Abstract
Mutator strains of Escherichia coli have been used to define mechanisms that account for the high fidelity of chromosome duplication and chromosome stability. Mutant strains defective in post-replicative mismatch repair display a strong mutator phenotype consistent with a role for correction of mismatches arising from replication errors. Inactivation of the gene (dam) encoding DNA adenine methyltransferase results in a mutator phenotype consistent with a role for DNA methylation in strand discrimination during mismatch repair. This review gives a personal perspective on the discovery of dam mutants in E. coli and their relationship to mismatch repair and mutator phenotypes.
Keywords: Alkylating agents, DNA repair, Escherichia coli, Mutator, Methylation, Recombination
1. Introduction
Chromosome replication requires a high degree of fidelity, and studies in Escherichia coli K-12 over the last fifty years or so have identified the major mechanisms by which this is achieved. The experimental approach used to solve the fidelity question has relied mainly on the isolation and characterization of mutator strains. A mutator phenotype (Mut−) is displayed by mutants that have an increased spontaneous mutation frequency relative to wild-type (Mut+). The underlying assumption is that such bacteria are impaired in systems that normally correct spontaneous replication errors and, in general, this assumption has been correct. It took some time, however, for this assumption to take hold given that the first E. coli mutator strain was described in 1954 [1] and systematic screening for mutator strains did not begin until 1970 [2].
In this review I have focused on a group of related mutator strains (and one in particular) that has been the subject of my research for the past few decades. I decided to present a personal view of the developments in this research area in the hope it offers insight into the history of these mutants and will be more entertaining than a formal scientific summary. The latter part of the review is a more conventional summary of mutator genes and their effects, and further details can be found in other reviews [3–6]. Genes discussed in this review are listed in Table 1 with a brief explanation of each.
Table 1.
E. coli genes considered in this article.
| Gene | Gene product |
|---|---|
| dam | DNA adenine methyltransferase |
| dcm | DNA cytosine methyltransferase |
| dnaB | Replicative helicase |
| dnaE | Catalytic alpha-subunit of DNA polymerase III holoenzyme |
| dnaG | DNA primase |
| dnaQ | Epsilon-subunit of DNA polymerase III holoenzyme |
| fpg | Synonym for MutM |
| hexAB | MutSL homologs |
| lig | DNA ligase |
| mutD | Allele of dnaQ resulting in defective proof-reading |
| mutH | Mismatch repair endonuclease |
| mutL | Mismatch repair protein |
| mutM | Glycosylase specific for oxidized guanine-cytosine basepairs |
| mutS | Detects base mispairs to initiate mismatch repair |
| mutT | Prevents incorporation of oxidized guanine into DNA |
| mutU | Allele of uvrD |
| mutY | Glycosylase specific for oxidized guanine-adenine basepairs |
| polA | DNA polymerase I |
| polC | Synonym for dnaE |
| recA | Promotes synapsis of homologous DNA strands |
| recBCD | Double-strand end-specific exonuclease |
| ruvA | With RuvB acts as a Holliday junction translocase |
| ruvB | With RuvA acts as a Holliday junction translocase |
| ruvC | Holliday junction resolvase |
| uvrD | Mismatch repair associated helicase |
2. DNA methylation mutants
I was appointed to my first faculty position as an Instructor at Rutgers Medical School (as it was known then) in Piscataway, NJ, in 1971. I had come to join N. Ronald Morris who had been researching DNA methylation in eukaryotes. It seemed worthwhile studying the problem in Escherichia coli which, unlike eukaryotes, had both 6-methyladenine (6-meA) and 5-methylcytosine (5-meC) in its DNA. The approach would be a standard one - isolate mutants lacking methylated bases and deduce their functions by examining their properties.
The assay we used to isolate methylation-deficient mutants was based on two prior observations. First, DNA isolated from E. coli grown in the presence of ethionine, a methionine analog, was found to be deficient in methylation because it was a substrate for the transfer of methyl groups from S-adenosyl-methionine (SAM) to such DNA in crude extracts prepared from wild-type cells grown without ethionine [7]. DNA isolated from untreated E. coli was not a substrate because the DNA was fully methylated. Second, Herb Boyer’s lab had located the gene (near his) for cytosine methylation on the E. coli K-12 map by using this assay on recombinants obtained from crosses between K-12 and B which does not have methylated cytosine in its DNA [8]. These findings suggested a way to detect mutants deficient in methylation - they would incorporate methyl groups into their DNA while wild-type cells would not. Accordingly, I treated my wild-type cells with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and combined the survivors in groups of ten. DNA was isolated from the pool and assayed for methyl group transfer from tritiated SAM into DNA. If a positive signal was obtained, the pool was split in two for re-testing and finally to individuals. This brute force screen led to the isolation of 14 DNA and 10 RNA methylation mutants from about 1500 survivors. The RNA mutants were easily identified by testing radioactivity in the alkali-treated nucleic acid supernatant fraction designed to remove RNA.
The 14 DNA methylation mutants were grown with tritiated methionine and the amount of 6-meA and 5-meC quantified. This led to the identification of three mutants lacking 6-meA and 11 lacking 5-meC [9]. Seven tRNA methylation mutants were also recovered and in collaboration with Dieter Soll’s lab at Yale were shown to be deficient in ribothymine (5 isolates), 7-methylguanine and 2-thio-5-methylaminoethyluracil [10]. At this point it was necessary to identify the genes involved by mapping the mutations and this was done first by conjugational crosses and then by transductional crosses [11]. I tested recombinant classes using the assay above, which was successful but laborious. The 6-meA and 5-meC mutants were designated dam (DNA adenine methylase) and dcm (DNA cytosine methylase), respectively, although in recent years I’ve been using methyltransferase instead of methylase. I had toyed with mad (methyladenine deficient) as the designation for the gene but dam won out. The Dam and Dcm methyltransferases methylate -GATC- and -CC(A/T)GG- sequences, respectively, of which there are 19,120 and 12,045, respectively, in the E. coli chromosome. Stan Hattman’s lab had also isolated dcm mutants at the same time by looking for E. coli mutants that would not protect phage lambda from the restriction system encoded by plasmid N3 [12]. For many years this was the only phenotype associated with dcm mutants. From this point on I will deal only with the dam mutants since these had not been previously isolated, and I had concentrated my efforts on them.
The mutations in a clean genetic background were tested for a variety of phenotypic traits. I had included in the isolation protocol the possibility that the mutations conferred a temperature-sensitive phenotype but none of the mutants were temperature-sensitive (Ts) for growth. This was somewhat disappointing but K. Brooks Low, then a junior faculty member in Therapeutic Radiology at Yale, consoled me by pointing out that recA mutants were not conditionally lethal but were still interesting to study. Microscopic observation showed that the dam cells were not uniform in size confirming that for a given optical density in broth cultures the viable count was always lower for the dam mutant than the wild type. In my previous work with dnaB and dnaG mutants I had also observed this when the cells were grown at the non-permissive temperature and this led me to look at the DNA sedimentation profile in alkaline sucrose gradients. There were single-strand breaks in the chromosomal DNA of the dam cells and these were amplified in dam polA (Ts) and dam lig (Ts) strains. These latter strains were inviable at the non-permissive temperature as were dam mutations in combination with recA and recBCD mutant alleles. It was clear that the dam mutants were defective in some kind of DNA repair but the best that could be done at the time was to exclude nucleotide excision repair since the uvr genes had no effect on dam phenotypes [13]. During the mapping of the dam gene, I noticed that my control plates for the dam mutants often had colonies on them while those of the wild type did not. The mutator phenotype of the dam mutants was quickly confirmed.
These results were published as my three-year appointment at Rutgers Medical School was coming to an end, and I was busy trying to find a new position. My wife was seven months pregnant when in June 1974 she drove our Volvo and I drove the U-Haul truck to Worcester, Massachusetts, where I was to take up a faculty position in the newly-formed University of Massachusetts Medical School. I had expected to be there for only a few years before continuing our nomadic existence but I have remained there ever since.
In my assistant professor position at UMass Medical School, I isolated more dam mutants by various means and all had the same range of phenotypes as those previously isolated. In order to confirm that these were associated with the dam mutation and not something else, advantage was taken of the inviability of dam recA mutants to isolate true revertants. These had none of the phenotypes associated with the dam strains. In addition to the true revertants there were also suppressed revertants which had mutations in the mutS or mutL genes (see below). These did not have the phenotypes associated with dam except for the mutator phenotype which was much stronger [14]. The interesting result was that suppressor mutations of the dam mutator phenotype were in mutator genes which had a stronger mutator phenotype.
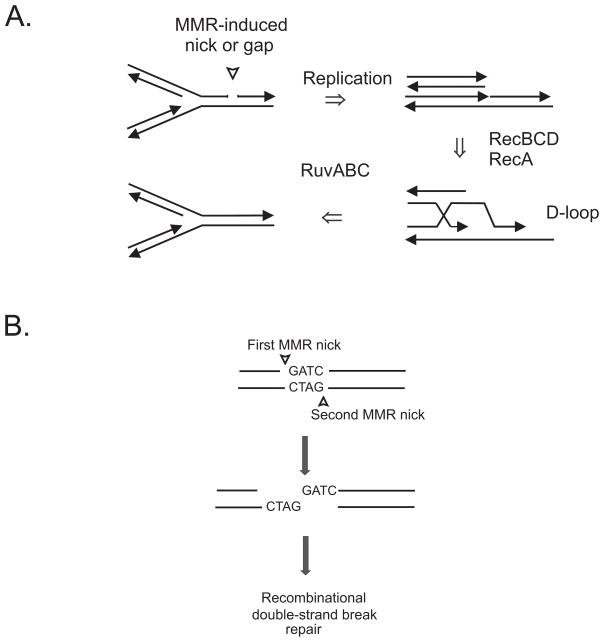
In 1974 the SOS hypothesis had not yet been formulated, and it was a few years later that we showed the dam mutants to be sub-induced for the SOS response. Of all the SOS genes only expression of the recA, ruvA and ruvB genes was necessary for dam cell survival. It was also shown subsequently that double-strand breaks were present in the DNA of dam bacteria. The evidence made it clear that mismatch repair is responsible for the formation of DNA breaks, that DNA ligase is required for repair of single-strand interruptions, and that homologous recombination is essential for double-strand break repair. What is still not known is how the double-strand breaks are formed, and Fig. 1 shows two possibilities. First, a replication fork encountering a gap or nick in duplex DNA will collapse (Fig. 1A) but can be repaired by homologous recombination. It is not known what fraction of collapsed forks is mended. Second, the presence of a nick on each strand at a GATC sequence is equivalent to a double-strand break (Fig. 1B) which requires a sister chromosome as a template for recombinational repair.
Fig. 1.
Formation of double-strand breaks in dam bacteria. (A) The encounter of a replication fork with a strand discontinuity due to mismatch repair (MMR) results in fork collapse and the formation of a double-stranded end. This end is a substrate for the RecBCD exonuclease which, upon digestion, can lead to the loading of RecA and the formation of a D-loop after strand invasion. Resolution of the Holliday junction by RuvABC can restore the fork. (B) Mismatch repair nicking of both strands at a GATC sequence results in a double-strand break that can be repaired using a sister chromosome as template. This type of recombinational repair also requires RecBCD, RecA, and RuvABC.
3. Mismatch repair
The existence of this repair system had been postulated by Holliday [15] to account for gene conversion in fungi. The formation of heteroduplex DNA (one DNA chain from the mutant, the other from wild type) leads to the creation of base mismatches and, depending on the direction of correction, could explain the excess or deficiency of recombinant classes observed as an excess of the phenotype conferred by one allele relative to that of the other. Studies in Streptococcus pneumoniae with heteroduplexes showed that transformation frequencies were, in part, dependent on mismatch correction. Furthermore, a mutant strain (hex) appeared deficient for this type of repair and had a mutator phenotype [16].
The mutagen 5-bromouracil (5-BU) is known to form base pairs with either adenine or guanine. Rydberg showed that in wild-type E. coli, 5-BU mutagenesis is suppressed at low concentrations suggesting that 5-BU mispairs were subject to correction. He devised an assay to isolate mutants defective in correction and these mapped to the mutH, mutL, mutS and uvrD (=mutU) genes. These mutants all had spontaneous mutation frequencies at least one-hundredfold greater than wild type [17]. It was subsequently shown that hexAB mutator strains of S. pneumoniae were in genes homologous to mutSL.
4. Dam methylation and mismatch repair
The connection between Dam methylation and mismatch repair was suggested by R. Wagner and M. Messelson; methylation was the way E. coli could discriminate between old and new DNA chains [18]. To investigate this possibility, advantage was taken of the ability to separate the strands of bacteriophage lambda using cesium chloride density gradient centrifugation, thereby allowing annealing of a wild-type strand with a complementary mutant strand (from a clear plaque mutant) to form a heteroduplex molecule [19]. In addition, it was possible to obtain strands that were unmethylated (propagated in a dam mutant) or fully methylated (grown in a Dam overproducer). Heteroduplexes were made that were unmethylated on both strands; methylated on the wild-type or mutant strand (hemimethylated); and methylated on both strands. Infection of a wild-type E. coli host with the hemimethylated heteroduplexes yielded progeny strongly biased to the configuration of the methylated strand. That is, if the wild-type strand was methylated the progeny formed mostly turbid plaques, while if the mutant strand was methylated the progeny were mostly clear-plaque formers. This biased directionality was not present if a mutL host was used, consistent with the idea that this strain was defective in mismatch repair. Directionality of repair was also reduced with the unmethylated heteroduplex.
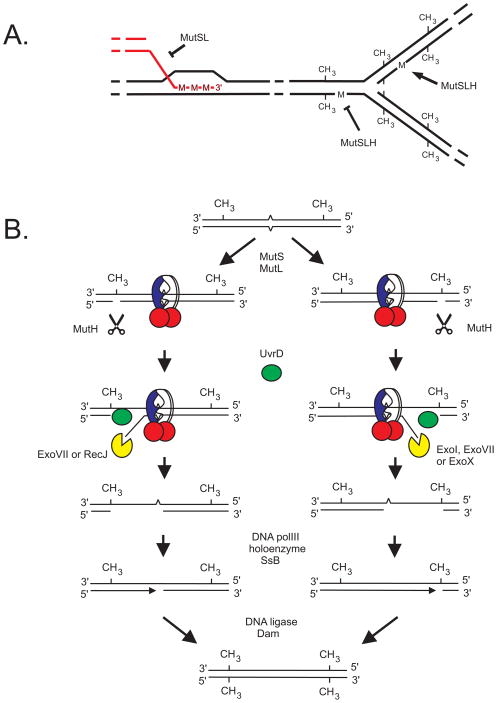
A surprising finding was that if both strands were methylated, there was no correction in a wild-type host indicating that only hemimethylated heteroduplex regions were subject to repair [19]. Since hemimethylated DNA occurs only transiently behind the replication fork, it was postulated that the mismatch repair system acted on replication errors at this location. The error occurs in the newly-synthesized unmethylated strand which is then targeted for repair using the parental methylated strand as template (Fig. 2A).
Fig. 2.
Functions of the mismatch repair system in E. coli. (A) The two known functions of mismatch repair are antirecombination (left) and correction of mismatches generated by replication (right). The initial step in antirecombination is the formation of a D-loop by insertion of a single strand into a duplex molecule by RecA protein. The invading strand is homeologous and therefore creates mismatches in the duplex DNA that are recognized by MutS and MutL. These proteins block the RecA reaction probably by reversing it. MutS and MutL have no effect on RecA catalysis if the two molecules are homologous. A mismatch (M) at the replication fork (right) in hemimethylated DNA is acted upon by mismatch repair, but a mismatch in fully methylated DNA is refractory. Lack of Dam methyltransferase results in correction of either the newly-synthesized strand or the parental strand. In the latter case, mutations are introduced into the genome. Overproduction of Dam methyltransferase results in premature methylation of DNA behind the fork and stabilization of mismatches by lack of repair. (B) A mismatch in hemimethylated DNA is recognized by the MutS protein (blue and white) which recruits MutL (red circles) and MutH (scissors), and the latter incises the unmethylated strand at a GATC sequence. UvrD helicase (green circle) loads at the nick created by MutH and unwinds the DNA strand towards the mismatch. Exonucleases (yellow) digest the unwound single strand including the mismatched nucleotide. Different exonucleases are used depending on the orientation of the mismatch relative to the GATC site (ExoVII or RecJ in one case, and ExoI, ExoVII, or ExoX in the other). The resultant gap is filled by the action of DNA polymerase III holoenzyme, and DNA ligase seals the nick. Dam methyltransferase acts at the hemimethylated GATC sites to fully methylate them.
Further evidence for this model was that dam mutants have a mutator phenotype because directionality of mismatch correction is lost and the error-containing strand is used as template to introduce mutations into the genome. In addition, overproduction of Dam methyltransferase in a wild-type E. coli host leads to an increased mutation rate because newly-synthesized DNA becomes methylated before it can be corrected and thereby leads to introduction of mutations into the chromosome [20, 21]. Results showing that mutations in the mutHLS genes suppress the lethality of 2-aminopurine to dam strains were also consistent with this model [22].
Although the mutHLS mutants display a strong mutator phenotype, that of the dam mutant is relatively weak. This is unexpected since the anticipation is that the mutation frequency should be the same as that of the mut strains. It is possible that a certain fraction of the dam population is lost due to the inability to repair all the single- and double-strand breaks that arise. This loss would be reflected in a lower mutator phenotype.
Fig. 2 illustrates the functions of the mismatch repair system in E. coli which are correction of replication errors and anti-recombination. Anti-recombination will be discussed in detail in the next section. The right side of Fig. 2A depicts a replication fork showing a transient section of hemimethylated DNA with a base pair mismatch that is susceptible to mismatch repair while a mismatch in fully methylated DNA is not. Fig. 2B summarizes the biochemical studies from Paul Modrich’s laboratory [23] on the correction of base mispairs in hemimethylated DNA at the replication fork. Base mismatches are recognized and bound by the MutS protein which recruits MutL and MutH. The latter is a latent endonuclease that, upon formation of the ternary complex, cleaves 5′ to the G at a nearby GATC sequence on the unmethylated strand. The MutH protein is released from the complex, and the UvrD helicase is recruited to the nick and unwinds the DNA from the nick towards the mismatch. This unwinding can occur in either the 5′ to 3′ or 3′ to 5′ direction depending on the relative orientation of the GATC to the mismatch. Specific exonucleases are used for each direction: ExoVII or RecJ in one case, and ExoI, ExoVII, or ExoX in the other. The resultant gap is filled by the action of DNA polymerase III holoenzyme, and DNA ligase seals the nick. Dam methyltransferase acts at the hemimethylated GATC sites to fully methylate them and to prevent further repair.
All eukaryotes and most bacteria unrelated to E. coli, do not have methylated GATC sequences in DNA. How is the newly synthesized strand recognized for targeting by the mismatch repair system in these organisms? This problem was faced by the streptococcal geneticists who were using heteroduplexes containing various base mismatches in transformation experiments. They found that the transformants fell into two groups yielding either high or low frequencies. It was proposed that mismatch repair resulted in low transformation frequencies while lack of repair resulted in high transformation frequencies. They also found that correction occurred only on the incoming strand and not on the recipient’s DNA chains. They proposed that the strand targeted for mismatch correction was that which had at least one end. Since the chromosomal DNA had no ends and the incoming DNA had two, it was the latter that was targeted [16]. This model can be extrapolated to the replication fork; the parental strands do not have an end nearby but the newly-synthesized strands have 3′-ends and therefore are targeted. This model applies to bacteria with circular chromosomes and eukaryotes with linear chromosomes.
A puzzling aspect of this general model is why E. coli and its relatives have abandoned it in favor of a methylation-based discrimination model. Furthermore, why doesn’t strand discrimination occur in a dam mutant?
5. Antirecombination, Dam methylation, mismatch repair and MNNG
Genetic crosses between E. coli and Salmonella typhimurium are sterile. However, if the recipient in these crosses is mismatch repair-deficient (mutS or mutL inactivation), bona fide recombinants are formed [24]. This result suggests that MutS and MutL prevent recombination between homeologous sequences, which are related but not identical. This antirecombination effect may be important in preventing recombination in the DNA of organisms that have repeated homeologous DNA sequences as such recombination could lead to genome instability. An initial step in homologous recombination is the insertion of the 3′-end of a single strand into duplex DNA to form a D-loop. This reaction is catalyzed by the RecA protein and is not affected by the presence of MutS and MutL. If the DNA molecules in this reaction are homeologous, however, then MutS and MutL will block the formation of RecA product (no D-loops are formed)[25]. This is shown diagrammatically in Fig. 2A where the D-loop formed by RecA action between homeologous substrates results in the formation of multiple mismatches and the reaction is aborted by MutS and MutL.
I was on sabbatical leave in 1980–1981 at the Medical Research Council Cell Mutation Unit at the University of Sussex where Peter Karran and I collaborated on a project on the sensitivity of dam mutants to methylating agents and to MNNG (N-methyl-N′-nitro-N-nitrosoguanidine) in particular. Peter had come to Sussex from the Lindahl lab, then in Gothenburg in Sweden, and was working on the biochemistry of enzymes acting on DNA modified by methylating agents. We showed that although dam mutants were rapidly killed by MNNG, dam mut strains were not, indicating that mismatch repair was responsible for the inviability. This work produced two important findings. First, the differential susceptibility of dam mutants to methylating agents indicated that O6-methylguanine (O6-meG) was the lesion recognized by the mismatch repair system. It was already known that this base leads to ambiguous coding such that O6-meG-T or O6-meG-C base pairs are formed after replication. The second important point was the proposal that mismatch repair of O6-meG-T would lead to formation of O6-meG-C but that this basepair would also be subjected to mismatch repair. Repeated mismatch repair attempts produce a futile repair cycle and eventually lead to cell death [26].
After my sabbatical leave I returned to Worcester to begin work on mutational specificity, and Peter moved to the Imperial Cancer Research Fund (now Cancer Research UK) Clare Hall laboratories when Thomas Lindahl became the director. Peter extended the E. coli work to mammalian cells that are sensitive to killing by MNNG but not when mismatch repair is inactivated. This research gained importance when (a) a subset of tumors from relapsed cancer patients treated with MNNG-like drugs were found to be mismatch repair-deficient [27], and (b) mismatch repair-deficiency was found to be associated with hereditary and sporadic colon cancer [28, 29].
In both E. coli dam mutants and mammalian cells, mismatch repair therefore sensitizes the cells to killing after exposure to MNNG. We showed that DNA double-strand breaks accumulate in MNNG-exposed dam mutants to a much higher level than in untreated dam cells but were not detectable in either case in dam mutants that were mismatch repair-deficient [30]. Unrepaired double-strand DNA breaks are known to be lethal to the cell, and these could account for the sensitivity to MNNG of the dam mutants. Such double-strand breaks might result from futile cycling but this possibility remains to be tested.
A second mechanism that may promote mismatch repair-mediated cell death of MNNG-exposed dam mutants relates to antirecombination. Double-strand breaks in E. coli are repaired by homologous recombination involving RecA activity. We found that MNNG-methylated homologous DNA was recognized as if it were homeologous in an in vitro reaction using RecA, MutS and MutL [31].
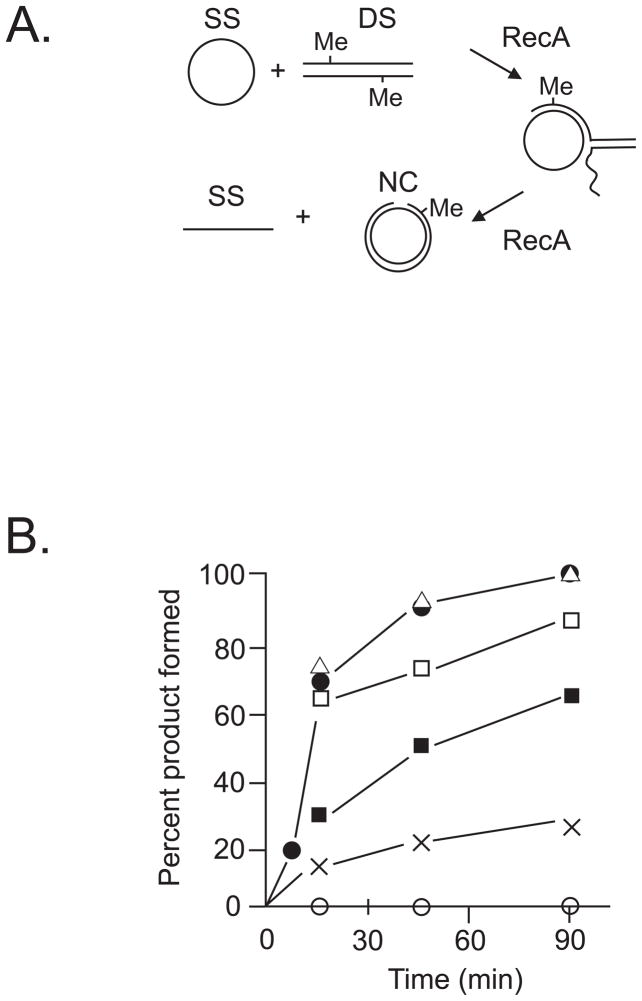
The RecA reaction (Fig. 3A) transfers a homologous strand from a duplex molecule (DS) to a single-stranded circular (SS) molecule to form a nicked circular (NC) product. When one of the two molecules (6kb in length) contains 10–20 O6-methylguanines, the RecA reaction proceeds at the same rate as if no methylated bases were present (Fig. 3A). With the addition of MutS, the rate of the RecA reaction is slowed and in the presence of both MutS and MutL the reaction can be blocked (Fig. 3B). The same concentrations of MutS and MutL have no effect on the RecA reaction in substrate molecules lacking O6-methylguanine. This result suggests that both double-strand-break formation by mismatch repair and the inhibition of double-strand break recombinational repair by MutS and MutL may contribute to the killing of dam mutants by MNNG. The possibility that double-strand breaks and/or inhibition of their recombinational repair are involved in mismatch repair-dependent killing of mammalian cells by MNNG has not yet been tested.
Fig. 3.
The RecA strand-transfer reaction. (A) A homologous strand from linear duplex DNA (DS) is transferred to a single-stranded circular (SS) molecule by RecA to form a double-stranded nicked circular (NC) product which can be easily detected. The linear duplex DNA contains O6-meG bases (Me). (B) Rate of formation of NC product when 10–20 O6-meG bases are present in DS DNA. No addition (triangles), 25 nM MutS (unfilled squares), 100 nM MutS (filled squares), 25 nM MutS and 50 nM MutL (crosses), 100 nM MutS and 100 nM MutL (unfilled circles). The reaction rate in the absence of O6-meG modification is denoted by the filled circles. MutS and MutL do not affect the RecA reaction rate in the absence of DNA modification. (Figure adapted from [31])
6. Other mutator genes in E. coli
In general, mutator strains are defective in functions that (a) prevent incorporation of mutagenic bases, (b) correct replication errors, or (c) remove potentially mutagenic bases from DNA. An example of a nucleotide-pool cleansing enzyme is MutT. It converts oxidized dGTP to oxidized dGMP, which is not used by DNA polymerases. Although this mutator strain was first discovered in 1954 [1], it was not until 1992 that its function was uncovered [32].
The dnaE (=polC) gene encodes the catalytic alpha subunit of DNA polymerase III holoenzyme, and dnaQ (=mutD) encodes the proof-reading epsilon exonuclease subunit. These two proteins are in close contact. Although mutations in the catalytic subunit are expected to increase or decrease the fidelity of base selection and incorporation and thereby impart a mutator phenotype, no such bona fide mutations have been isolated. The only mutator allele of dnaE so far examined appears to affect the activity of the epsilon proof-reading exonuclease subunit. Mutator alleles affecting this subunit include the dominant mutD5 allele, which decreases correction of replication errors. However, the large number of errors generated in this strain also overwhelms MutHLS mismatch repair correction leading to a mutator phenotype due to transversions, transitions and frameshifts.
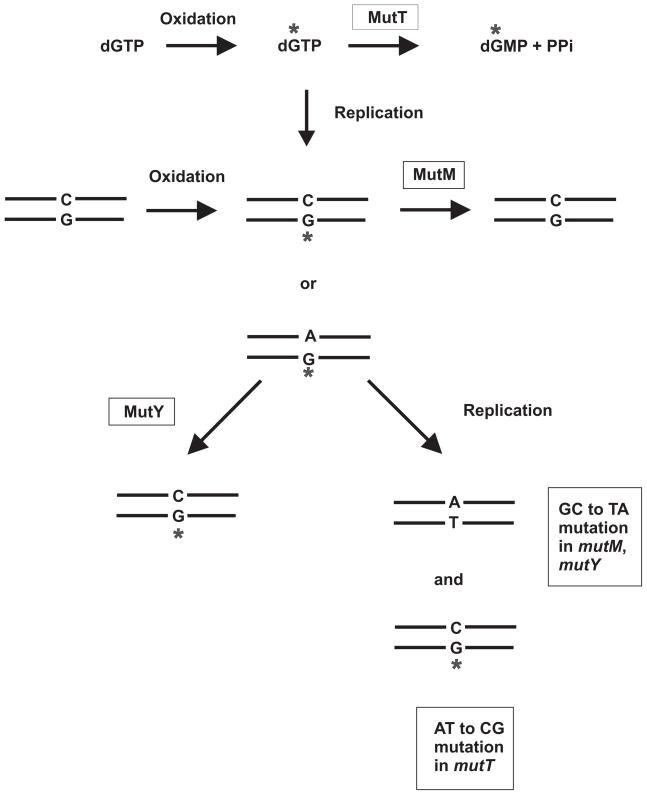
The oxidation product 8-oxoguanine can be formed in DNA either by incorporation as a deoxyribonucleotide triphosphate precursor (d*GTP) or by the oxidation of guanine residues in DNA (Fig. 4). The d*GTP that escapes the cleansing action of MutT can be incorporated opposite either cytosine or adenine. The *G-C base pair is a substrate for the MutM glycosylase which removes the oxidized base and leads to its replacement with G. The *G-A base pair is acted upon by MutY, which removes the A residue and replaces it with C. The *G-C pair is then acted upon by MutM (Fig. 4). If the *G-A base pair is replicated before repair, an A-T will result in one daughter molecule and a *G-C in the other, the latter again a substrate for MutM. This pathway explains the observed GC to TA changes observed in mutM mutY double mutants and the AT to CG specificity of the mutT strain. Loss of both MutM and MutY results in a strong mutator phenotype (about the same level as the MutHLS mismatch repair system), indicating the importance of this system [5, 6].
Fig. 4.
Action of MutT, MutM and MutY proteins. Guanine is readily oxidized leading to various products including 8-oxoguanine (designated *G). The MutT protein converts d*GTP to d*GMP thereby preventing its utilization by DNA polymerase. Incorporation of d*GTP into DNA results in base pairing with either A or C. *G-C base pairs are substrates for the MutM glycosylase which removes the *G. *G-A base pairs are substrates for MutY glycosylase which removes the A residue resulting in a *G-C base pair which is acted upon by MutM. Replication of a *G-A base pair leads to a G to T transversion of the original G-C or an A to C transversion of the original A-T. (Figure adapted from [6])
The four systems described above (pool cleansing, exonucleolytic proofreading, MutHLS correction and MutY/MutM action) constitute the major mechanisms to prevent or correct base mismatches in DNA. Other strains displaying a mild mutator phenotype have also been characterized, as have effects of overproducing gene products, but the magnitude of these effects is generally small and these are described in more detail elsewhere [6].
Our knowledge of mutator strains is still largely derived from studies with E. coli. The sequencing of genomes of many other bacteria and Archaea has largely confirmed the existence of the error correction pathways described in E. coli, but other more exotic mutator genes probably await discovery. For example, organisms living in high temperature, low pH, or high pressure environments presumably have developed special mechanisms preventing damage to their DNA that might elicit a mutator phenotype if not working correctly.
7. Mutator genes in populations of pathogens and commensals
The frequency of mutator strains in a population of E. coli K-12 (the laboratory strain) is less than one in 100,000. In contrast, this frequency increases to at least one percent in pathogenic or commensal E. coli strains [33, 34]. The relatively high frequency of mutator strains in these populations has led to speculation that they may be important when the population is subjected to stress. Bacterial populations in nature are probably constantly changing and the reservoir of mutator bacteria may help to select variants more suited to the new environment. For example, on leaving the alimentary tract of an animal, the E. coli population would need to adapt quickly to extra-intestinal life and the presence of a pre-existing mutant population may facilitate this change. If the mutator subpopulation serves such a function, the variants would need to rapidly lose their mutator phenotype. The mechanism by which this could occur is not known.
Acknowledgments
My research on DNA methylation, mismatch repair and recombination has been funded over the years by the American Cancer Society, the National Science Foundation and the National Institutes of Health (currently GM 63790).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Treffers HP, Spinelli V, Belser NO. A factor (or mutator gene) influencing mutation rates in Escherichia coli. Proc Natl Acad Sci U S A. 1954;40:1064–1071. doi: 10.1073/pnas.40.11.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liberfarb RM, Bryson V. Isolation, characterization, and genetic analysis of mutator genes in Escherichia coli B and K-12. J Bacteriol. 1970;104:363–375. doi: 10.1128/jb.104.1.363-375.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox EC. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:548–555. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- 4.Smith KC. Spontaneous mutagenesis: experimental, genetic and other factors. Mutat Res. 1992;277:139–162. doi: 10.1016/0165-1110(92)90002-q. [DOI] [PubMed] [Google Scholar]
- 5.Miller JH. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- 6.Horst JP, Wu TH, Marinus MG. Escherichia coli mutator genes. Trends Microbiol. 1999;7:29–36. doi: 10.1016/s0966-842x(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 7.Gold M, Hurwitz J. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. VI. Further studies on the properties of the deoxyribonucleic acid methylation reaction. J Biol Chem. 1964;239:3866–3874. [PubMed] [Google Scholar]
- 8.Mamelak L, Boyer HW. Genetic control of the secondary modification of deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1970;104:57–62. doi: 10.1128/jb.104.1.57-62.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marinus MG, Morris NR. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973;114:1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marinus MG, Morris NR, Soll D, Kwong TC. Isolation and partial characterization of three Escherichia coli mutants with altered transfer ribonucleic acid methylases. J Bacteriol. 1975;122:257–265. doi: 10.1128/jb.122.1.257-265.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marinus MG. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973;127:47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- 12.Hattman S, Schlagman S, Cousens L. Isolation of a mutant of Escherichia coli defective in cytosine-specific deoxyribonucleic acid methylase activity and in partial protection of bacteriophage lambda against restriction by cells containing the N-3 drug-resistance factor. J Bacteriol. 1973;115:1103–1107. doi: 10.1128/jb.115.3.1103-1107.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marinus MG, Morris NR. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974;85:309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- 14.McGraw BR, Marinus MG. Isolation and characterization of Dam+ revertants and suppressor mutations that modify secondary phenotypes of dam-3 strains of Escherichia coli K-12. Mol Gen Genet. 1980;178:309–315. doi: 10.1007/BF00270477. [DOI] [PubMed] [Google Scholar]
- 15.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 16.Claverys JP, Lacks SA. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986;50:133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rydberg B. Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli. Mutat Res. 1978;52:11–24. doi: 10.1016/0027-5107(78)90091-x. [DOI] [PubMed] [Google Scholar]
- 18.Wagner R, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976;73:4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pukkila PJ, Peterson J, Herman G, Modrich P, Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983;104:571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman GE, Modrich P. Escherichia coli K-12 clones that overproduce dam methylase are hypermutable. J Bacteriol. 1981;145:644–646. doi: 10.1128/jb.145.1.644-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marinus MG, Poteete A, Arraj JA. Correlation of DNA adenine methylase activity with spontaneous mutability in Escherichia coli K-12. Gene. 1984;28:123–125. doi: 10.1016/0378-1119(84)90095-7. [DOI] [PubMed] [Google Scholar]
- 22.Glickman BW, Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980;77:1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- 24.Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 25.Worth L, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci U S A. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karran P, Marinus MG. Mismatch correction at O6-methylguanine residues in E. coli DNA. Nature. 1982;296:868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- 27.Li GM. The role of mismatch repair in DNA damage-induced apoptosis. Oncol Res. 1999;11:393–400. [PubMed] [Google Scholar]
- 28.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 29.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 30.Nowosielska A, Marinus MG. DNA mismatch repair-induced double-strand breaks. DNA Repair (Amst) 2008;7:48–56. doi: 10.1016/j.dnarep.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calmann MA, Evans JE, Marinus MG. MutS inhibits RecA-mediated strand transfer with methylated DNA substrates. Nucleic Acids Res. 2005;33:3591–3597. doi: 10.1093/nar/gki673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 33.LeClerc JE, Li B, Payne WL, Cebula TA. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 34.Taddei F, Matic I, Godelle B, Radman M. To be a mutator, or how pathogenic and commensal bacteria can evolve rapidly. Trends Microbiol. 1997;5:427–428. doi: 10.1016/S0966-842X(97)01157-8. [DOI] [PubMed] [Google Scholar]