Abstract
The development of expressed sequence tags (ESTs) from pea has provided a useful source for mining novel simple sequence repeat (SSR) markers. In the present research, in order to find EST-derived SSR markers, 18 552 pea ESTs from the National Center for Biotechnology Information (NCBI) database were downloaded and assembled into 10 086 unigenes. A total of 586 microsatellites in 530 unigenes were identified, indicating that merely 5.25% of sequences contained SSRs. The most abundant SSRs within pea were tri-nucleotide repeat motifs, and among all the tri-nucleotide repeats, the motif GAA was the most abundant type. In total, 49 SSRs were used for primer design. EST-SSR loci were subsequently screened on 10 widely adapted varieties in China. Of these, nine loci showed polymorphic profiles that revealed two to three alleles per locus. The polymorphism information content value ranged from 0.18 to 0.58 with an average of 0.41. Furthermore, transferable analysis revealed that some of these loci showed transferability to faba bean. Because of their polymorphism and transferability, these nine novel EST-SSRs will be valuable tools for marker-assisted breeding and comparative mapping of pea in the future.
Keywords: Pea, Expressed sequence tag (EST), Simple sequence repeat (SSR), Microsatellite
1. Introduction
In all kinds of molecular markers, simple sequence repeat (SSR) marker has become one of the most important molecular markers, due to co-dominant inheritance, multi-allelic nature, relative abundance, extensive genome coverage, and ease detection by polymerase chain reaction (PCR) (Powell et al., 1996). However, the development of traditional ‘anonymous’ SSRs from genomic DNA is costly and time-consuming (Squirrell et al., 2003; Ellis and Burke, 2007). Recently, with the development of functional genomics, a huge number of expressed sequence tags (ESTs) were deposited in a public sequence database (Kong et al., 2007), providing a potentially rich source of SSRs (Ellis and Burke, 2007). Deriving from EST, EST-SSRs have some intrinsic advantages over genomic SSRs in their direct association with transcribed genes, low expense for development, and high level of transferability to related species (Varshney et al., 2005). To date, the development of SSR through searching the database of EST (dbEST) has become a fast, efficient, and low-cost option for many plants (Tangphatsornruang et al., 2008).
Pea (Pisum sativum L.) is one of the most popular legumes in the world. Its production ranks second among the cool season pulses in the world and can be used as a vegetable, pulse, and feed. Despite its long history of domestication and economical importance, to our knowledge, only a limited number of SSRs have been developed so far (Burstin et al., 2001; Loridon et al., 2005). There still has been no report of the development of EST-SSR markers from pea. Here, we report the isolation and identification of EST-SSRs in P. sativum, including their frequency and distribution, polymorphism, and transferability to the related species Vicia faba.
2. Materials and methods
2.1. Plant materials
Ten pea varieties cultivated across China were used to evaluate the marker polymorphism. These varieties included ‘Zhewan 1’, ‘Xiaoshanbaihua’, ‘Zhongwan 4’, ‘Zhongwan 6’, ‘Tengfei 5’, ‘Qizhen 77’, ‘Zhenzhulu’, ‘Taizhong 11’, ‘Zhongjia 604’, and ‘Shijiadacaiwan’. Three V. faba L. varieties, including ‘Dabaidou’, ‘Xiaoqingdou’, and ‘Linyu 1’, were used for transferability studies. Genomic DNA from each variety was extracted from young seedlings grown in a glasshouse. A total of 0.1 g leaf material was used for each repeat and isolated using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. The DNA concentration was estimated by agarose gel electrophoresis using DNA standard.
2.2. Database search and primer definition
P. sativum L. ESTs were acquired by searching GenBank (up to November 2009). Those ESTs were assembled into unigenes using DNASTAR software and the parameters for clustering were set at a minimum of 95% identity in 40-bp overlap. The unigenes were used for identifying SSRs via simple sequence repeat identification tool (SSRIT) software (http://www.gramene.org/gramene/searches/ssrtool). The criteria for SSRs identification were 7, 5, 4, 3 repeat units for di-, tri-, tetra-, penta-, and higher order nucleotides, respectively. The putative function of the EST-SSR markers was identified by basic local alignment search tool X (BLASTX) analysis at the National Center for Biotechnology Information (NCBI), with the threshold of 10−10 for the expect value (E-value).
The primers were designed using the Primer Premier 5.0 software with length of 17–24 bp, annealing temperature of 50–60 °C, and product sizes ranging from 100 to 400 bp. The forward primers of each pair were labeled with 6-carboxyfluorescein (6-FAM) fluorescent dye.
2.3. PCR reaction and genotyping
All PCR amplifications were carried out in a 20-μl reaction mixture containing 10–20 ng of genomic DNA, 1×PCR buffer, 1 U of Taq DNA polymerase (TaKaRa, Dalian, Liaoning, China), 2 mmol/L MgCl2, 0.2 μmol/L of each primer, and 0.2 mmol/L of deoxynucleotide triphosphates (dNTPs).
PCR reactions were performed on PTC-225 Peltier Thermal Cycler (MJ Research, Waltham, MA, USA) with an initial 5 min of denaturation at 94 °C, followed by 30 cycles of 94 °C for 30 s, appropriate annealing temperature for 30 s, 72 °C for 1 min, and a final extension at 72 °C for 10 min.
The PCR reaction products were diluted and detected on MegaBACE 1000 DNA analysis system (Amersham Biosciences, Piscataway, NJ, USA) at the Center of Analysis and Measurement in Zhejiang University, China. Sizes of amplified fragments were analyzed using ET550-R size standard (GE Healthcare, Piscataway, NJ, USA) and Genetic Profiler 2.0 (GE Healthcare).
2.4. Statistical analysis
Polymorphism information content (PIC) was calculated using the formula developed by Anderson et al. (1993).
3. Results
3.1. EST-SSRs in pea
A total of 18 552 pea ESTs were obtained from the dbEST of NCBI. After removing clustering and assembling of the ESTs, 10 086 unigenes were generated and then used to search for SSRs. The search revealed that only a subset of 530 unigenes contains 586 microsatellites, suggesting that merely 5.25% of sequences contained SSRs (Table 1). Among all the non-redundant ESTs, 42 unigenes contained two SSRs, 4 contained three SSRs, and 2 contained four SSRs. The putative position of the EST-SSRs was identified by BLASTX analysis at NCBI. The results showed that 34.98% of SSRs were in the coding DNA sequence, 9.90% in 5′ untranslated region (UTR), and 25.43% in 3′ UTR. There were 29.69% SSRs with unknown position.
Table 1.
Summary of in silico mining of unigene sequences of pea
| Parameter | Number |
| Total ESTs | 18 552 |
| Total unigene sequence searched | 10 086 |
| Total SSRs identified | 586 |
| Sequences containing one SSR | 482 |
| Sequences containing two SSRs | 42 |
| Sequences containing three SSRs | 4 |
| Sequences containing four SSRs | 2 |
| Total ESTs containing SSRs | 530 |
3.2. Frequencies and distribution of EST-SSRs
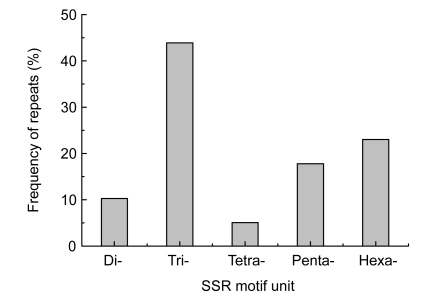
The most abundant SSRs within pea were tri-nucleotide repeats, accounting for 43.90%, followed by 23.00%, 17.77%, 10.28%, and 5.05% for hexa-, penta-, di-, and tetra-nucleotide repeats (Fig. 1). The repeat unit numbers of SSR loci were from 3 to 25. The different types of all SSR units were showed in Table 2. CT/AG was the most common motif, accounting for 23.73% among dimeric SSRs, while GAA/TTC was the most abundant type (19, 7.54%) among the tri-nucleotide repeats. Out of all repeat motifs, the length of SSR was from 14 to 50 bp, with the mean of 17.07 bp.
Fig. 1.
Frequency distribution of different repeat type (2–6 motif units) microsatellites identified in unigene sequences of pea
Table 2.
Occurrence of different type SSRs in pea ESTs
| SSR motif | Number of repeats |
Total | ||||||||||||
| 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ≥15 | ||
| Di-nucleotides | ||||||||||||||
| CT/AG | − | − | − | − | 3 | 4 | 2 | 2 | 1 | − | − | − | 2 | 14 |
| TC/GA | − | − | − | − | 5 | 2 | 1 | 2 | − | − | − | 1 | − | 11 |
| AG/CT | − | − | − | − | 2 | 2 | 4 | − | − | − | − | − | − | 8 |
| TA/TA | − | − | − | − | 1 | 3 | 1 | − | 2 | 1 | − | − | − | 8 |
| Others | − | − | − | − | 9 | 3 | 4 | 1 | 1 | − | − | − | − | 18 |
| Tri-nucleotides | ||||||||||||||
| GAA/TTC | − | − | 13 | 5 | − | 1 | − | − | − | − | − | − | − | 19 |
| CAA/TTG | − | − | 11 | 3 | − | − | − | − | − | − | − | − | − | 14 |
| ACA/TGT | − | − | 7 | 4 | − | 1 | − | − | − | − | − | − | − | 12 |
| Others | − | − | 124 | 49 | 23 | 5 | 2 | 2 | − | − | − | − | 2 | 207 |
| Tetra-nucleotides | ||||||||||||||
| TATG/CATA | − | 3 | − | − | − | − | − | − | − | − | − | − | − | 3 |
| Others | − | 23 | − | 2 | 1 | − | − | − | − | − | − | − | − | 26 |
| Penta-nucleotides | ||||||||||||||
| TTTTA/TAAAA | 5 | − | − | − | − | − | − | − | − | − | − | − | − | 5 |
| Others | 85 | 12 | − | − | − | − | − | − | − | − | − | − | − | 97 |
| Hexa-nucleotides | 111 | 17 | 2 | 2 | − | − | − | − | − | − | − | − | − | 132 |
3.3. Polymorphism of EST-SSRs
Forty-nine of the all SSR loci were selected randomly to test the polymorphism of the EST-SSRs in pea. Of these, 37 (75.51%) were found to have products with the expected size range, 5 exhibited unexpected products, and 7 had no significant products. Among the successful amplifiable primers, nine pairs showed polymorphism. The number of alleles per locus varied from 2 to 3, with the average 2.3. The PIC values varied from 0.18 to 0.58, with mean value 0.41 (Table 3).
Table 3.
Characteristics of nine polymorphic EST-derived SSRs for Pisum sativum L.
| Primer name | Accession No. of putative homology | SSR motif | Primer sequence 5′→3′ | Allele size range (bp) | Allele number | Putative function | PIC | Allele number and size range in Vicia faba L. (bp)* |
| P66 | 32542470-13; 2542362-1 | (tatt)4 | F: GCCGAGGTACAAAAGAAGT; R: CTGGAAACCAAGAAAAGTG | 323 | 2 | 0.18 | 0 | |
| P133 | 32544169-1; 32542559 | (aac)6 | F: CAATGATGGGTGGAAGATG; R: AGGCAGTGATTCAGACGGT | 337 | 2 | N-rich protein | 0.32 | 2 (336–339) |
| P248 | 32543080-1 | (ttc)7 | F: GAGCAGCATTTTGTTGGA; R: CTGGAGGAGGCTTTCATT | 178 | 2 | Sucrose transport protein SUT1 | 0.48 | 2 (178–181) |
| P251 | 32543524-1 | (gaa)5 | F: ATCCAGAACTCACAACAT; R: TAGAATCAAAACACGACC | 242 | 2 | P54 protein | 0.32 | 0 |
| P314 | 2537373-1 | (aat)7 | F: AAGAGAGGTGTGGTTCA; R: ATTTCGTTTTGGTTACG | 254 | 2 | Unnamed protein product | 0.50 | 1 (246) |
| P402 | 90646231-1 | (tc)10 | F: CAACAACACAAATCCAT; R: AGTCTCACAACAGCACC | 352 | 2 | Unnamed protein product | 0.50 | 1 (349) |
| P636 | 32542612-1 | (aac)5 | F: ATGAAGCACATGAAAAAT; R: TGGTGAGGAGGAAACTAT | 212 | 3 | Unknown protein | 0.34 | 0 |
| P1109 | 32545076-1 | (ttgat)3 | F: CTCCATCTCAAGAAATCC; R: CACATAACTAAAAAACCC | 383 | 3 | Histone H1 subtype 7 | 0.50 | 0 |
| P1188 | 90646520-1 | (gca)5 | F: CTCTCCCTTTTCATTCCAT; R: TTTCGCTTGTCTCCTTGTT | 155 | 3 | unnamed protein product | 0.58 | 0 |
Allele number and size range of cross-species amplification products in Vicia faba L.
BLASTX searches showed that seven of those polymorphic SSR-associated ESTs matched to the known genes involved in N-rich proteins, sucrose transport protein SUT1, P54 protein, histone H1, and three unnamed protein products. One represented an unknown protein and one represented a novel sequence (Table 3).
3.4. Transferability of EST-SSRs
Three faba bean cultivars were employed to perform cross species amplification in order to evaluate the transferability of the nine EST-SSRs in related species. The results revealed that four SSRs showed successful amplification and three showed polymorphism in faba bean.
4. Discussion
In this study, an abundant number of pea ESTs (18 552) obtained from NCBI were used to mine for SSRs. The results indicate that pea ESTs provide an effective resource to search for SSR markers. A total of 586 (5.25%) potential unigenes-contained microsatellite motifs were found, at a higher frequency than that previously reported in some plant ESTs (Cardle et al., 2000; Poncet et al., 2006). However, the overall frequency and the frequency of different repeat motifs might depend on not only the redundancy level of the sequence, but also the criteria and the datasets for searching for SSR (Yan et al., 2008). In general, about 5% of ESTs contained SSRs in many plant species when the minimum repeat length was set to be 20 bp (Varshney et al., 2005).
The type and abundance of different motif repeats have been reported to show variable and uneven distribution in different plants. In the present study, tri-nucleotide repeats were the most abundant, similar to previous studies with other plants, including wheat, cereal, grape, and so forth (Cardle et al., 2000; Gupta et al., 2003). The reason for the abundance of tri-nucleotide SSRs in plants might be attributed to an absence of frameshift mutations due to the variety of tri-nucleotide repeats (Metzgar et al., 2000). Among the tri-nucleotide repeats, GAA was observed most frequently in this study, in agreement with that reported on SSR in Fagaceae (Ueno et al., 2008). Interestingly, the (GAA)n SSRs were also reported in a high frequency in other plants, including Arabidopsis and soybean (Tian et al., 2004). It seems that GAA might be the most abundant EST-SSR motif in dicots.
It has been reported that EST-SSR markers show lower polymorphism compared to genomic SSR markers (Saha et al., 2006). In our research, a total of 9 EST-SSR markers have been found polymorphic from 49 EST-SSRs, giving a polymorphism rate of 18%, compared with the 72% polymorphism rate found by Burstin et al. (2001). The reason for the low level of polymorphism from EST-derived SSRs might be due to the possible selection against alterations in the conserved sequences of EST-SSRs (Scott et al., 2000). Among the nine EST-SSR markers, seven loci matched to the known genes, which were involved in different functional types of proteins. When associating with the coding regions of the genome, these EST-SSRs would be directly assisted in marker trait associations (Eujayl et al., 2002; Thiel et al., 2003).
However, the low level of polymorphism of EST-SSRs may be compensated for by their potential of interspecific transferability (Thiel et al., 2003). The transferability of EST-SSR markers has also been reported in some bean species (Gutierrez et al. 2005; Yu and Li, 2008). The SSR markers from Medicago truncatula exhibited transferability to faba bean, chickpea, and pea (Gutierrez et al., 2005). The EST-SSR markers from chickpea also showed high transferability to the six Cicer species and seven legume genera (P. mungo, P. sativum, G. max, T. alexandrinum, L. esculenta, C. cajan, and M. truncatula) (Choudhary et al., 2009). The present results show that three loci of nine polymorphic P. sativum EST-SSR markers can be amplified in faba bean, which suggests that it may assist in reducing the costs of marker development, and promote genetic analysis among the bean species.
In conclusion, the present research is the first report about the development of EST-SSRs of the pea. Because of their polymorphism and transferability, these nine novel EST-SSRs will be valuable tools for marker-assisted breeding and comparative mapping of pea in the future.
Footnotes
Project supported by the Zhejiang Provincial Science and Technology Program (No. 2007C32013) and the Zhejiang Provincial Natural Science Foundation (No. Y3090660) of China
References
- 1.Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME. Optimizing parental selection for genetic linkage maps. Genome. 1993;36(1):181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- 2.Burstin J, Deniot G, Potier J, Weinachter C, Aubert G, Baranger A. Microsatellite polymorphism in Pisum sativum . Plant Breed. 2001;120(4):311–317. doi: 10.1046/j.1439-0523.2001.00608.x. [DOI] [Google Scholar]
- 3.Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics. 2000;156:847–854. doi: 10.1093/genetics/156.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhary S, Sethy NK, Shokeen B, Bhatia S. Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor Appl Genet. 2009;118(3):591–608. doi: 10.1007/s00122-008-0923-z. [DOI] [PubMed] [Google Scholar]
- 5.Ellis JR, Burke JM. EST-SSRs as a resource for population genetic analyses. Heredity. 2007;99(2):125–132. doi: 10.1038/sj.hdy.6801001. [DOI] [PubMed] [Google Scholar]
- 6.Eujayl I, Sorrells ME, Baum M, Wolters P, Powell W. Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet. 2002;104(2-3):399–407. doi: 10.1007/s001220100738. [DOI] [PubMed] [Google Scholar]
- 7.Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, Balyan HS. Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Genet Genom. 2003;270(4):315–323. doi: 10.1007/s00438-003-0921-4. [DOI] [PubMed] [Google Scholar]
- 8.Gutierrez MV, Vaz Patto MC, Huguet T, Cubero JI, Moreno MT, Torres AM. Cross-species amplification of Medicago truncatula microsatellites across three major pulse crops. Theor Appl Genet. 2005;110(7):1210–1217. doi: 10.1007/s00122-005-1951-6. [DOI] [PubMed] [Google Scholar]
- 9.Kong Q, Xiang C, Yu Z, Zhang C, Liu F, Peng C, Peng X. Mining and charactering microsatellites in Cucumis melo expressed sequence tags from sequence database. Mol Ecol Notes. 2007;7(2):281–283. doi: 10.1111/j.1471-8286.2006.01580.x. [DOI] [Google Scholar]
- 10.Loridon K, McPhee J, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G, Rameau C, Baranger A, Coyne C, Lejeune-Hènaut I, et al. Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.) Theor Appl Genet. 2005;111(6):1022–1031. doi: 10.1007/s00122-005-0014-3. [DOI] [PubMed] [Google Scholar]
- 11.Metzgar D, Bytof J, Wills C. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10(1):72–80. doi: 10.1101/gr.10.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S, de Kochko A, Hamon P. SSR mining in coffee tree EST databases: potential use of EST-SSRs as markers for the Coffea genus. Mol Genet Genom. 2006;276(5):436–449. doi: 10.1007/s00438-006-0153-5. [DOI] [PubMed] [Google Scholar]
- 13.Powell W, Machray GC, Provan J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996;1(7):215–222. doi: 10.1016/1360-1385(96)86898-1. [DOI] [Google Scholar]
- 14.Saha MC, Cooper JD, Rouf Mian MA, Chekhovskiy K, May GD. Tall fescue genomic SSR markers: development and transferability across multiple grass species. Theor Appl Genet. 2006;113(8):1449–1458. doi: 10.1007/s00122-006-0391-2. [DOI] [PubMed] [Google Scholar]
- 15.Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ. Analysis of SSRs derived from grape ESTs. Theor Appl Genet. 2000;100(5):723–726. doi: 10.1007/s001220051344. [DOI] [Google Scholar]
- 16.Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W. How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol. 2003;12(6):1339–1348. doi: 10.1046/j.1365-294X.2003.01825.x. [DOI] [PubMed] [Google Scholar]
- 17.Tangphatsornruang S, Sraphet S, Singh R, Okogbenin E, Fregene M, Triwitayakorn K. Development of polymorphic markers from expressed sequence tags of Manihot esculenta Crantz. Mol Ecol Resour. 2008;8(3):682–685. doi: 10.1111/j.1471-8286.2007.02047.x. [DOI] [PubMed] [Google Scholar]
- 18.Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor Appl Genet. 2003;106(3):411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 19.Tian AG, Wang J, Cui P, Han YJ, Xu H, Cong LJ, Guang XG, Wang XL, Jiao YZ, Wang BJ, et al. Characterization of soybean genomic features by analysis of its expressed sequence tags. Theor Appl Genet. 2004;108(5):903–913. doi: 10.1007/s00122-003-1499-2. [DOI] [PubMed] [Google Scholar]
- 20.Ueno S, Taguchi Y, Tsumura Y. Microsatellite markers derived from Quercus mongolica var. crispula (Fagaceae) inner bark expressed sequence tags. Genes Genet Syst. 2008;83(2):179–187. doi: 10.1266/ggs.83.179. [DOI] [PubMed] [Google Scholar]
- 21.Varshney RK, Graner A, Sorrells ME. Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 2005;23(1):48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Yan QL, Zhang YH, Li HB, Wei CH, Niu LL, Guan S, Li SG, Du LX. Identification of microsatellites in cattle unigenes. J Genet Genomics. 2008;35(5):261–266. doi: 10.1016/S1673-8527(08)60037-5. [DOI] [PubMed] [Google Scholar]
- 23.Yu H, Li Q. Exploiting EST databases for the development and characterization of EST-SSRs in the pacific oyster (Crassostrea gigas) J Hered. 2008;99(2):208–214. doi: 10.1093/jhered/esm124. [DOI] [PubMed] [Google Scholar]