Abstract
Aminoflavone (AF), the active component of a novel anticancer agent (AFP464) in phase I clinical trials, is a ligand of the aryl hydrocarbon receptor (AhR). AhR dimerizes with HIF-1β/ARNT, which is shared with HIF-1α, a transcription factor critical for the response of cells to oxygen deprivation. To address whether pharmacological activation of the AhR pathway might be a potential mechanism for inhibition of HIF-1, we tested the effects of AF on HIF-1 expression. AF inhibited HIF-1α transcriptional activity and protein accumulation in MCF-7 cells. However, inhibition of HIF-1α by AF was independent from a functional AhR pathway. Indeed, AF inhibited HIF-1α expression in AhR100 cells, in which the AhR pathway is functionally impaired, yet did not induce cytotoxicity, providing evidence that these effects are mediated by distinct signaling pathways. Moreover, AF was inactive in MDA-MB-231 cells, yet inhibited HIF-1α in MDA-MB-231 cells transfected with the SULT1A1 gene. AF inhibited HIF-1α mRNA expression by approximately 50%. Notably, actinomycin-D completely abrogated the ability of AF to down-regulate HIF-1α mRNA, indicating that active transcription was required for the inhibition of HIF-1α expression. Finally, AF inhibited HIF-1α protein accumulation and the expression of HIF-1-target genes in MCF-7 xenografts.
These results demonstrate that AF inhibits HIF-1α in an AhR-independent fashion and they unveil additional activities of AF that may be relevant for its further clinical development.
Keywords: HIF-1, aminoflavone, hypoxia, AhR, mRNA transcription
Introduction
Hypoxia, a decrease in oxygen levels, is a hallmark of solid tumors. The response of mammalian cells to hypoxia is mediated, at least in part, by a family of transcription factors known as Hypoxia Inducible Factors (HIF) (1). HIF-1 is a heterodimer consisting of a constitutively expressed β subunit and an oxygen regulated α subunit (2) which is ubiquitinated and degraded under normoxic conditions (3). In contrast under hypoxic conditions the HIF-α subunit is stabilized and translocates to the nucleus, where it dimerizes with HIF-1β (also known as aryl hydrocarbon receptor nuclear translocator, ARNT) and activates transcription of genes involved in key steps of tumorigenesis, including angiogenesis, metabolism, proliferation, metastasis and differentiation (4). Overexpression of HIF-α has been reported in more than 70% of human cancers (5–11) and is associated with poor patients prognosis, making HIF-1 an attractive target for the development of novel cancer therapeutics (12).
Aminoflavone (NSC 686288) is the active component of a pro-drug (AFP464) in phase I clinical trials. AFP464 is rapidly converted to AF, in plasma or tissue culture, by nonspecific plasma esterases. AF has a unique COMPARE pattern of cytotoxicity in the NCI60 (13, 14), with activity only in a subset of cell lines, including MCF-7 breast cancer cells (15–18). The sensitivity of cancer cell lines to AF has been associated with its ability to act as a ligand of the aryl hydrocarbon receptor (AhR), which upon dimerization with HIF-1β/ARNT activates transcription by binding to the xenobiotic response element (XRE) in the promoters of target genes, including but not limited to cytochrome P450 1A1 (CYP1A1). Indeed, the presence of a functional AhR pathway and the induction of CYP1A1 expression by AF appear to be essential for its anti-proliferative activity in MCF-7 cells (18–20).
Several studies have suggested the existence of a crosstalk between the AhR and HIF-1 pathways (21–26). However, whether pharmacological activation of the AhR pathway may be a viable approach to inhibit HIF-1 remains poorly understood. We demonstrate that AF inhibits HIF-1α expression, both in vitro and in MCF-7 xenografts, in an AhR-independent fashion. Notably, AF partially inhibited HIF-1α mRNA expression, yet almost completely blocked HIF-1α protein accumulation. The ability of actinomycin-D to completely revert inhibition of HIF-1α mRNA expression by AF is consistent with the existence of a transcription-dependent pathway that may regulate HIF-1α mRNA expression and its translation.
Materials and methods
Cell lines and reagents
Human cells lines were maintained in RPMI-1640 supplemented with L-glutamine and 5% heat-inactivated fetal bovine serum (Hyclone) and grown at 37°C in 5% CO2 and ambient oxygen (normoxia). Hypoxia was achieved in an Invivo2 400 hypoxic workstation (Ruskinn Technologies) delivering 1% oxygen in 5% CO2 at 37°C. AhR-deficient MCF-7 (AhR100) (27) were kindly provided by Dr. David Vistica (STB, NCI, Frederick, MD). MDA-MB-231 stably transfected with a SULT1A1 cDNA, MDA/SULT1A1 (28), were kindly provided by Dr David C Spink, (Wadsworth Center, Albany, NY). All cell lines were obtained from Developmental Therapeutics Program (DTP) and were validated according to information provided on the DTP web site (http://dtp.nci.nih.gov/branches/btb/characterizationNCI60.html). Aminoflavone (AF, NSC 686288) was from the Drug Synthesis and Chemistry Branch, DTP, NCI.
Sulforhodamine B assay (SRB)
Cells, seeded into 96-well plates, were treated with AF for additional 72 hours and cell viability was assessed as previously described (29).
Immunoblot analysis
Total protein lysates were obtained as described previously (30). Antibodies used are: HIF-1α and p21 (BD- Transduction Laboratories), HIF-1β, SULT1A1, HIF-2α and AhR (Novus Biologicals), actin (Chemicon International), γH2AX (Cell Signaling, Inc.).
Real-Time PCR
VEGF, HIF-1α, p21, CA9, LOX, actin and CYP1A1 expression was measured by real-time PCR as described previously (30). Primers and probes used are listed in Supplementary Table 1. VEGF primers and probe were described previously (30). 18S rRNA was used as an internal control.
HIF-1α Protein Translational Assay
MCF-7, treated for 12 hours with AF (0.25 μM), were with 35S-methionine/cysteine (MP Biomedicals) as previously described (31). Total 35S incorporation was monitored by both size fractionation of total lysates and TCA precipitation.
Luciferase activity
MCF-7 were transfected using Effectene (Qiagen) with pGL2-TK-HRE, pGL3-control (30), NF-κB-Luc, AP-1-luc (Dr. Nancy Colburn, National Cancer Institute Frederick, MD) and XRE-Luc (by Dr. F.J. Gonzalez, NCI, NIH, Bethesda, MD). Efficiency of transfection was assessed by co-transfection with pQTK-Renilla (Promega). Results are expressed as fold increase of luciferase levels relative to normoxic untreated controls.
Transfection with siRNA for HIF-1α, SULT1A1 and AhR
SiRNA targeting SULT1A1 (s5`-CAGAGGGAGTGTGCGAATCAA), AhR (5`-TTGGATTAAATTAGTTTGTGA), HIF-1 α (5`-AGGACAAGTCACCACAGGA) and negative control (NC) were purchased from Qiagen.
Animal studies
Studies were conducted in an AAALAC-accredited facility with an approved animal protocol. MCF-7 (1 × 107) were injected subcutaneously (s.c.) into the flank of female athymic nude (NCr/nu) mice (Animal Production Area, NCI-Frederick). Beta-estradiol cypionate (3 mg/kg) was administered intramuscularly every 7 days. Tumor size was determined by collecting length and width measurements and calculating the tumor weight (mg) as [tumor length × (tumor width)2]/2, where the tumor length is the longest dimension (mm) and the tumor width is the narrowest dimension (mm). AF (saline/0.05% Tween 80) was dosed i.p. Five mice per group were treated daily for 4 days with AF (60 mg/kg) or vehicle control. When mice were sacrificed (day 4), tumors from each animal were harvested and used to analyze mRNA and protein expression, as described previously (32).
Tissue HIF-1α assay
Tissue lysates were prepared as described previously (32) and used for the quantitative determination of HIF-1α using electrochemiluminescence assay (Meso-Scale). HIF-1α concentration (pg/ml) was determined using a recombinant protein (R&D Systems) as standard.
Statistical analysis
Results are either representative or average of at least three independent experiments performed. Statistical analysis was performed using ANOVA test and t test (Prism, GraphPad).
RESULTS
AF inhibits HIF-1α transcriptional activity in MCF-7 breast cancer cells
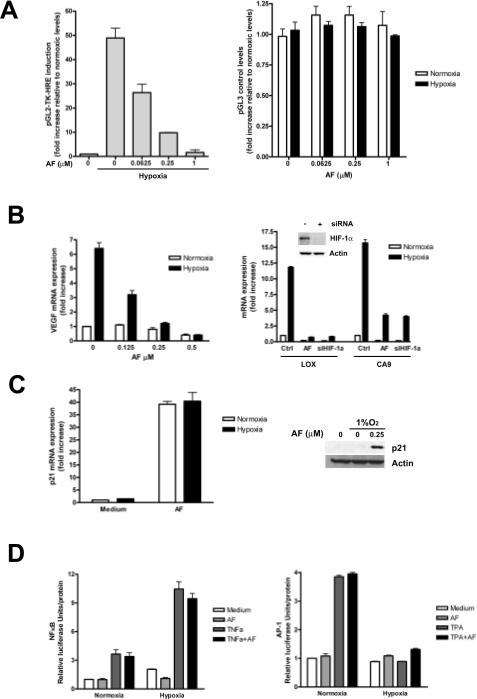
To test whether AF inhibited HIF-1 activity, MCF-7 cells were transiently transfected either with pGL2-TK-HRE, containing the luciferase reporter gene under control of 3 copies of a hypoxia response element (HRE) or with a control vector (pGL3 control). Hypoxia increased HRE-dependent luciferase expression by 49-fold, relative to cells cultured under normoxic conditions (Figure 1A, left panel). AF inhibited luciferase expression in a dose-dependent manner, but did not affect constitutive luciferase expression (Figure 1A, right panel) suggesting that inhibition of luciferase by AF was HIF-1 dependent. AF also inhibited endogenous HIF-1 transcriptional activity, as indicated by inhibition of hypoxic induction of VEGF (Figure 1B, left panel), CA9 and LOX mRNA expression (Figure 1B, right panel), similar to the effects of siRNA targeting HIF-1α. In contrast, AF caused up to 40-fold increase of p21 mRNA expression, under both normoxic and hypoxic conditions (Figure 1C, left panel), indicating that AF differentially affects gene expression in MCF-7 cells.
Figure 1. AF inhibits HIF-1α transcriptional activity.
A. MCF-7 were transfected with pGL2-TK-HRE (left panel) or pGL3-Control (right panel) and cultured under normoxia or hypoxia in the absence or presence of increasing concentrations of AF. B. MCF-7 cells were cultured under normoxia or hypoxia for 16 hours in the absence or presence of increasing concentrations of AF and VEGF mRNA levels were measured (left panel). Right panel, MCF-7 were cultured under normoxic or hypoxic conditions for 16 hours either in the absence or presence of AF (0.25 μM) or after transfection with a siRNA targeting HIF-1α (inset shows HIF-1α down-regulation following siRNA trasfection). Levels of LOX and CA9 mRNA are expressed as fold increase relative to untreated normoxic controls. C. MCF-7 were cultured under normoxic or hypoxic conditions for 16 hours in the absence or presence of AF (0.25 μM). Levels of p21 mRNA and protein were assessed. D. MCF-7, transfected with NF-κB-luc (left panel) or AP-1-luc (right panel) were treated for 16 hours under normoxic or hypoxic conditions in the absence or presence of AF (0.25 μM), TNFα (30 ng/ml) or TPA (10 ng/ml), as indicated.
Notably, neither NF-κB-(Fig 1D, left panel) nor AP-1-dependent transcriptional activities (right panel) were inhibited by AF, further demonstrating that AF specifically inhibits HIF-1.
AF inhibits HIF-1α and HIF-2α protein accumulation in a cell-type dependent fashion
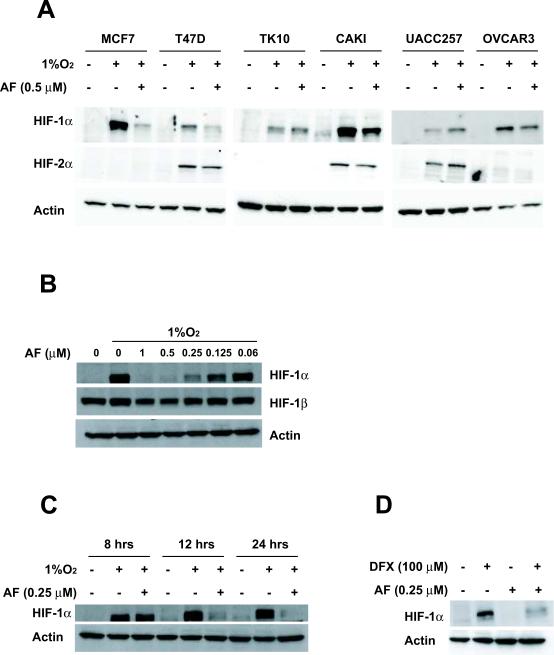
To address whether AF inhibited HIF-1α and HIF-2α protein accumulation, we tested 6 cell lines from the NCI60 panel that were reported to be sensitive to AF. Hypoxia increased HIF-1α protein expression in all the cell lines examined, while HIF-2α was induced to detectable levels in three cell lines (T47D, CAKI, UACC257). AF (0.5μM) inhibited HIF-1α protein accumulation, albeit to different extent, in MCF-7, T47D, CAKI and OVCAR3. In contrast, HIF-2α was slightly decreased by AF in T47D and CAKI cells but not in UACC257 (Figure 2A), suggesting that AF inhibits HIF-1α and HIF-2α protein accumulation in a cell type specific fashion, although HIF-2α appears to be slightly less susceptible to AF inhibition, relative to HIF-1α.
Figure 2. AF inhibits HIF-1α protein accumulation in human cancer cell lines.
A. Cancer cell lines were incubated for 16 hours in the presence or absence of AF (0.5μM) under normoxic or hypoxic conditions. Protein levels for HIF-1α, HIF-2α and actin were assessed. B. MCF-7 were incubated for 16 hours under normoxic or hypoxic conditions in the absence or presence of increasing concentrations of AF, as indicated. Protein levels of HIF-1α, HIF-1β and actin were assessed by western blot. C–D. MCF-7 cells were cultured under normoxic conditions in the absence or presence of AF (0.25 μM, C–D) or DFX (100 μM, D) for 16 hours. Protein levels of HIF-1α and actin were measured.
Further experiments demonstrated that AF inhibited HIF-1α protein accumulation in MCF-7 cells in a dose-dependent fashion, with approximately 80% decrease at 0.25 μM (Figure 2B). By contrast, HIF-1β was only inhibited by 15% and actin levels were not changed in the presence of up to 1 μM of AF. Kinetic experiments demonstrated that AF (0.25 μM) caused little or no inhibition of HIF-1α protein at 8 hours, but completely abrogated its accumulation after 12–24 hours of treatment (Figure 2C). AF also inhibited HIF-1α protein accumulation induced by the iron chelator desferrioxamine (100 μM) (Figure 2D), suggesting that its effects are not restricted to hypoxic signaling. By contrast, AF induced p21 protein accumulation in MCF-7 cells cultured under either normoxia (data not shown) or hypoxia (Figure 1C, right panel), consistent with the induction of p21 mRNA expression and demonstrating a differential effect on distinct target proteins.
A functional AhR pathway is not required for inhibition of HIF-1α expression by AF
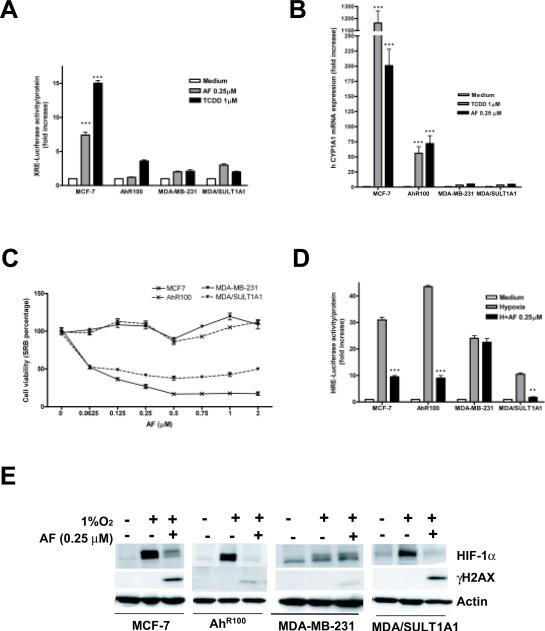
Previous studies have indicated that AF is a ligand of AhR (18). Indeed, AF caused a 7- to 8-fold increase in XRE-dependent luciferase expression (Figure 3A) and induced up to 200-fold higher levels of CYP1A1 mRNA expression in MCF-7 cells (Figure 3B), demonstrating that AF was able to induce AhR-dependent transcriptional activity. TCDD, used as positive control, induced a 15-fold increase in XRE-dependent luciferase expression (Figure 3A) and up to 1160-fold higher levels of CYP1A1 mRNA expression (Figure 3B), relative to untreated cells.
Figure 3. AF inhibits HIF-1α in an AhR-independent fashion.
A. MCF-7, AhR100, MDA-MB-231 and MDA/SULT1A1 were transfected with XRE-luc and then treated with TCDD (1 μM) or AF (0.25 μM) for 16 hours. *** = p<0.001, relative to control. B. MCF-7, AhR100, MDA-MB-231 and MDA/SULT1A1 were cultured under normoxia for 16 hours in absence or presence of AF (0.25 μM) or TCDD (1 μM). CYP1A1 mRNA expression was analyzed. *** = p<0.001, relative to control. C. MCF-7, AhR100, MDA-MB-231 and MDA/SULT1A1 were treated with increasing concentrations of AF as indicated for 72 hours. Cell viability was assessed. D. MCF-7, AhR100, MDA-MB-231 and MDA/SULT1A1 were transfected with pGL2-TK-HRE and then treated for 16 hours under normoxic or hypoxic conditions in the absence or presence of AF (0.25 μM). *** = p<0.001, ** = p<0.005, relative to hypoxia. E. MCF-7, AhR100, MDA-MB-231 and MDA/SULT1A1 were cultured under normoxia or under hypoxic conditions in the absence or presence of AF (0.25 μM) for 16 hours. Levels of HIF-1α, actin and γH2AX were measured
To address whether inhibition of HIF-1α by AF required a functional AhR pathway, we took advantage of AhR100 cells, MCF-7-derived cells that express low levels of AhR and are resistant to the cytotoxic effects of AF (18). Indeed, AF caused cytotoxicity in parental MCF-7 cells but not in AhR100 cells, even at concentrations as high as 2 μM (Figure 3C). Consistent with a functional impairment of the AhR pathway, TCDD induction of XRE-dependent luciferase expression was decreased by 75% in AhR100 cells, relative to parental MCF-7 cells, and induction of CYP1A1 mRNA expression was decreased by 95% (Figure 3A–B). More importantly, AF failed to induce XRE-dependent luciferase expression in AhR100 cells and only modestly induced CYP1A1 mRNA expression (35% of the levels induced in wild type MCF-7 cells) (Figure 3A–B). However, AF inhibited hypoxic induction of HIF-1 transcriptional activity irrespective of a functional AhR, as demonstrated by inhibition of HRE-dependent luciferase expression in both MCF-7 and AhR100 cells (Figure 3D). Accordingly, AF also completely inhibited hypoxic induction of HIF-1α protein accumulation in AhR100 cells (Figure 3E), demonstrating that inhibition of HIF-1α expression by AF is independent from a functional AhR pathway.
Aminoflavone inhibits HIF-1α in MDA/SULT1A1, but not in MDA-MB-231 parental breast cancer cells
The restricted spectrum of AF activity in the NCI60 cells has been attributed to a requirement for its intracellular activation (18) by pathway(s) yet to be completely elucidated. A potential correlation between sensitivity to AF and expression of SULT1A1 has also been suggested, consistent with the ability of SULT1A1 to induce the formation of AF metabolites that mediate DNA damage (33). Indeed, AF exerted anti-proliferative effects in MDA/SULT1A1, but not in MDA-MB-231 parental cells (Figure 3C). However, both MDA-MB-231 and MDA/SULT1A1 cells express little or no AhR transcriptional activity, as indicated by minimal if any induction of XRE-dependent luciferase (Figure 3A) or CYP1A1 mRNA expression by either AF or TCDD (Figure 3B).
We then tested whether AF inhibited HIF-1 transcriptional activity in MDA-MB-231 and MDA-SULT1A1 cells. AF completely inhibited HIF-1-dependent luciferase expression in MDA/SULT1A1, but did not affect its expression in MDA-MB-231 parental cells, demonstrating that exogenous expression of SULT1A1 was sufficient to mediate AF-dependent inhibition of HIF-1 activity (Figure 3D). Accordingly, AF almost completely inhibited hypoxic induction of HIF-1α protein in MDA/SULT1A1 cells, but not in MDA-MB-231 (Figure 3E and Supplementary Figure 1).
Next, we evaluated expression of HIF-1α in MCF-7 cells transfected with either negative control (NC) siRNA or siRNA targeting SULT1A1 or AhR (Supplementary Figure 2A–B). AF inhibited hypoxic induction of HIF-1α protein by 75% in cells transfected with NC siRNA and by 50% in cells transfected with AhR siRNA, relative to hypoxia treated cells (Supplementary Figure 2B). In contrast, down-regulation of SULT1A1 almost completely prevented inhibition of HIF-1α by AF (15% inhibition, compared to hypoxia) (Supplementary Figure 2B), demonstrating that SULT1A1 expression was implicated in HIF-1α inhibition by AF and further supporting the conclusion that inhibition of HIF-1α by AF is independent from a functional AhR pathway.
Inhibition of HIF-1α by AF is independent from DNA damage
Induction of DNA damage by AF, measured by phosphorylation of H2AX, paralleled results obtained in cytotoxicity assay. Indeed, AF induced significantly higher levels of γH2AX in sensitive MCF-7 and MDA/SULT1A1 cells than in resistant AhR100 and MDA-MB-231 cells (Figure 3E). However, AF was equally able to inhibit hypoxic induction of HIF-1α protein in AhR100 and MDA/SULT1A1 cells, demonstrating a complete dissociation between induction of DNA damage and HIF-1α inhibition (Figure 3E).
We have previously demonstrated that agents that inhibit topoisomerase I or II may affect HIF-1α protein translation (31, 34). Consistent with the finding that induction of DNA damage by AF does not appear to involve topoisomerases (35), AF was able to inhibit HIF-1α protein expression in cells transfected with siRNA targeting topo I or topo IIα (Supplementary Figure 3B). Furthermore, AF inhibited hypoxic induction of HIF-1α protein accumulation in the presence of aphidicolin, which blocks DNA polymerase and prevents DNA damage (Supplementary Figure 4), further demonstrating that AF inhibited HIF-1α by a DNA damage independent pathway.
AF does not affect HIF-1α degradation, but decreases the rate of HIF-1α translation
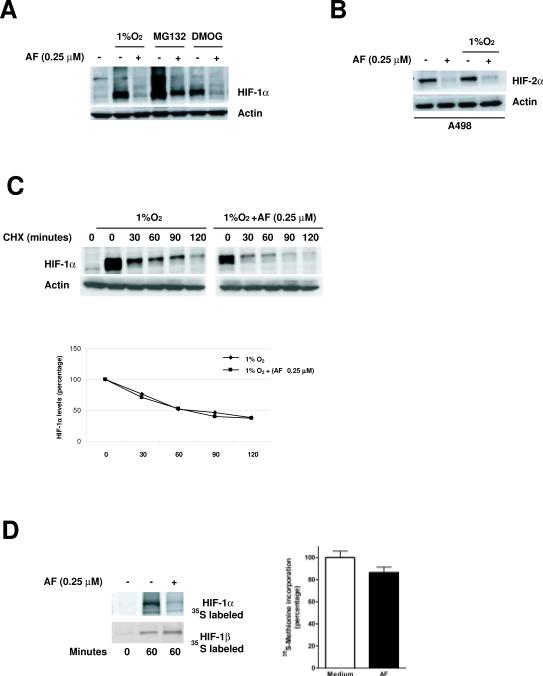
HIF-1α steady state is the result of a balance between protein translation and its degradation. To investigate whether AF affected HIF-1α degradation, we performed experiments in the presence of inhibitors of either proteasome activity (MG-132) or prolyl hydroxylase enzymes (DMOG, a global inhibitor of 2-oxoglutarate-dependent dioxygenase enzymes), which cause an accumulation of HIF-1α under normoxic conditions. As shown in Figure 4A, AF inhibited HIF-1α protein accumulation regardless of inhibition of the proteasome or prolyl hydroxylases, suggesting that AF did not affect degradation of HIF-1α protein. Consistent with these results, AF also inhibited normoxic accumulation of HIF-2α in a sensitive VHL-deficient renal cancer cell line (A498, which do not express HIF-1α), indicating that a functional VHL pathway, essential for normoxic degradation of the HIF-α subunit, is not required for the inhibition of HIF-α by AF (Figure 4B). Finally, experiments conducted in the presence of cycloheximide failed to demonstrated significant differences in HIF-1α protein half-life between cells cultured in the absence or presence of AF (half-life ~60 min) (Figure 4C). Taken together, these results demonstrate that AF does not affect HIF-1α protein degradation.
Figure 4. AF does not affect HIF-1α protein half-life but decreases its translation.
A. MCF-7 were cultured, in the absence or presence of AF (0.25 μM), either under hypoxia or in the presence of MG132 (20 μM) or DMOG (100 μM) for 16 hours. HIF-1α and actin levels were analyzed. B. A498 were treated for 16 hours with AF (0.25 μM) under normoxic or hypoxic conditions and HIF-2α and actin levels were assessed. C. MCF-7 were cultured under hypoxia in the absence or presence of AF (0.25 μM) for 12 hours (time 0), when CHX (40 μg/ml) was added for the indicated times. HIF-1α levels were quantified by densitometry (bottom panel). Values were normalized to the expression of actin and expressed as percentage relative to time 0 (arbitrarily considered equal to 100%). D. MCF-7 were treated under normoxic conditions in the absence or presence of AF (0.25 μM) for 12 hours. Newly synthesized HIF-1α and HIF-1β levels (left panel) were assessed as described in Material and Methods. Total incorporation of 35S (right panel) from TCA-precipitated cellular proteins was measured as an indication of total protein synthesis.
We then assessed whether AF inhibited HIF-1α translation by evaluating the rate of HIF-1α protein synthesis in normoxic MCF-7 cells in the absence or presence of AF. Cells were pulse-labeled with [35S] methionine for 60 minutes at which point HIF-1α was immuno-precipitated and analyzed by PAGE and autoradiography. As shown in Figure 4D (left panel), AF significantly decreased 35S-labeled HIF-1α accumulation relative to untreated cells, suggesting that it may affect its rate of translation. Importantly, under the same experimental conditions AF did not significantly affect the synthesis of HIF-1β or that of total proteins (Figure 4D, right panel).
Active transcription is required for inhibition of HIF-1α mRNA expression by AF
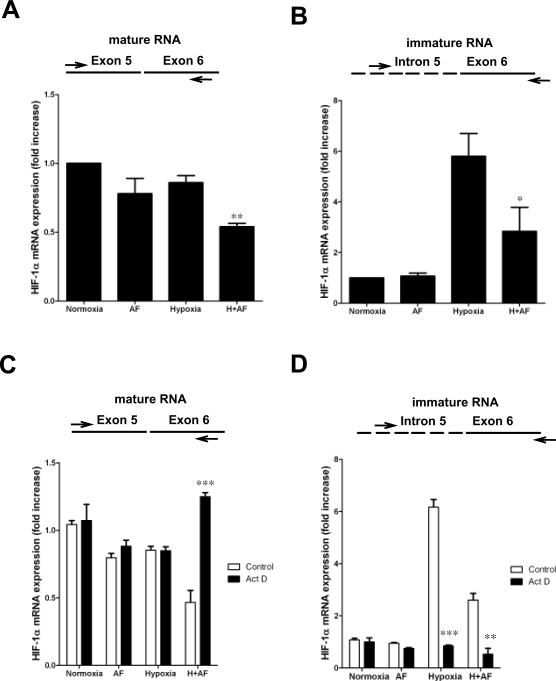
Regulation of HIF-1α translation under hypoxic conditions is still poorly understood and recent evidence suggests that levels of HIF-1α mRNA may be a crucial factor affecting the rate of HIF-1α translation (36). AF, under hypoxic but not normoxic conditions, caused a 50% decrease of HIF-1α mRNA expression, relative to normoxic cells (p=0.005) (Figure 5A). Next, to address whether AF inhibited HIF-1α mRNA transcription, we measured the levels of HIF-1α pre-mRNA. Unexpectedly, we found that hypoxia induced up to 5.8 fold higher levels of HIF-1α pre-mRNA, relative to normoxic cells (Figure 5B). Notably, despite the increase of HIF-1α pre-mRNA under hypoxic conditions, which was confirmed using different set of primers (data not shown), hypoxia only marginally affected the levels of HIF-1α mRNA, suggesting possible abnormalities in HIF-1α mRNA maturation and/or processing. AF did not affect HIF-1α pre-mRNA expression under normoxic conditions, yet reduced the hypoxic induction to levels 2–3-fold higher than those present in normoxic cells.
Figure 5. Active transcription is required for inhibition of HIF-1α mRNA expression by AF.
A–B. MCF-7 were cultured under normoxia or hypoxia for 16 hours in the absence or presence of AF (0.25 μM). HIF-1α mRNA (A) and HIF-1α pre-mRNA (B) expression was analyzed. ** = p<0.005, * = p<0.01, relative to medium. C–D. MCF-7 were cultured under normoxia or hypoxia for 16 hours in the absence or presence of AF (0.25 μM) and/or actinomycin D (5μg/ml). HIF-1α mRNA (C) and HIF-1α pre-mRNA (D) expression was assessed. *** = p<0.001, ** = p<0.005, relative to medium.
To further address the mechanism by which AF affected HIF-1α mRNA levels, we performed experiments in the presence of actinomycin-D, an inhibitor of transcription. We found that hypoxic induction of HIF-1α pre-mRNA, both in the absence and presence of AF, was completely abrogated by addition of actinomycin-D (Figure 5D), demonstrating that it was dependent on active transcription. Surprisingly, we also found that inhibition of mature HIF-1α mRNA expression by AF, observed in hypoxic cells, was completely reversed by addition of actinomycin-D, demonstrating that induction rather than inhibition of transcription is required for HIF-1α mRNA down-regulation in the presence of AF (Figure 5C). Consistent with these results, addition of actinomycin-D also partially reversed the inhibitory effect of AF on HIF-1α protein accumulation (Supplementary Fig 5), suggesting that the effects on HIF-1α mRNA expression were causally related to the inhibition of HIF-1α protein.
Taken together, these results demonstrate that active transcription, in hypoxic cells treated with AF, is required for the inhibition of HIF-1α mRNA expression.
AF inhibits HIF-1α expression in MCF-7 xenografts
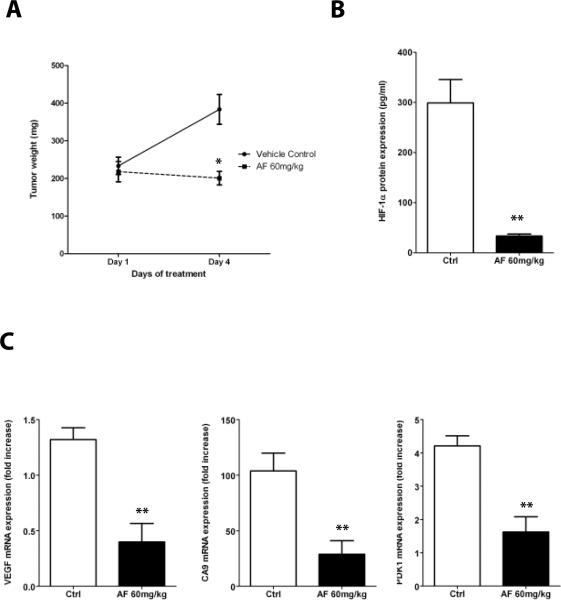
To test whether AF was able to inhibit HIF-1α expression in tumor xenografts, MCF-7 cells were implanted subcutaneously in female athymic nude mice. When tumors reached approximately 200 mg, mice (n=5/group) were randomized to receive either vehicle control or AF (60mg/kg, ip) daily for four days. As shown in Figure 6A, AF exerted a cytostatic effect on tumor growth (p<0.01), relative to vehicle-treated mice. Notably, AF inhibited HIF-α protein accumulation in tumor lysates by 90% (p<0.005), relative to vehicle treated mice (Figure 6B) and mRNA expression of the HIF-1-target genes VEGF, CA9 and PDK-1 by approximately 70% (p<0.05, Figure 6C), demonstrating that AF inhibits HIF-1α expression and activity in MCF-7 xenografts.
Figure 6. AF inhibits HIF-1α expression and transcriptional activity in MCF-7 xenografts.
A. MCF-7 were implanted into nude mice (n = 5/group) and allowed to grow up to ~ 200mg, when treatment with AF (60 mg/kg daily × 4 days i.p.) was started. Tumor weight was measured as described in Materials and Methods, (Mann-Whitney test; *, p < 0.01). B. Quantitative determination of HIF-1α protein levels (**, p <0.05). C. mRNA expression of HIF-1-target genes (VEGF, CA9 and PDK-1) in tumor lysates from mice treated with vehicle or AF were assessed (p < 0.05). Values are expressed as fold change relative to levels detected in mRNA harvested from normoxic MCF-7.
Because of the potentially confounding effects of tumor size on HIF-1α expression, we also conducted experiments in which MCF-7 tumors bearing animals were only treated for two days with either vehicle control or AF (120mg/kg, ip). As shown in Supplementary figure 6, AF did not affect tumor growth under these conditions, yet it significantly inhibited HIF-1α protein accumulation and VEGF mRNA expression (Supplementary figure 6B–C, respectively), suggesting that inhibition of HIF-1α and HIF-1-target genes by AF is independent from its cytostatic/cytotoxic activity.
Discussion
Hypoxia Inducible Factor 1 (HIF-1) has emerged over the last several years as an attractive target for the development of novel cancer therapeutics. We and others have identified small molecule inhibitors of HIF-1α that act by distinct mechanisms of action, including, but not limited to, inhibition of HIF-1α translation (31, 34, 37, 38), inhibition of HIF-1 DNA binding (39) or transcriptional activity (40–42), inhibition of protein-protein interaction (43), increased protein degradation (44, 45), or inhibition of mitochondrial respiration (46). A number of HIF-1 inhibitors identified at the National Cancer Institute share the property of inducing DNA damage, including topotecan (30) and NSC 644221 (34), although DNA damage and HIF-1 inhibition appear to be concomitant but independent events.
AF induces DNA damage and acts as a ligand of AhR. AF exerts anti-proliferative activity in a fairly limited number of human cancer cell lines and activation of AhR appears to be required for its conversion to DNA damaging species, at least in part, by transcriptional activation of CYP1A1 and SULT1A1, two XRE target genes (18). In this manuscript we provide evidence that AF inhibits HIF-1α expression in MCF-7 cells in an AhR independent fashion, as indicated by the following results: a) AF inhibited HIF-1α protein accumulation in both MCF-7 and AhR100 cells, in which the AhR pathway is functionally impaired; b) AF did not inhibit HIF-1α in the resistant MDA-MB-231 breast cancer cells, yet it did in MDA/SULT1A1, MDA-MB-231 cells transfected with SULT1A1and c) siRNA targeting SULT1A1 significantly blocked the ability of AF to inhibit HIF-1α in MCF-7 cells (Supplementary Figure 2). Along with the ability of AF to exert anti-proliferative activity in MCF-7 and MDA-SULT1A1, but not in AhR100 or MDA-MB-231, these results suggest that a) inhibition of HIF-1α by AF is independent from a functional AhR pathway and b) there is no correlation between cytotoxicity and HIF-1α inhibition. Indeed, AF was completely ineffective in inducing anti-proliferative effects in AhR100 cells yet it was able to inhibit HIF-1α expression. In addition, AF induced significantly lower levels of γH2AX, a marker of DNA damage, in AhR100 and MDA-MB-231 cells, which did not correlate with its ability to inhibit HIF-1α. These results not only demonstrate a dissociation between cytotoxicity and HIF-1 inhibition, but they also raise the possibility that cancer cells found to be “resistant” to the cytotoxic effects of AF may be sensitive to HIF-1α inhibition.
A significant number of small molecule inhibitors of HIF-1α identified thus far appears to affect HIF-1 translation (31, 34, 37, 38), yet regulation of HIF-1α translation under hypoxic conditions is still poorly understood. We were then intrigued by the fact that AF also appeared to inhibit HIF-1α synthesis. Further experiments demonstrated that AF inhibited HIF-1α mRNA expression by approximately 50%, under hypoxic, but not normoxic conditions. The magnitude of HIF-1α mRNA inhibition was unlikely to account for the almost complete inhibition of HIF-1α protein accumulation, raising the possibility that additional mechanisms were implicated. Using primers that specifically detect HIF-1α pre-mRNA we discovered that hypoxia (in the absence or presence or AF) induced higher levels of transcript, relative to normoxic cells. The increased levels of HIF-1α pre-mRNA did not seem to correlate with the decreased expression of HIF-1α mRNA in the presence of AF. Notably, experiments conducted in the presence of actinomycin D, demonstrated that active transcription was required for the down-regulation of HIF-1α mRNA expression by AF. The discrepancy between accumulation of HIF-1α pre-mRNA and decrease of HIF-1α mRNA levels might have suggested a potential effect of AF plus hypoxia on HIF-1α mRNA processing and/or maturation. However, results obtained in the presence of actinomycin-D argue against this conclusion and are consistent with transcriptional induction of either a) a repressor, which in turn is responsible for inhibition of HIF-1α mRNA expression or b) non coding RNA species, which may account for both inhibition of HIF-1α mRNA expression and translation. The latter possibility is consistent with a) the mechanism of action of AF, which implicates protein nucleic acid complexes (33, 35), b) the lack of correlation between magnitude of HIF-1α mRNA inhibition and inhibition of HIF-1α translation and c) the ability of topotecan, a DNA damaging agent that also inhibits HIF-1α translation, to increase the levels of anti-sense transcripts of the HIF-1α genomic sequence (47). Several miRs have been recently identified that indeed target the 3′-UTR of HIF-1α mRNA leading to inhibition of HIF-1α protein levels (48–50). It is then plausible that the effects of DNA damaging agents on HIF-1 protein translation may at least in part implicate non-coding RNA species that target HIF-1α mRNA expression and/or HIF-1α mRNA translation. Further experiments are required to characterize the spectrum of miR induced by agents that concomitantly inhibit HIF-1α and induce DNA damage to formally demonstrate the existence of a mechanistic link.
The proposed mechanism of HIF-1α inhibition by AF is conceivably associated with effects on other genes and pathways. However, AF did not inhibit NF-kB or AP-1 transcriptional activities and potently induced p21 mRNA and protein expression, ruling out a global effect on mRNA expression and/or protein translation. Moreover, gene array experiments (using the Affymetrix human 133 2.0 plus chip), indicated that 1.78% of genes were induced more than 2 fold and 0.69% of genes were inhibited more than 2 fold in MCF-7 cells treated with AF (0.25μM) for 16 hours, demonstrating that AF does not cause a global inhibition of gene expression (data not shown).
Evidence of inhibition of HIF-1α expression in xenograft tissue (Fig 6) further corroborates potential translational implications of the findings described in this mansucript. Inhibition of HIF-1α in AhR100 cells also raises the possibility that AF might modulate HIF-1-dependent pathways even in cells that are not sensitive to its cytotoxic effects, a feature that would be emphasized by more protracted schedules of administration, as opposed to the ones generally used for cytotoxic agents. Finally, modulation of both mRNA expression and mRNA translation induced by AF may potentially contribute to its anti-cancer actvitiy and unveils novel properties of AF that may help its clinical development.
Supplementary Material
Table 1.
Primers used for Real time PCR.
| Human CYP1A1 | Forward | 5`GATTGGGCACATGCTGACG-3` |
| Reverse | 5`-TGCTGGCTCATCCTTGACAG-3` | |
| Human p21 | Forward | 5`ACGCGACTGTGATGCGC-3` |
| Reverse | 5`-AAGTTCCATCGCTCACGGG-3` | |
| Human CA 9 | Forward | 5`-GAGGCCTGGCCGTGTTG-3` |
| Reverse | 5`-AACTGCTCATAGGCACTGTTTTCTT-3` | |
| Human LOX | Forward | 5`-TGCTTGGTGGAGACTGAGATACC-3` |
| Reverse | 5`-AATCACGTGAGGGAAGGAGAAA-3` | |
| Human HIF-1α (intron 5-exon 6) | Forward | 5`-TGCTTTTTTTTTCCCTAGCATTGT-3` |
| Reverse | 5`-TGGTTACTGTTGGTATCATATACGTGAA-3` | |
| Human HIF-1α (exon5-exon 6) | Forward | 5`-TAGCCGAGGAAGAACTATGAACATAA-3` |
| Reverse | 5`-TGAGGTTGGTTACTGTTGGTATCATATA-3` | |
| Probe | 5'-TTGCACTGCACAGGCCACATTCAC-3' |
Acknowledgments
The authors would like to thank members of the Melillo's laboratory, Yves Pommier and Robert H. Shoemaker for helpful discussion. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported by the Developmental Therapeutics Program, DCTD, of the National Cancer Institute, NIH.
Reference List
- 1.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Hypoxia and cancer. Cancer Metastasis Rev. 2007;26:223–4. doi: 10.1007/s10555-007-9058-y. [DOI] [PubMed] [Google Scholar]
- 3.Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–43. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 4.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 5.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5. [PubMed] [Google Scholar]
- 6.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–85. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koukourakis MI, Bentzen SM, Giatromanolaki A, Wilson GD, Daley FM, Saunders MI, Dische S, Sivridis E, Harris AL. Endogenous markers of two separate hypoxia response pathways (hypoxia inducible factor 2 alpha and carbonic anhydrase 9) are associated with radiotherapy failure in head and neck cancer patients recruited in the CHART randomized trial. J Clin Oncol. 2006;24:727–35. doi: 10.1200/JCO.2005.02.7474. [DOI] [PubMed] [Google Scholar]
- 8.Birner P, Schindl M, Obermair A, Plank C, Breitenecker G, Oberhuber G. Overexpression of hypoxia-inducible factor 1alpha is a marker for an unfavorable prognosis in early-stage invasive cervical cancer. Cancer Res. 2000;60:4693–6. [PubMed] [Google Scholar]
- 9.Birner P, Schindl M, Obermair A, Breitenecker G, Oberhuber G. Expression of hypoxia-inducible factor 1alpha in epithelial ovarian tumors: its impact on prognosis and on response to chemotherapy. Clin Cancer Res. 2001;7:1661–8. [PubMed] [Google Scholar]
- 10.Aebersold DM, Burri P, Beer KT, Laissue J, Djonov V, Greiner RH, Semenza GL. Expression of hypoxia-inducible factor-1alpha: a novel predictive and prognostic parameter in the radiotherapy of oropharyngeal cancer. Cancer Res. 2001;61:2911–6. [PubMed] [Google Scholar]
- 11.Bos R, van der GP, Greijer AE, Shvarts A, Meijer S, Pinedo HM, Semenza GL, van Diest PJ, van der WE. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97:1573–81. doi: 10.1002/cncr.11246. [DOI] [PubMed] [Google Scholar]
- 12.Melillo G. Targeting hypoxia cell signaling for cancer therapy. Cancer Metastasis Rev. 2007;26:341–52. doi: 10.1007/s10555-007-9059-x. [DOI] [PubMed] [Google Scholar]
- 13.Monks A, Scudiero DA, Johnson GS, Paull KD, Sausville EA. The NCI anti-cancer drug screen: a smart screen to identify effectors of novel targets. Anticancer Drug Des. 1997;12:533–41. [PubMed] [Google Scholar]
- 14.Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, Plowman J, Boyd MR. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J Natl Cancer Inst. 1989;81:1088–92. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- 15.Akama T, Shida Y, Sugaya T, Ishida H, Gomi K, Kasai M. Novel 5-aminoflavone derivatives as specific antitumor agents in breast cancer. J Med Chem. 1996;39:3461–9. doi: 10.1021/jm950938g. [DOI] [PubMed] [Google Scholar]
- 16.Akama T, Ishida H, Kimura U, Gomi K, Saito H. Structure-activity relationships of the 7-substituents of 5,4'-diamino-6,8,3'-trifluoroflavone, a potent antitumor agent. J Med Chem. 1998;41:2056–67. doi: 10.1021/jm970728z. [DOI] [PubMed] [Google Scholar]
- 17.Akama TO, Okazaki Y, Ito M, Okuizumi H, Konno H, Muramatsu M, Plass C, Held WA, Hayashizaki Y. Restriction landmark genomic scanning (RLGS-M)-based genome-wide scanning of mouse liver tumors for alterations in DNA methylation status. Cancer Res. 1997;57:3294–9. [PubMed] [Google Scholar]
- 18.Loaiza-Perez AI, Kenney S, Boswell J, Hollingshead M, Alley MC, Hose C, Ciolino HP, Yeh GC, Trepel JB, Vistica DT, Sausville EA. Aryl hydrocarbon receptor activation of an antitumor aminoflavone: basis of selective toxicity for MCF-7 breast tumor cells. Mol Cancer Ther. 2004;3:715–25. [PubMed] [Google Scholar]
- 19.Kuffel MJ, Schroeder JC, Pobst LJ, Naylor S, Reid JM, Kaufmann SH, Ames MM. Activation of the antitumor agent aminoflavone (NSC 686288) is mediated by induction of tumor cell cytochrome P450 1A1/1A2. Mol Pharmacol. 2002;62:143–53. doi: 10.1124/mol.62.1.143. [DOI] [PubMed] [Google Scholar]
- 20.Whitlock JP., Jr. Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–25. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 21.Chan WK, Yao G, Gu YZ, Bradfield CA. Cross-talk between the aryl hydrocarbon receptor and hypoxia inducible factor signaling pathways. Demonstration of competition and compensation. J Biol Chem. 1999;274:12115–23. doi: 10.1074/jbc.274.17.12115. [DOI] [PubMed] [Google Scholar]
- 22.Zhang N, Walker MK. Crosstalk between the aryl hydrocarbon receptor and hypoxia on the constitutive expression of cytochrome P4501A1 mRNA. Cardiovasc Toxicol. 2007;7:282–90. doi: 10.1007/s12012-007-9007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi H, Chun YS, Kim SW, Kim MS, Park JW. Curcumin inhibits hypoxia-inducible factor-1 by degrading aryl hydrocarbon receptor nuclear translocator: a mechanism of tumor growth inhibition. Mol Pharmacol. 2006;70:1664–71. doi: 10.1124/mol.106.025817. [DOI] [PubMed] [Google Scholar]
- 24.Gassmann M, Kvietikova I, Rolfs A, Wenger RH. Oxygen- and dioxin-regulated gene expression in mouse hepatoma cells. Kidney Int. 1997;51:567–74. doi: 10.1038/ki.1997.81. [DOI] [PubMed] [Google Scholar]
- 25.Nie M, Blankenship AL, Giesy JP. Interactions between aryl hydrocarbon receptor (AhR) and hypoxia signaling pathways. Environ Toxicol Pharmacol. 2001;10:17–27. doi: 10.1016/s1382-6689(01)00065-5. [DOI] [PubMed] [Google Scholar]
- 26.Takacova M, Holotnakova T, Vondracek J, Machala M, Pencikova K, Gradin K, Poellinger L, Pastorek J, Pastorekova S, Kopacek J. Role of aryl hydrocarbon receptor in modulation of the expression of the hypoxia marker carbonic anhydrase IX. Biochem J. 2009 doi: 10.1042/BJ20080952. [DOI] [PubMed] [Google Scholar]
- 27.Ciolino HP, Dankwah M, Yeh GC. Resistance of MCF-7 cells to dimethylbenz(a)anthracene-induced apoptosis is due to reduced CYP1A1 expression. Int J Oncol. 2002;21:385–91. [PubMed] [Google Scholar]
- 28.Spink BC, Katz BH, Hussain MM, Pang S, Connor SP, Aldous KM, Gierthy JF, Spink DC. SULT1A1 catalyzes 2-methoxyestradiol sulfonation in MCF-7 breast cancer cells. Carcinogenesis. 2000;21:1947–57. doi: 10.1093/carcin/21.11.1947. [DOI] [PubMed] [Google Scholar]
- 29.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 30.Rapisarda A, Uranchimeg B, Scudiero DA, Selby M, Sausville EA, Shoemaker RH, Melillo G. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62:4316–24. [PubMed] [Google Scholar]
- 31.Rapisarda A, Uranchimeg B, Sordet O, Pommier Y, Shoemaker RH, Melillo G. Topoisomerase I-mediated inhibition of hypoxia-inducible factor 1: mechanism and therapeutic implications. Cancer Res. 2004;64:1475–82. doi: 10.1158/0008-5472.can-03-3139. [DOI] [PubMed] [Google Scholar]
- 32.Rapisarda A, Zalek J, Hollingshead M, Braunschweig T, Uranchimeg B, Bonomi CA, Borgel SD, Carter JP, Hewitt SM, Shoemaker RH, Melillo G. Schedule-dependent inhibition of hypoxia-inducible factor-1alpha protein accumulation, angiogenesis, and tumor growth by topotecan in U251-HRE glioblastoma xenografts. Cancer Res. 2004;64:6845–8. doi: 10.1158/0008-5472.CAN-04-2116. [DOI] [PubMed] [Google Scholar]
- 33.Meng LH, Shankavaram U, Chen C, Agama K, Fu HQ, Gonzalez FJ, Weinstein J, Pommier Y. Activation of aminoflavone (NSC 686288) by a sulfotransferase is required for the antiproliferative effect of the drug and for induction of histone gamma-H2AX. Cancer Res. 2006;66:9656–64. doi: 10.1158/0008-5472.CAN-06-0796. [DOI] [PubMed] [Google Scholar]
- 34.Creighton-Gutteridge M, Cardellina JH, Stephen AG, Rapisarda A, Uranchimeg B, Hite K, Denny WA, Shoemaker RH, Melillo G. Cell type-specific, topoisomerase II-dependent inhibition of hypoxia-inducible factor-1alpha protein accumulation by NSC 644221. Clin Cancer Res. 2007;13:1010–8. doi: 10.1158/1078-0432.CCR-06-2301. [DOI] [PubMed] [Google Scholar]
- 35.Meng LH, Kohlhagen G, Liao ZY, Antony S, Sausville E, Pommier Y. DNA-protein cross-links and replication-dependent histone H2AX phosphorylation induced by aminoflavone (NSC 686288), a novel anticancer agent active against human breast cancer cells. Cancer Res. 2005;65:5337–43. doi: 10.1158/0008-5472.CAN-05-0003. [DOI] [PubMed] [Google Scholar]
- 36.Young RM, Wang SJ, Gordan JD, Ji X, Liebhaber SA, Simon MC. Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J Biol Chem. 2008;283:16309–19. doi: 10.1074/jbc.M710079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mabjeesh NJ, Escuin D, LaVallee TM, Pribluda VS, Swartz GM, Johnson MS, Willard MT, Zhong H, Simons JW, Giannakakou P. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell. 2003;3:363–75. doi: 10.1016/s1535-6108(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Qian DZ, Tan YS, Lee K, Gao P, Ren YR, Rey S, Hammers H, Chang D, Pili R, Dang CV, Liu JO, Semenza GL. Digoxin and other cardiac glycosides inhibit HIF-1alpha synthesis and block tumor growth. Proc Natl Acad Sci U S A. 2008;105:19579–86. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong D, Park EJ, Stephen AG, Calvani M, Cardellina JH, Monks A, Fisher RJ, Shoemaker RH, Melillo G. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 2005;65:9047–55. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 40.Kung AL, Zabludoff SD, France DS, Freedman SJ, Tanner EA, Vieira A, Cornell-Kennon S, Lee J, Wang B, Wang J, Memmert K, Naegeli HU, Petersen F, Eck MJ, Bair KW, Wood AW, Livingston DM. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Kaluz S, Kaluzova M, Stanbridge EJ. Proteasomal inhibition attenuates transcriptional activity of hypoxia-inducible factor 1 (HIF-1) via specific effect on the HIF-1alpha C-terminal activation domain. Mol Cell Biol. 2006;26:5895–907. doi: 10.1128/MCB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeo EJ, Ryu JH, Cho YS, Chun YS, Huang LE, Kim MS, Park JW. Amphotericin B blunts erythropoietin response to hypoxia by reinforcing FIH-mediated repression of HIF-1. Blood. 2006;107:916–23. doi: 10.1182/blood-2005-06-2564. [DOI] [PubMed] [Google Scholar]
- 43.Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci U S A. 2009;106:17910–5. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem. 2002;277:29936–44. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- 45.Qian DZ, Kachhap SK, Collis SJ, Verheul HM, Carducci MA, Atadja P, Pili R. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res. 2006;66:8814–21. doi: 10.1158/0008-5472.CAN-05-4598. [DOI] [PubMed] [Google Scholar]
- 46.Lin X, David CA, Donnelly JB, Michaelides M, Chandel NS, Huang X, Warrior U, Weinberg F, Tormos KV, Fesik SW, Shen Y. A chemical genomics screen highlights the essential role of mitochondria in HIF-1 regulation. Proc Natl Acad Sci U S A. 2008;105:174–9. doi: 10.1073/pnas.0706585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baranello L, Bertozzi D, Fogli MV, Pommier Y, Capranico G. DNA topoisomerase I inhibition by camptothecin induces escape of RNA polymerase II from promoter-proximal pause site, antisense transcription and histone acetylation at the human HIF-1alpha gene locus. Nucleic Acids Res. 2010;38:159–71. doi: 10.1093/nar/gkp817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cha ST, Chen PS, Johansson G, Chu CY, Wang MY, Jeng YM, Yu SL, Chen JS, Chang KJ, Jee SH, Tan CT, Lin MT, Kuo ML. MicroRNA-519c Suppresses Hypoxia-Inducible Factor-1{alpha} Expression and Tumor Angiogenesis. Cancer Res. 2010 doi: 10.1158/0008-5472.CAN-09-2448. [DOI] [PubMed] [Google Scholar]
- 49.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–86. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68:5540–5. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.