting of the points to Michaelis-Menten kinetics. The KM and Vmax values for 1.5 μM, 2.0 μM and 2.5 μM oxidized cytochrome c4 are plotted in (F).
Discussion
Although initially thought to be confined to proteobacteria (7, 11), the cbb3 oxygen reductases are widely distributed among bacterial phyla (5, 10). Although there are exceptions, most archaea lack c-type cytochromes (55), and no example of a cbb3-type oxidase in an archaea has been reported. Nearly all studies have focused on the role of the C-family (cbb3) oxygen reductases in the aerobic respiratory chains of proteobacteria(8), exceptions being the cbb3-type oxidases from Rhodothermus marinus (yet to be confirmed in the genome sequence) (32) and from Sulfurihydrogenibium azorense (10).
In most cases, the natural electron donors for these C-family oxygen reductases are not known. The current work has identified the di-heme cytochrome c4 as a natural electron donor to the cbb3-type oxygen reductase from the γ-proteobacterium, V. cholerae. Cytochrome c4 is one of five cytochrome c's encoded in the genome of V. cholerae that are predicted to be soluble, periplasmic proteins (44). Cytochromes c4 and c5 appear to be among the major cytochrome c's expressed under aerobic growth conditions (44). Heterologous expression in E. coli was successful, generating soluble cytochromes c4 and c5. Cytochrome c4 from V. cholerae has spectroscopic and electrochemical properties similar to the cytochrome c4's from other organisms. These include the high midpoint potentials (240 mV and 340 mV, vs SHE) of the two hemes, the split α-band and the low ratio of the peak heights of the α- and β-bands (50, 56, 57).
Cytochrome c4 can clearly donate electrons to the cbb3-type oxidase, supporting steady state reduction of O2 at a rate of at least 300 e−1/s. Rates attained using either cytochrome c5 or horse heart cytochrome c were a 10- to 20-fold less under comparable conditions. The one caveat is that the steady state rate of cytochrome c4 oxidase activity does not saturate and quite high concentrations (100 μM) are required to reach the turnover of 300 s−1. The lack of saturation (Figure 4A) is unusual and not expected. By comparison, the steady state turnover of H. pylori cytochrome c553 by the H. pylori cbb3-type oxidase is 250 s−1 with a Km of 0.9 μM. The nearly linear dependence of oxygen reductase activity as a function of the concentration of cytochrome c4 (Figure 4A) can be qualitatively explained being due to the influence of oxidized cytochrome c4 (39). There is a significant concentration of oxidized cytochrome c4 present at all times during the oxygen reductase assay and the presence of ascorbate even as high as 100 mM does not alleviate this problem. The data do show that the true Vmax for the oxidation of cytochrome c4 must be at least 300 s−1, but the true Km cannot be measured using this assay. In the steady state assay, the velocity is measured in the presence of 100 μM reduced cytochrome c4 with about 25 μM oxidized c4 also being present. The stopped flow experiments cannot be performed under these conditions, but the data show that the values of both Vmax and KM depend on the concentration of oxidized cytochrome c4, and the stopped-flow data are consistent with the steady state kinetics. It is quite conceivable that the true Km is in the range of 1 to 10 μM, but further work will be needed to determine this.
Since the oxidized and reduced forms of cytochrome c often have similar affinities for their redox partners, it is to be expected that oxidized cytochrome c4 would competitively inhibit the activity between reduced cytochrome c4 and the cbb3-type oxidase. However, product inhibition can only explain the increase in the KM but not the increase in Vmax that is observed as the concentration of the product increases (Figure 5). Whether this kinetic behavior reflects an in vitro artifact or an unusual form of allosteric regulation is not clear. This unusual behavior might be related to the fact that cytochrome c4 is a two-electron carrier. The cyclic voltammetry shows that the di-heme cytochrome c4 exhibits two substantially different midpoint potentials, but the extent of cooperativity between the two hemes is not known. In principle, there could be two different binding sites on cytochrome c4 for the cbb3 oxygen reductase, each with a distinct KM and Vmax. In any event, further characterization of the oxidation kinetics of the diheme cytochrome c4 is needed to address this question.
At this point, it is safe to conclude that cytochrome c4 is definitely a substrate for the cbb3-type oxidase whereas cytochrome c5 is a very poor substrate, at best, under in vitro conditions.
The roles of cytochrome c4 in other bacteria
Analysis of sequenced genomes demonstrates that the distribution of cytochrome c4 is limited, with the majority of homologs being found within the β- and γ- proteobacterial clades. Cytochrome c4 is also sporadically distributed within the α- proteobacterial clade as well as a few other bacterial phyla. Bacteria within the β- and γ-proteobacterial clades which contain a cbb3 oxygen reductase are very likely to also contain a cytochrome c4. Within these groups of bacteria, it is reasonable to assume that cytochrome c4 is an electron donor to the cbb3 oxygen reductase. It is noted that the cbb3 oxygen reductases within the β- and γ- proteobacterial clades all contain the auxiliary subunit CcoP, a di-heme cytochrome c which is the likely electron acceptor from cytochrome c4. A large number of the cbb3 oxygen reductases outside the β- and γ- proteobacterial clades are missing the CcoP subunit and others have an alternative subunit, CcoR (10). Indeed, many organisms that are outside the β- and γ-proteobacterial clades which encode a cbb3 oxygen reductase, do not contain cytochrome c4 and, therefore, must be using a different electron donor to this enzyme.
Other γ-proteobacteria which contain cbb3 oxygen reductases include Azotobacter vinlandii (58), and Pseudomonas stutzeri (33–37). Knock-out mutants of both cytochromes c4 and c5 in A. vinlandii support a role for both of these cytochromes in aerobic respiration, though there are no data concerning a specific role with the cbb3 oxygen reductase. The cytochrome c4 from P. stutzeri has been extensively characterized (49, 59–62), but its physiological role has not yet been defined.
Acidothiobacillus ferrooxidans is a γ-proteobacterium whose genome includes several cytochrome c4's but which does not contain a cbb3-type oxidase (63–65). The cytochrome c4's are strongly implicated in the aerobic respiratory chain (66–68). A. ferrooxidans is an obligate chemolithotropic bacterium which grows at acidic pH (pH 1.5 to 4) and uses either ferrous iron or reduced sulfur compounds as an electron source. The cyc1 gene encodes a di-heme c4-type cytochrome c552 within an operon which includes an A-family oxygen reductase as well as the blue copper protein rusticyanin (69). These respiratory components comprise the electron transport chain between the ferrous iron oxidase and O2. The cytochrome c4 forms a complex with rusticyanin which is likely the immediate electron donor to the A-family oxygen reductase (68, 70). The structure of the Cyc1 cytochrome c4 has been determined and the complex with rusticyanin has been modeled computationally (64, 70–72). Two additional genes encoding cytochrome c4's, cycA1 and cycA2, are in operons encoding two separate bc1 complexes (66). It is suggested that when growing on reduced sulfur compounds, CycA2 shuttles electrons from the bc1 complex encoded by the petII operon to one or several terminal reductases (66). When growing on Fe+2, CycA1 is proposed to be part of an aerobic respiratory chain which runs in the reverse direction to generate NADH: CycA→bc1 (petI)→Q→Complex I (66, 67).
The β-proteobacterial clade includes Neissseria meningitidis and Neisseria gonorrhoeae, which are each human pathogens and are notable also for the fact that the cbb3 oxygen reductase is the only respiratory oxidase encoded in their genomes (42, 43). Knock-out mutations have implicated cytochrome c4 along with cytochrome cx (a homologue of cytochrome c552 from T. thermophilus) as being an electron donor to the cbb3 oxygen reductase in N. meningitidis (73). Cytochrome c5 is required for electron transfer to the nitrite reductase in N. meningitidis (73). Knock-out mutations have also indicated a role of cytochrome c4 along with cytochrome c5 as substrates of the cbb3 oxygen reductase in N. gonorrhoeae, but not cytochrome c2 (42). Interestingly, the CcoP subunit of the cbb3-type oxidase in N. gonorrhoeae has an extra cytochrome c-domain which is homologous to cytochrome c5, and this domain is implicated in electron transfer to the nitrite reductase which is tethered to the outer membrane (74).
Outside the β- and γ- proteobacterial clades, there is limited information about the native electron donors for the cbb3 oxygen reductase. One exception is H. pylori, in which the monoheme cytochrome c553 (39) is the substrate for the cbb3 oxygen reductase (38). H. pylori is an ε-proteobacterium. Closely related ε-proteobacteria which contain cbb3 oxygen reductases and which might also use homologues of cytochrome c553 as substrates are Campylobacter jejuni (41, 75) and Wolinella succinogenes (76, 77).
The cbb3 oxygen reductases have been biochemically characterized from several α-proteobacteria: Rhodobacter sphaeroides (5, 12–17), Paracoccus denitrificans (18), Rhodobacter capsulatus (19–23) and Bradyrhizobium japonicum (25–28, 29, 30, 31), Rhodothermus marinus (32). Using knock-out mutants, it was concluded that both the A- and C-family oxygen reductases from R. sphaeroides can utilize either cytochrome c2 or membrane-anchored cytochrome cy (21).
Conclusion
Cytochrome c4 has been shown to be the native electron donor for the C-family (cbb3) oxygen reductase in V. cholerae. Within the γ- and β-protobacterial clades, there is a strong correlation for the co-existence of a C-family oxygen reductase and a cytochrome c4, suggesting that in these organisms, it is likely that cytochrome c4 is a natural electron donor. This does not exclude other electron donors from also being substrates within these organisms. Outside the γ-and β-protobacterial clades there is clear evidence that other cytochrome c's are substrates for C-family oxygen reductase, though in most cases, there are no data to identify these electron donors.
Acknowledgements
We thank Dr. SangMoon Lhee and Dr. Antony R. Crofts of Department of Biochemistry, University of Illinois at Urbana-Champaign for technical assistance.
This work was supported by grants from the National Institutes of Health (HL16101) to R. B. G.
Abbreviations
The abbreviations used are
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PEG
pyrolytic graphite-edge
- TMPD
N,N,N',N'-tetramethyl-p-phenylenediamine
- TMBZ
3,3`,5,5`-tetramethylbenzidine
References
- (1).Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yang K, Borisov VB, Konstantinov AA, Gennis RB. The fully oxidized form of the cytochrome bd quinol oxidase from E. coli does not participate in the catalytic cycle: direct evidence from rapid kinetics studies. FEBS Lett. 2008;582:3705–3709. doi: 10.1016/j.febslet.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hemp J, Christian C, Barquera B, Gennis RB, Martinez TJ. Helix Switching of a Key Active-Site Residue in the Cytochrome cbb3 Oxidases. Biochemistry. 2005;44:10766–10775. doi: 10.1021/bi050464f. [DOI] [PubMed] [Google Scholar]
- (4).Hemp J, Gennis RB. Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling. Results Probl Cell Differ. 2008;45:1–31. doi: 10.1007/400_2007_046. [DOI] [PubMed] [Google Scholar]
- (5).Hemp J, Han H, Roh JH, Kaplan S, Martinez TJ, Gennis RB. Comparative genomics and site-directed mutagenesis support the existence of only one input channel for protons in the C-family (cbb3 oxidase) of heme-copper oxygen reductases. Biochemistry. 2007;46:9963–9972. doi: 10.1021/bi700659y. [DOI] [PubMed] [Google Scholar]
- (6).Pereira MM, Sousa FL, Verissimo AF, Teixeira M. Looking for the minimum common denominator in haem-copper oxygen reductases: Towards a unified catalytic mechanism. Biochim Biophys Acta. 2008;1777:929–934. doi: 10.1016/j.bbabio.2008.05.441. [DOI] [PubMed] [Google Scholar]
- (7).Pereira MM, Santana M, Teixeira M. A Novel Scenario for the Evoluation of Haem-copper Oxygen Reductases. Biochim. Biophys. Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- (8).Pitcher RS, Watmough NJ. The bacterial cytochrome cbb3 oxidases. Biochim Biophys Acta. 2004;1655:388–399. doi: 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- (9).Pitcher RS, Brittain T, Watmough NJ. Cytochrome cbb(3) oxidase and bacterial microaerobic metabolism. Biochem Soc Trans. 2002;30:653–658. doi: 10.1042/bst0300653. [DOI] [PubMed] [Google Scholar]
- (10).Ducluzeau AL, Ouchane S, Nitschke W. The cbb3 oxidases are an ancient innovation of the domain bacteria. Mol Biol Evol. 2008;25:1158–1166. doi: 10.1093/molbev/msn062. [DOI] [PubMed] [Google Scholar]
- (11).Cosseau C, Batut J. Genomics of the ccoNOQP-encoded cbb3 oxidase complex in bacteria. Arch Microbiol. 2004;181:89–96. doi: 10.1007/s00203-003-0641-5. [DOI] [PubMed] [Google Scholar]
- (12).Toledo-Cuevas M, Barquera B, Gennis RB, Wikström M, García-Horsman JA. The cbb3-type Cytochrome c Oxidase from Rhodobacter sphaeroides, a Proton-pumping Heme-copper Oxidase. Biochim. Biophys. Acta. 1998;1365:421–434. doi: 10.1016/s0005-2728(98)00095-4. [DOI] [PubMed] [Google Scholar]
- (13).Garcia-Horsman JA, Berry E, Shapleigh JP, Alben JO, Gennis RB. A Novel Cytochrome c Oxidase from Rhodobacter sphaeroides that Lacks CuA. Biochemistry. 1994;33:3113–3119. doi: 10.1021/bi00176a046. [DOI] [PubMed] [Google Scholar]
- (14).Sharma V, Puustinen A, Wikstrom M, Laakkonen L. Sequence analysis of the cbb3 oxidases and an atomic model for the Rhodobacter sphaeroides enzyme. Biochemistry. 2006;45:5754–5765. doi: 10.1021/bi060169a. [DOI] [PubMed] [Google Scholar]
- (15).Rauhamaki V, Baumann M, Soliymani R, Puustinen A, Wikstrom M. Identification of a histidine-tyrosine cross-link in the active site of the cbb3-type cytochrome c oxidase from Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 2006;103:16135–16140. doi: 10.1073/pnas.0606254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sharma V, Wikstrom M, Laakkonen L. Modeling the active-site structure of the cbb3-type oxidase from Rhodobacter sphaeroides. Biochemistry. 2008;47:4221–4227. doi: 10.1021/bi702088r. [DOI] [PubMed] [Google Scholar]
- (17).Rauhamaki V, Bloch DA, Verkhovsky MI, Wikstrom M. Active site of cytochrome cbb3. J Biol Chem. 2009;284:11301–11308. doi: 10.1074/jbc.M808839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).de Gier J-WL, Schepper M, Reijnders WNM, van dyck SJ, Slotboom DJ, Warne A, Saraste M, Krab K, Finel M, Stouthamer AH, van Spanning RJM, van der Oost J. Structural and Functional Analysis of aa3-type and cbb3-type Cytochrome c Oxidases of Paracoccus denitrificans Reveals Significant Differences in Proton-pump Design. Mole. Microbiol. 1996;20:1247–1260. doi: 10.1111/j.1365-2958.1996.tb02644.x. [DOI] [PubMed] [Google Scholar]
- (19).Peters A, Kulajta C, Pawlik G, Daldal F, Koch H-G. Stability of the cbb3-Type Cytochrome Oxidase Requires Specific CcoQ-CcoP Interactions. J. Bacteriol. 2008;190:5576–5586. doi: 10.1128/JB.00534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kulajta C, Thumfart JO, Haid S, Daldal F, Koch HG. Multi-step Assembly Pathway of the cbb(3)-type Cytochrome c Oxidase Complex. J Mol Biol. 2006;355:989–1004. doi: 10.1016/j.jmb.2005.11.039. [DOI] [PubMed] [Google Scholar]
- (21).Daldal F, Mandaci S, Winterstein C, Myllykallio H, Duyck K, Zannoni D. Mobile Cytochrome c2 and Membrane-Anchored Cytochrome cy Are Both Efficient Electron Donors to the cbb3- and aa3-Type Cytochrome c Oxidases During Respiratory Growth of Rhodobacter sphaeroides. J. Bacteriology. 2001;183:2013–2024. doi: 10.1128/JB.183.6.2013-2024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Koch HG, Winterstein C, Saribas AS, Alben JO, Daldal F. Roles of the ccoGHIS Gene Products in the Biogenesis of the ccb(3)-Type Cytochrome c Oxidase. J. Mol. Biol. 2000;297:49–65. doi: 10.1006/jmbi.2000.3555. [DOI] [PubMed] [Google Scholar]
- (23).Gray KA, Grooms M, Myllykallio H, Moomaw C, Slaughter C, Daldal F. Rhodobacter capsulatus Contains a Novel cb-type Cytochrome c Oxidase Without a CuA Center. Biochemistry. 1994;33:3120–3127. doi: 10.1021/bi00176a047. [DOI] [PubMed] [Google Scholar]
- (24).Hemp J, Robinson DE, Ganesan KB, Martinez TJ, Kelleher NL, Gennis RB. Evolutionary Migration of a Post-Translationally Modified Active-Site Residue in the Proton-Pumping Heme-Copper Oxygen Reductases. Biochemistry. 2006;45:15405–15410. doi: 10.1021/bi062026u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Arslan E, Kannt A, Thony-Meyer L, Hennecke H. The symbiotically essential cbb(3)-type oxidase of Bradyrhizobium japonicum is a proton pump. FEBS Lett. 2000;470:7–10. doi: 10.1016/s0014-5793(00)01277-1. [DOI] [PubMed] [Google Scholar]
- (26).Zufferey R, Arsian E, Thöny-Meyer L, Hennecke H. How Replacements of the 12 Conserved Histidines of Subunit I Affect Assembly, Cofactor Binding, and Enzymatic Activity of the Bradyrhizobiuim japonicum cbb3-type Oxidases. J. Biological Chemistry. 1998;273:6452–6459. doi: 10.1074/jbc.273.11.6452. [DOI] [PubMed] [Google Scholar]
- (27).Arslan E, Schulz H, Zufferey R, Kunzler P, Thony-Meyer L. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem Biophys Res Commun. 1998;251:744–7. doi: 10.1006/bbrc.1998.9549. [DOI] [PubMed] [Google Scholar]
- (28).Zufferey R, Presig O, Hennecke H, Thöny-Meyer L. Assembly and Function of the Cytochrome cbb3 Oxidase Subunits in Bradyrhizobium japonicum. J. Biol. Chem. 1996;271:9114–9119. doi: 10.1074/jbc.271.15.9114. [DOI] [PubMed] [Google Scholar]
- (29).Preisig O, Zufferey R, Thöny-Meyer L, Appleby CA, Hennecke H. A High-Affinity cbb3-Type Cytochrome Oxidase Terminates the Symbiosis-Specific Respiratory Chain of Bradyrhizobium japonicum. J. Bact. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Todorovic S, Verissimo A, Wisitruangsakul N, Zebger I, Hildebrandt P, Pereira MM, Teixeira M, Murgida DH. SERR-spectroelectrochemical study of a cbb3 oxygen reductase in a biomimetic construct. J Phys Chem B. 2008;112:16952–16959. doi: 10.1021/jp807862m. [DOI] [PubMed] [Google Scholar]
- (31).Verissimo AF, Sousa FL, Baptista AM, Teixeira M, Pereira MM. Thermodynamic redox behavior of the heme centers of cbb3 heme-copper oxygen reductase from Bradyrhizobium japonicum. Biochemistry. 2007;46:13245–13253. doi: 10.1021/bi700733g. [DOI] [PubMed] [Google Scholar]
- (32).Pereira MM, Carita JN, Anglin R, Saraste M, Teixeira M. Heme Centers of Rhodothermus marinus Respiratory Chain. Characterization of Its cbb3 Oxidase. J. Bioenerg. Biomemb. 2000;32:143–152. doi: 10.1023/a:1005555829301. [DOI] [PubMed] [Google Scholar]
- (33).Pitcher RS, Brittain T, Watmough NJ. Complex interactions of carbon monoxide with reduced cytochrome cbb3 oxidase from Pseudomonas stutzeri. Biochemistry. 2003;42:11263–11271. doi: 10.1021/bi0343469. [DOI] [PubMed] [Google Scholar]
- (34).Pitcher RS, Cheesman MR, Watmough NJ. Molecular and spectroscopic analysis of the cytochrome cbb(3) oxidase from Pseudomonas stutzeri. J Biol Chem. 2002;277:31474–31483. doi: 10.1074/jbc.M204103200. [DOI] [PubMed] [Google Scholar]
- (35).Stavrakis S, Koutsoupakis K, Pinakoulaki E, Urbani A, Saraste M, Varotsis C. Decay of the transient Cu(B)-CO complex is accompanied by formation of the heme Fe-CO complex of cytochrome cbb(3)-CO at ambient temperature: evidence from time-resolved Fourier transform infrared spectroscopy. J Am Chem Soc. 2002;124:3814–3815. doi: 10.1021/ja0169825. [DOI] [PubMed] [Google Scholar]
- (36).Forte E, Urbani A, Saraste M, Sarti P, Brunori M, Giuffre A. The cytochrome cbb3 from Pseudomonas stutzeri displays nitric oxide reductase activity. Eur J Biochem. 2001;268:6486–6491. doi: 10.1046/j.0014-2956.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- (37).Urbani A, Gemeinhardt S, Warne A, Saraste M. Properties of the detergent solubilised cytochrome c oxidase (cytochrome cbb(3)) purified from Pseudomonas stutzeri. FEBS Lett. 2001;508:29–35. doi: 10.1016/s0014-5793(01)03006-x. [DOI] [PubMed] [Google Scholar]
- (38).Tsukita S, Koyanagi S, Nagata K, Koizuka H, Akashi H, Shimoyama T, Tamura T, Sone N. Characterization of a cb-type cytochrome c oxidase from Helicobacter pylori. J Biochem (Tokyo) 1999;125:194–201. doi: 10.1093/oxfordjournals.jbchem.a022259. [DOI] [PubMed] [Google Scholar]
- (39).Koyanagi S, Nagata K, Tamura T, Tsukita S, Sone N. Purification and characterization of cytochrome c-553 from Helicobacter pylori. J Biochem. 2000;128:371–375. doi: 10.1093/oxfordjournals.jbchem.a022763. [DOI] [PubMed] [Google Scholar]
- (40).Weingarten RA, Grimes JL, Olson JW. Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl Environ Microbiol. 2008;74:1367–1375. doi: 10.1128/AEM.02261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Smith MA, Finel M, Korolik V, Mendz GL. Characteristics of the Aerobic Respiratory Chains of the Microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch Microbiol. 2000;174:1–10. doi: 10.1007/s002030000174. [DOI] [PubMed] [Google Scholar]
- (42).Li Y, Hopper A, Overton T, Squire DJ, Cole J, Tovell N. Organisation of the electron transfer chain to oxygen in the obligate human pathogen, Neisseria gonorrhoeae: roles for cytochromes c4 and c5, but not cytochrome c2, in oxygen reduction. J Bacteriol. 2010;92:2395–2406. doi: 10.1128/JB.00002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Deeudom M, Rock J, Moir J. Organization of the respiratory chain of Neisseria meningitidis. Biochem Soc Trans. 2006;34:139–142. doi: 10.1042/BST0340139. [DOI] [PubMed] [Google Scholar]
- (44).Braun M, Thöny-Meyer L. Cytochrome c Maturation and the Physiological Role of c-Type Cytochromes in Vibrio cholerae. J Bacteriology. 2005;187:5996–6004. doi: 10.1128/JB.187.17.5996-6004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Reincke B, Thony-Meyer L, Dannehl C, Odenwald A, Aidim M, Witt H, Ruterjans H, Ludwig B. Heterologous expression of soluble fragments of cytochrome c552 acting as electron donor to the Paracoccus denitrificans cytochrome c oxidase. Biochim Biophys Acta. 1999;1411:114–120. doi: 10.1016/s0005-2728(99)00037-7. [DOI] [PubMed] [Google Scholar]
- (46).Fee JA, Chen Y, Todaro TR, Bren KL, Patel KM, Hill MG, Gomez-Moran E, Loehr TM, Ai J, Thony-Meyer L, Williams PA, Stura E, Sridhar V, McRee DE. Integrity of thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: biochemical, spectral, and structural characterization of the recombinant protein. Protein Sci. 2000;9:2074–2084. doi: 10.1110/ps.9.11.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ubbink M, Van Beeumen J, Canters GW. Cytochrome c550 from Thiobacillus versutus: cloning, expression in Escherichia coli, and purification of the heterologous holoprotein. J Bacteriol. 1992;174:3707–3714. doi: 10.1128/jb.174.11.3707-3714.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Thomas PE, Ryan D, Leven W. An Improved Straining Procedure for the Detection of the Peroxidase Activity of Cytochrome P-450 on Sodium Dodecyl Sulfate Polyacrylamide Gels. Anal. Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- (49).Conrad LS, Karlsson JJ, Ulstrup J. Electron transfer and spectral alpha-band properties of the di-heme protein cytochrome c4 from Pseudomonas stutzeri. Eur J Biochem. 1995;231:133–141. doi: 10.1111/j.1432-1033.1995.0133f.x. [DOI] [PubMed] [Google Scholar]
- (50).Pettigrew GW, Brown KR. Free and membrane-bound forms of bacterial cytochrome c4. Biochem J. 1988;252:427–435. doi: 10.1042/bj2520427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ogawa K, Sonoyama T, Takeda T, Ichiki S, Nakamura S, Kobayashi Y, Uchiyama S, Nakasone K, Takayama SJ, Mita H, Yamamoto Y, Sambongi Y. Roles of a short connecting disulfide bond in the stability and function of psychrophilic Shewanella violacea cytochrome c (5) Extremophiles. 2007;11:797–807. doi: 10.1007/s00792-007-0099-5. [DOI] [PubMed] [Google Scholar]
- (52).Zu Y, Fee JA, Hirst J. Breaking and re-forming the disulfide bond at the high-potential, respiratory-type Rieske [2Fe-2S] center of thermus thermophilus: characterization of the sulfhydryl state by protein-film voltammetry. Biochemistry. 2002;41:14054–14065. doi: 10.1021/bi026589r. [DOI] [PubMed] [Google Scholar]
- (53).Zu Y, Di Bernardo S, Yagi T, Hirst J. Redox properties of the [2Fe-2S] center in the 24 kDa (NQO2) subunit of NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 2002;41:10056–10069. doi: 10.1021/bi026026f. [DOI] [PubMed] [Google Scholar]
- (54).Shinkarev VP, Crofts AR, Wraight CA. In situ kinetics of cytochromes c1 and c2. Biochemistry. 2006;45:7897–7903. doi: 10.1021/bi060172u. [DOI] [PubMed] [Google Scholar]
- (55).Bertini I, Cavallaro G, Rosato A. Cytochrome c: Occurrence and Functions. Chem Rev. 2006;106:90–115. doi: 10.1021/cr050241v. [DOI] [PubMed] [Google Scholar]
- (56).Branca RM, Bodo G, Varkonyi Z, Debreczeny M, Osz J, Bagyinka C. Oxygen and temperature-dependent structural and redox changes in a novel cytochrome c(4) from the purple sulfur photosynthetic bacterium Thiocapsa roseopersicina. Arch Biochem Biophys. 2007;467:174–184. doi: 10.1016/j.abb.2007.07.031. [DOI] [PubMed] [Google Scholar]
- (57).Leitch FA, Brown KR, Pettigrew GW. Complexity in the redox titration of the dihaem cytochrome c4. Biochim Biophys Acta. 1985;808:213–218. doi: 10.1016/0005-2728(85)90001-5. [DOI] [PubMed] [Google Scholar]
- (58).Rey L, Maier RJ. Cytochrome c terminal oxidase pathways of Azotobacter vinelandii: analysis of cytochrome c4 and c5 mutants and up-regulation of cytochrome c-dependent pathways with N2 fixation. J Bacteriol. 1997;179:7191–7196. doi: 10.1128/jb.179.22.7191-7196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Karlsson JJ, Rostrup TE, Ulstrup J. pH and ionic strength effects on electron transfer rate constants and reduction potentials of the bacterial di-heme protein Pseudomonas stutzeri cytochrome c4. Acta Chem Scand. 1996;50:284–288. doi: 10.3891/acta.chem.scand.50-0284. [DOI] [PubMed] [Google Scholar]
- (60).Raffalt AC, Schmidt L, Christensen HE, Chi Q, Ulstrup J. Electron transfer patterns of the di-heme protein cytochrome c(4) from Pseudomonas stutzeri. J Inorg Biochem. 2009;103:717–722. doi: 10.1016/j.jinorgbio.2009.01.004. [DOI] [PubMed] [Google Scholar]
- (61).Kadziola A, Larsen S. Crystal structure of the dihaem cytochrome c4 from Pseudomonas stutzeri determined at 2.2A resolution. Structure. 1997;5:203–216. doi: 10.1016/s0969-2126(97)00179-2. [DOI] [PubMed] [Google Scholar]
- (62).Thuesen MH, Norgaard A, Hansen AM, Caspersen MB, Christensen HE. Expression of recombinant Pseudomonas stutzeri di-heme cytochrome c4 by high-cell-density fed-batch cultivation of Pseudomonas putida. Protein Expr Purif. 2003;27:175–181. doi: 10.1016/s1046-5928(02)00575-2. [DOI] [PubMed] [Google Scholar]
- (63).Giudici-Orticoni MT, Leroy G, Nitschke W, Bruschi M. Characterization of a new dihemic c(4)-type cytochrome isolated from Thiobacillus ferrooxidans. Biochemistry. 2000;39:7205–7211. doi: 10.1021/bi992846p. [DOI] [PubMed] [Google Scholar]
- (64).Abergel C, Nitschke W, Malarte G, Bruschi M, Claverie JM, Giudici-Orticoni MT. The structure of Acidithiobacillus ferrooxidans c(4)-cytochrome: a model for complex-induced electron transfer tuning. Structure. 2003;11:547–555. doi: 10.1016/s0969-2126(03)00072-8. [DOI] [PubMed] [Google Scholar]
- (65).Quatrini R, Appia-Ayme C, Denis Y, Jedlicki E, Holmes DS, Bonnefoy V. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-394. article 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Bruscella P, Appia-Ayme C, Levican G, Ratouchniak J, Jedlicki E, Holmes DS, Bonnefoy V. Differential expression of two bc1 complexes in the strict acidophilic chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans suggests a model for their respective roles in iron or sulfur oxidation. Microbiology. 2007;153:102–110. doi: 10.1099/mic.0.2006/000067-0. [DOI] [PubMed] [Google Scholar]
- (67).Elbehti A, Brasseur G, Lemesle-Meunier D. First evidence for existence of an uphill electron transfer through the bc(1) and NADH-Q oxidoreductase complexes of the acidophilic obligate chemolithotrophic ferrous ion-oxidizing bacterium Thiobacillus ferrooxidans. J Bacteriol. 2000;182:3602–3606. doi: 10.1128/jb.182.12.3602-3606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Malarte G, Leroy G, Lojou E, Abergel C, Bruschi M, Giudici-Orticoni MT. Insight into molecular stability and physiological properties of the diheme cytochrome CYC41 from the acidophilic bacterium Acidithiobacillus ferrooxidans. Biochemistry. 2005;44:6471–6481. doi: 10.1021/bi048425b. [DOI] [PubMed] [Google Scholar]
- (69).Appia-Ayme C, Guiliani N, Ratouchniak J, Bonnefoy V. Characterization of an operon encoding two c-type cytochromes, an aa(3)-type cytochrome oxidase, and rusticyanin in Thiobacillus ferrooxidans ATCC 33020. Appl Environ Microbiol. 1999;65:4781–4787. doi: 10.1128/aem.65.11.4781-4787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Giudici-Orticoni MT, Guerlesquin F, Bruschi M, Nitschke W. Interaction-induced redox switch in the electron transfer complex rusticyanin-cytochrome c(4) J Biol Chem. 1999;274:30365–30369. doi: 10.1074/jbc.274.43.30365. [DOI] [PubMed] [Google Scholar]
- (71).Mukhopadhyay BP, Ghosh B, Bairagya HR, Nandi TK, Chakrabarti B, Bera AK. Molecular modeling of the ternary complex of Rusticyanin-cytochrome c4-cytochrome oxidase: an insight to possible H-bond mediated recognition and electron transfer reaction in T. ferrooxidans. J Biomol Struct Dyn. 2008;25:543–551. doi: 10.1080/07391102.2008.10507201. [DOI] [PubMed] [Google Scholar]
- (72).Mukhopadhyay BP, Ghosh B, Bairagya HR, Bera AK, Nandi TK, Das SB. Modeling study of Rusticyanin-Cytochrome C(4) complex: an insight to possible H-bond mediated recognition and electron--transfer process. J Biomol Struct Dyn. 2007;25:157–164. doi: 10.1080/07391102.2007.10507164. [DOI] [PubMed] [Google Scholar]
- (73).Deeudom M, Koomey M, Moir JW. Roles of c-type cytochromes in respiration in Neisseria meningitidis. Microbiology. 2008;154:2857–2864. doi: 10.1099/mic.0.2008/020339-0. [DOI] [PubMed] [Google Scholar]
- (74).Hopper A, Tovell N, Cole J. A physiologically significant role in nitrite reduction of the CcoP subunit of the cytochrome oxidase cbb3 from Neisseria gonorrhoeae. FEMS Microbiol Lett. 2009;301:232–240. doi: 10.1111/j.1574-6968.2009.01824.x. [DOI] [PubMed] [Google Scholar]
- (75).Fagerquist CK, Bates AH, Heath S, King BC, Garbus BR, Harden LA, Miller WG. Sub-speciating Campylobacter jejuni by proteomic analysis of its protein biomarkers and their post-translational modifications. J Proteome Res. 2006;5:2527–2538. doi: 10.1021/pr050485w. [DOI] [PubMed] [Google Scholar]
- (76).Kern M, Eisel F, Scheithauer J, Kranz RG, Simon J. Substrate specificity of three cytochrome c haem lyase isoenzymes from Wolinella succinogenes: unconventional haem c binding motifs are not sufficient for haem c attachment by NrfI and CcsA1. Mol Microbiol. 75:122–137. doi: 10.1111/j.1365-2958.2009.06965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Moura I, Liu MY, Costa C, Liu MC, Pai G, Xavier AV, LeGall J, Payne WJ, Moura JJ. Spectroscopic characterization of a high-potential monohaem cytochrome from Wolinella succinogenes, a nitrate-respiring organism. Redox and spin equilibria studies. Eur J Biochem. 1988;177:673–682. doi: 10.1111/j.1432-1033.1988.tb14422.x. [DOI] [PubMed] [Google Scholar]