Abstract
The purpose of this study was to investigate the characteristics of transfer RNA (tRNA) responsible for the association between tRNA genes and genes of apparently foreign origin (genomic islands) in five high-light adapted Prochlorococcus strains. Both bidirectional best BLASTP (basic local alignment search tool for proteins) search and the conservation of gene order against each other were utilized to identify genomic islands, and 7 genomic islands were found to be immediately adjacent to tRNAs in Prochlorococcus marinus AS9601, 11 in P. marinus MIT9515, 8 in P. marinus MED4, 6 in P. marinus MIT9301, and 6 in P. marinus MIT9312. Monte Carlo simulation showed that tRNA genes are hotspots for the integration of genomic islands in Prochlorococcus strains. The tRNA genes associated with genomic islands showed the following characteristics: (1) the association was biased towards a specific subset of all iso-accepting tRNA genes; (2) the codon usages of genes within genomic islands appear to be unrelated to the codons recognized by associated tRNAs; and, (3) the majority of the 3′ ends of associated tRNAs lack CCA ends. These findings contradict previous hypotheses concerning the molecular basis for the frequent use of tRNA as the insertion site for foreign genetic materials. The analysis of a genomic island associated with a tRNA-Asn gene in P. marinus MIT9301 suggests that foreign genetic material is inserted into the host genomes by means of site-specific recombination, with the 3′ end of the tRNA as the target, and during the process, a direct repeat of the 3′ end sequence of a boundary tRNA (namely, a scar from the process of insertion) is formed elsewhere in the genomic island. Through the analysis of the sequences of these targets, it can be concluded that a region characterized by both high GC content and a palindromic structure is the preferred insertion site.
Keywords: Genomic islands, Prochlorococcus, Transfer RNA (tRNA), Palindromic structure, Codon usage
1. Introduction
The marine unicellular cyanobacterium Prochlorococcus, possibly the smallest and most abundant photosynthetic organism on the earth, dominates the euphotic zone in tropical and subtropical oligotrophic waters between 40° N and 40° S (Partensky et al., 1999; Hess et al., 2001; Tian et al., 2005; Zinser et al., 2007). Considering its wide distribution and strong adaptability, it is a suitable organism to explore microdiversity. Comparisons of the genome sequences of certain closely related Prochlorococcus strains have revealed the intimate link between their genomic divergence and adaptability to different oceanic niches (Rocap et al., 2003). Horizontal gene transfer (HGT), a process in which one organism transfers genetic material to another organism that is not its offspring (Syvanen, 1994; Koonin, 2009), plays an important role in giving rise to extremely dynamic cyanobacteria genomes (Zhaxybayeva et al., 2006).
HGT is now recognized as a major force shaping the evolutionary histories of prokaryotes (Koonin et al., 2001; Zhaxybayeva et al., 2006; Boto, 2010). In many prokaryotes, horizontal transfer genes (HTGs) contribute 1.6%–32.6% of the genes (Nelson et al., 1999; Garcia-Vallvé et al., 2000; Koonin et al., 2001; Nakamura et al., 2004; Choi and Kim, 2007). Recent studies have shown that a new type of mobile element known as a genomic island (GI), clusters of genes of apparently foreign origin in a prokaryotic genome, is acquired through HGT (Hacker and Carniel, 2001; Hsiao et al., 2003). A large amount of GIs are created by a site-specific recombination mechanism, which plays a crucial role and therefore is significantly useful in exploring the formation of GIs. Mediated by transfer RNA (tRNA) and initiated at the 3′ ends of the tRNA genes, this kind of site-specific recombination mechanism creates some short and direct repeats identical or nearly identical to the 3′ ends (Reiter et al., 1989; Cheetham and Katz, 1995; Williams, 2002; Baar et al., 2003; Tuanyok et al., 2008).
Currently, four hypotheses have been proposed from the perspectives of tRNA gene characterization on why the tRNA genes are frequently used as recombination sites of GIs. The first one holds that the complementarity of 5′ and 3′ ends of tRNA will bring about a pair of inverted repeats that presumably tend to be recognizable to an integrase, which, in turn, can integrate foreign genetic materials into genomes (Reiter et al., 1989). However, evidence has shown that in the λ phage, the distance between a pair of inverted repeats is 7 nucleotides (nt), but in tRNA it is at least 50–60 nt, a long separation that does not favor DNA recombination (Hou, 1999). Consequently, the first hypothesis is not approximate. The second hypothesis assumes that it is the multiple copies of tRNA in bacterial genomes that lead to repeated insertions of GIs into tRNA genes (Cheetham and Katz, 1995). The third hypothesis proposes that the conserved CCA end sequence at the 3′ end provides a cleavage site of initial recognition for the integrase, and after cleavage, the 3′ end of specific tRNA transcript will hybridize with one of the two disengaged DNA strands to form a stable RNA-DNA hybrid (Hou, 1999). The fourth hypothesis suggests that because a specific tRNA gene is associated with a GI, it is given preference to read the codons carried by this GI (Ritter et al., 1995).
In this study, to investigate whether tRNA genes are insertional hotspots of GIs and the cause of such insertions in Prochlorococcus strains, we identified the GIs in the most closely related five Prochlorococcus strains by multiple genomic comparisons, including Prochlorococcus marinus AS9601 (PMB), P. marinus MIT9515 (PMC), P. marinus MED4 (PMED4), P. marinus MIT9301 (PMG), and P. marinus MIT9312 (PMI), in view of cutting down the interference by large rearrangements such as translocation or inversion in the identification of GIs and maximizing the colinearity of compared genomes. Through the analysis of the tRNA genes associated with GIs in the five strains, we found that some of these tRNA genes demonstrate interesting characteristics that are in discordance with the hypotheses described above. Therefore, we discuss our observations on the basis of sequence analysis, and provide our perception of the location of real insertion site in tRNA and the cause of frequent insertion into tRNA genes as far as the five Prochlorococcus strains are concerned.
2. Materials and methods
2.1. Work platform of data analysis and sources of genomes
BioBIKE (Elhai et al., 2009), a web-based environment, was employed as our work platform. It combines a biological knowledge base, a graphical programming interface, and an extensible set of tools. The genomes used in this study and their sources are listed in Table A1 (Rocap et al., 2003; Coleman et al., 2006; Kettler et al., 2007).
2.2. Identification of Prochlorococcus GIs
We utilized a pipeline developed in BioLisp within BioBIKE to identify GIs in five Prochlorococcus strains, including PMED4, PMC, PMB, PMG, and PMI. The procedure was as follows: first, the candidate orthologs were obtained in BioBIKE through bidirectional best BLASTP (basic local alignment search tool for proteins) search with threshold E-value 10−6 (Altschul et al., 1997). Then they were analyzed through conservation of gene order, and if two candidates appear in the same order in closely related genomes, they are assigned to one orthologous group. Second, we identified the alien genes by examining whether each gene of one Prochlorococcus strain has orthologs in the genomes of the other four strains. A gene was operationally defined as alien if there were orthologous genes in at most one of the four other Prochlorococcus genomes, and a region that was composed of one or more continuous alien genes formed a GI. Therefore, 31 regions (191 genes) in PMED4, 52 regions (297 genes) in PMC, 21 regions (211 genes) in PMB, 27 regions (168 genes) in PMG, and 16 regions (208 genes) in PMI were identified as GIs (Liu and Zhu, 2010).
2.3. Monte Carlo simulation of the insertion of foreign genetic materials into the genomes
A program written in BioLisp within BioBIKE was developed to simulate the insertion of foreign genetic materials into a genome. The model for the insertion of a GI adjacent to tRNA is as follows: an insertion (X) is thrown randomly at a genome, and therefore follows a uniform distribution (X~U[1, L], where L means the length of a host genome). In every round of simulation, one insertion was shot at the host genome, and we examined whether the insertion was inside a gene, next to tRNA, or in the intergenic sequences. Excluding the insertions inside the genes (because the model supposes that they would lead to the death of the organism), we counted the number of remaining insertions that were next to tRNA genes and in the intergenic sequences, and calculated the expected ratios (f) of the former to the latter (f=N tRNA/N inter, where N tRNA is the number of insertions next to tRNA and N inter is the number of insertions in the intergenic sequences). The Chi-square (χ 2) test with one degree of freedom (df) was used to assess the significance of the insertions of GIs into tRNA genes:
| χ2=(Nobs−f∙Ntotal)2/(f∙Ntotal) |
, where N obs is the observed number of GIs associated with tRNA genes in a given genome and N total is the total number of GIs in a given genome.
2.4. Sequence search within GIs
To obtain the direct repeat sequences of the tRNA 3′ ends in GIs, we used iterative search, a type of BioBIKE function, to find all sequences related to an initial query with less than four mismatches (or 12 mismatches in the case of the longer direct repeat sequence in tRNA-Pro). For the other sequences, we used BLAST (Altschul et al., 1997; Wang et al., 2005) to search all similar sequences related to an initial query with threshold E-value 10−6.
2.5. Sequence alignment and construction of phylogenetic trees
Using program ClustalW under default settings in MEGA Version 4.0 (Tamura et al., 2007), we performed the multiple sequence alignment of the sequences within GIs and removed the unconserved regions of alignment manually. In addition, we constructed the phylogenetic trees through MEGA version 4.0 employing the neighbor-joining (NJ) method and unweighted pair group method with arithmetic mean (UPGMA), whose substitution model of nucleotide was p-distance.
2.6. Nucleic acid folding
The secondary structures of single-stranded DNA sequences were determined using the MFOLD 3.2 program (Zuker, 2003).
3. Results and discussion
3.1. tRNA as an insertion hotspot
According to our sequence analysis of the five closely related Prochlorococcus strains, there are 7 GIs immediately adjacent to tRNA in PMB, 11 in PMC, 8 in PMED4, 6 in PMG, and 6 in PMI (Table A2). The observed ratios of the GIs inserted into tRNA to those into intergenic sequences are 0.33 in PMB, 0.21 in PMC, 0.26 in PMED4, 0.22 in PMG, and 0.37 in PMI. In order to assess whether GIs appear adjacent to tRNA genes more frequently than expected by chance alone, we implemented 100 000 replications of simulation, and found that the expected ratios of the number of insertions next to tRNA genes to the number of intergenic sequences are 0.0661, 0.0600, 0.0801, 0.0568, and 0.0828 in PMB, PMC, PMED4, PMG, and PMI, respectively (Table 1). According to the χ 2 test, the observed insertions are significant at P=0.01 level, which proved that tRNA gene loci are the insertion hotspots in the genome of Prochlorococcus. Our results confirm, from a statistical perspective, many earlier observations in prokaryotes (Reiter et al., 1989; Parreira and Gyles, 2003; van Aartsen, 2008). This previous work has revealed that tRNA loci are not only central components in translation, but also commonly serve as insertion sites for mobile elements in bacteria because there is an attB (bacterial attachment site) within some tRNA genes such as Arg and Pro (Reiter et al., 1989; Semsey et al., 2002), and therefore, the presence of these tRNA genes gives rise to variable genomic regions and the observed divergence of Prochlorococcus genomes.
Table 1.
Numbers of insertions into tRNA genes in PMB, PMC, PMED4, PMG, and PMI
| Organism | TGa | Insertion hitsb |
Exp/TG | Obs/TG | |
| Exp. | Obs. | ||||
| PMB | 21 | 1.4 | 7* | 0.0661 | 0.33 |
| PMC | 52 | 3.1 | 11* | 0.0600 | 0.21 |
| PMED4 | 31 | 2.5 | 8* | 0.0801 | 0.26 |
| PMG | 27 | 1.5 | 6* | 0.0568 | 0.22 |
| PMI | 16 | 1.3 | 6* | 0.0828 | 0.37 |
TG is the total GIs identified in a given genome
The number of insertions associated with tRNA in the given genome. The expected number (Exp.) is the number of GIs that would arise if the random insertions were associated with tRNA in the given genome. The observed number (Obs.) is the number of GIs associated with tRNA in the given genome
Statistical significance to the level P<0.01 in χ 2 test
3.2. Characteristics discordant with three hypotheses in the case of tRNA genes associated with GIs
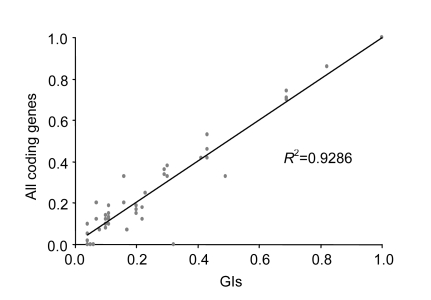
When we observed the inserted sites of the GIs in the five Prochlorococcus strains, we found that the GIs associated with specific tRNA genes (tRNA-Ala, tRNA-Arg, tRNA-Pro, and tRNA-Thr) favor to insert into certain instead of all iso-accepting tRNA genes, although they are homologous (Table 2). We also found that in a total of 16 tRNA genes associated with GIs, 11 do not have CCA ends at their 3′ ends. That is, the second and the third hypotheses mentioned above can hardly explain these observations. We further computed two kinds of codon usages that are defined as the ratio of the number of occurrences of a codon corresponding to tRNA associated with GIs to the sum of all synonymous codons in genomes and GIs (Xu et al., 2008). The results of our computation showed a strong positive correlation (R 2=0.93) in the codon usages between genomes and GIs (Fig. 1). At the same time, most of the codons corresponding to the tRNA associated with GIs are rarely used in GIs and genomes. These findings are inconsistent with the fourth hypothesis which proposes that the codons corresponding to these tRNA genes associated with GIs tend to be used within GI genes.
Table 2.
Total number of GI insertions into the tRNA genes corresponding to the same codons in the five strains
| tRNA | Iso-accepting tRNAa | Numberb | Insertion hitsc |
|
| Exp. | Obs. | |||
| Ala* | Ala-GCC | 5 | 2.50 | 5 |
| Ala-GCA | 5 | 2.50 | 0 | |
| Arg* | Arg-CGT | 5 | 0.75 | 0 |
| Arg-CGG | 5 | 0.75 | 0 | |
| Arg-AGG | 5 | 0.75 | 0 | |
| Arg-AGA | 5 | 0.75 | 3 | |
| Asn | Asn-AAC | 5 | 3.00 | 3 |
| Cys | Cys-TGC | 5 | 1.00 | 1 |
| Gly | Gly-GGC | 5 | 0.50 | 1 |
| Gly-GGA | 5 | 0.50 | 0 | |
| Leu | Leu-CTT | 5 | 0.25 | 0 |
| Leu-TTG | 5 | 0.25 | 0 | |
| Leu-TTA | 5 | 0.25 | 1 | |
| Leu-CTA | 5 | 0.25 | 0 | |
| Lys | Lys-AAA | 5 | 1.00 | 1 |
| Met | Met-ATG | 13 | 3.00 | 3 |
| Phe | Phe-TTC | 5 | 1.00 | 1 |
| Pro* | Pro-CCC | 5 | 2.50 | 0 |
| Pro-CCA | 5 | 2.50 | 5 | |
| Ser | Ser-TCG | 5 | 2.75 | 4 |
| Ser-AGC | 5 | 2.75 | 3 | |
| Ser-TCC | 5 | 2.75 | 0 | |
| Ser-TCA | 5 | 2.75 | 4 | |
| Thr* | Thr-ACG | 5 | 1.75 | 2 |
| Thr-ACC | 10 | 3.50 | 5 | |
| Thr-ACA | 5 | 1.75 | 0 | |
| Tyr | Tyr-TAC | 5 | 5.00 | 5 |
The iso-accepting tRNA in the five strains. The letters in front of a hyphen stand for the amino acid carried by tRNA, and the letters behind a hyphen stand for the codon corresponding to tRNA
The total number of each iso-accepting tRNA in the five strains
The number of insertions in the given iso-accepting tRNA. The expected number (Exp.) is the number of the random insertions in the given iso-accepting tRNA. The observed number (Obs.) is the number of GIs associated with the given iso-accepting tRNA
Statistical significance to the level P<0.05 in χ 2 test
Fig. 1.
Codon usages corresponding to the tRNA genes associated with the GIs among both GIs and genomes
3.3. Determination of insertion sites of GIs associated with tRNA
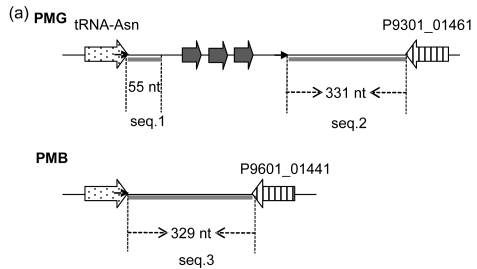
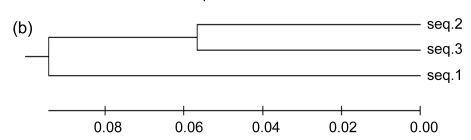
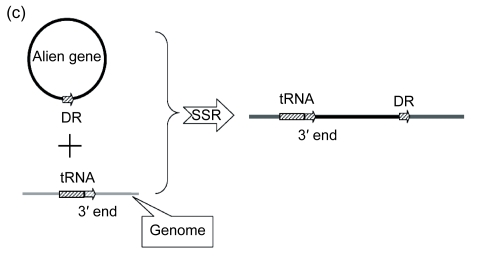
The mechanism of the introgression of foreign genes into host genomes can provide an important clue to the determination of insertion sites. In some cases, the 3′ end sequence of a boundary tRNA is repeated elsewhere in GIs, always as a direct repeat; therefore, it assists one to probe into the underlying mechanisms. Here, we took the GI associated with tRNA-Asn in PMG as an example (Fig. 2a). There are three genes present, including P9301_01431, P9301_01441, and P9301_001451. To obtain the remnant information of the GI insertion into tRNA-Asn, we analyzed the regions flanking the GI (namely seq.1 and seq.2 in Fig. 2a) and found that seq.1, seq.2, and seq.3 in PMG and PMB are homologous. As is shown in Fig. 2b, if seq.1 was native, seq.2 should have been more homologous with it than with seq.3, but in fact, seq.2 has a similarity of 72.4% with it, and 83% with seq.3. That is, the sequence between the 3′ end of tRNA-Asn and its direct repeat comes from some other organism, which has a segment similar to seq.2. This suggests that foreign genetic materials are introduced into the host genomes by site-specific recombination using the 3′ end of tRNA as the target and the direct repeats are generated at the time the GIs are formed (Fig. 2c).
Fig. 2.
Schematic representation of the insertion of foreign genetic materials into tRNA by site-specific recombination exemplified by the GI between tRNA-Asn and P9301_01461 in PMG
(a) Schematic representation of the GI between tRNA-Asn and P9301_01461. The location and orientation of the genes flanking the GI are indicated by spotted and striped thick arrows, and the corresponding orthologs are indicated by the same pattern. Gray thick arrows indicate the genes (left to right: P9301_01431, P9301_01441, P9301_01451) within the GI. Direct repeats are indicated by thin arrows. Gray thick lines indicate homologous sequences. Seq.1, seq.2, and seq.3 indicate homologous sequences; (b) The analyses of similarities of seq.1, seq.2, and seq.3 using UPGMA method provided by MEGA 4.0. The scale is in nucleotide substitutions per site; (c) Schematic representation of the insertion of foreign genetic materials into host genome. DR: direct repeat; SSR: site-specific recombination
3.4. Characteristics of the 3′ end of the tRNA related with GI insertion
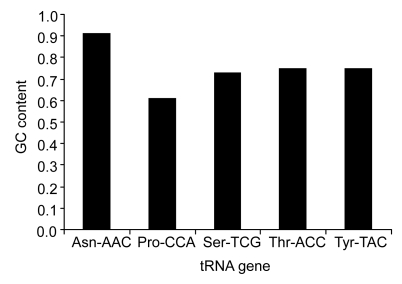
We analyzed the tRNA genes (Asn-AAC, Pro-CCA, Ser-TCG, Thr-ACC, Tyr-TAC, and Cys-TGC) in the five Prochlorococcus strains and found a general characteristic: high GC contents at all the 3′ ends inserted by GIs (Fig. 3).
Fig. 3.
GC contents of the repeated sequences in tRNA genes associated with GIs
The letters in front of a hyphen stand for the amino acid carried by tRNA, and the letters behind a hyphen stand for the codon corresponding to tRNA
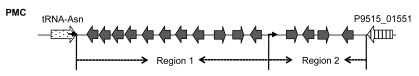
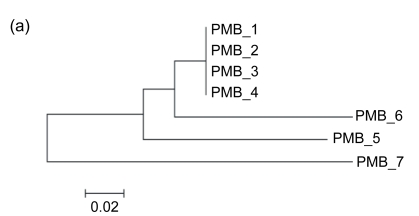
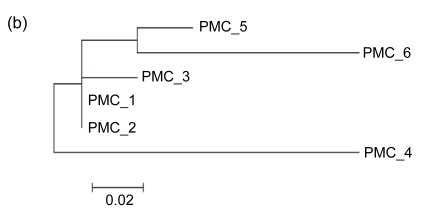
We also noticed that, although the direct repeats are identical with the 3′ ends and therefore have high GC contents, they are not preferred in the insertion. According to our observations, the GI associated with tRNA-Asn in PMC is separated by a direct repeat into two regions, which means that it is formed by the foreign materials acquired from two insertions (Fig. 4). We determined the time order of the insertions so as to make out whether the target site is the 3′ end or its direct repeat. In general, due to their low selective pressure, intergenic sequences should reflect the DNA composition of the donor and the host genomes more explicitly than the sequences of coding genes. Our calculations showed that the GC fractions of the intergenic sequences in Region 1, Region 2, and genome are 29.9%, 18.2%, and 22.7%, respectively. Also, in order to get an impression of the variability in GC measurments, we carried out 100 replications of simulation, joining all intergenic sequences in genome and calculating a randomly selected 782 nt within them. The mean GC content of the 100 simulations was 22.6%, with a standard deviation of 3.2%. The expected GC fraction in the genome deviates far more from Region 1 than from Region 2. It is well-known that the earlier a foreign genetic material introgresses, the more similar its DNA composition is to the host genome due to its amelioration in the recipient organism. Therefore, Region 2 was inserted into the recipient genome earlier than Region 1. The time order implicated that the 3′ end is the target site, and otherwise Region 1 would have been inserted earlier. Moreover, according to the phylogenetic trees, the direct repeats closer to tRNA-Pro are more similar to the 3′ end sequence of tRNA-Pro (Figs. 5–7). It also shows that the 3′ ends are preferred in insertions, because the earlier an insertion occurs, the higher degree of mutation the direct repeat formed by the insertion demonstrates due to the evolutionary force. In other words, only when a 3′ end is taken as the insertion target will its direct repeats form a pattern of ascending order in terms of the similarity to it.
Fig. 4.
Schematic representation of the GI between tRNA-Asn and P9515_01551 in PMC
The location and orientation of genes flanking the GI are indicated by thick arrows. The genes within the GI are indicated by gray thick arrows. Direct repeats are indicated by thin arrows. The GI is separated by the direct repeats into two regions (Region 1 and Region 2)
Fig. 5.
Schematic representations of the GIs associated with tRNA-Pro in PMB, PMC, PMED4, PMG, and PMI
The flanking genes are labeled with names in the figure. The location and orientation of the genes flanking the GIs are indicated by spotted and striped thick arrows, and the corresponding orthologs are indicated by the same pattern. Gray thick arrows indicate genes within GIs. Direct repeats are indicated by thin arrows. “1” indicates the position of the repeated sequences of 3′ ends of tRNA genes, and the other numbers indicate the positions of the direct repeats
Fig. 7.
Phylogenetic tees deduced from the 3′ ends of tRNA-Pro genes associated with GIs and their direct repeats using NJ method provided by MEGA 4.0 in PMB (a), PMC (b), PMED4 (c), PMG (d), and PMI (e), respectively
The positions of these 3′ ends and their direct repeats are shown in Fig. 5. The scale is in nucleotide substitutions per site
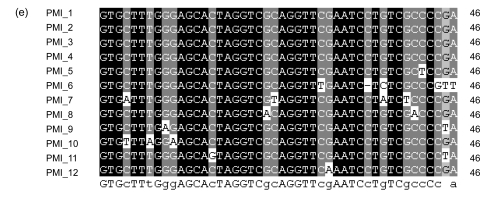
Fig. 6.
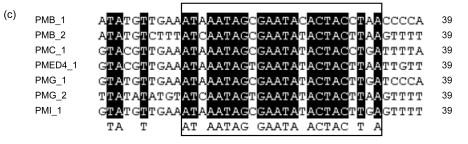
Alignments of the 3′ ends of tRNA-Pro genes associated with GIs and their direct repeats in PMB (a), PMC (b), PMED4 (c), PMG (d), and PMI (e), respectively
The positions of these 3′ ends and their direct repeats are shown in Fig. 5
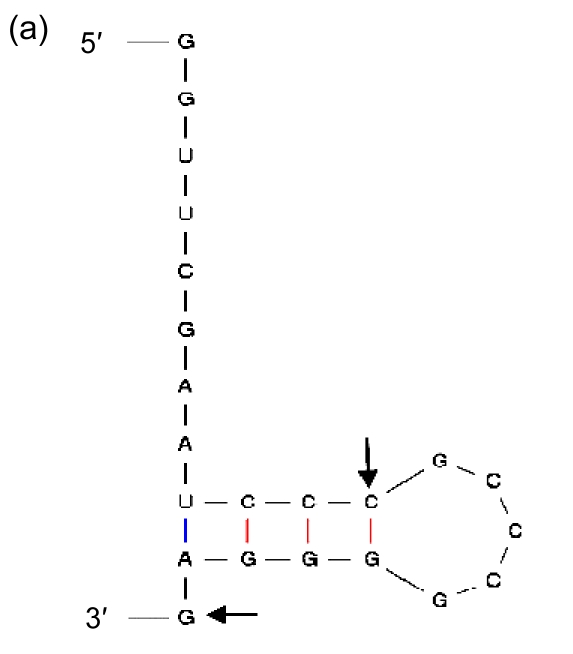
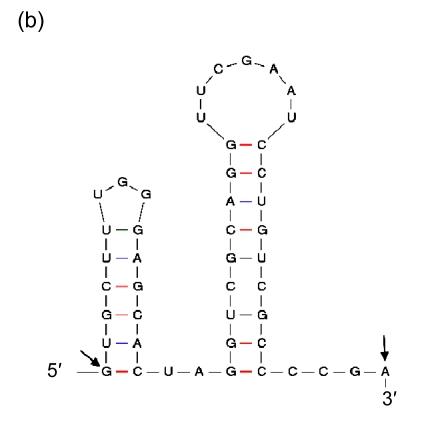
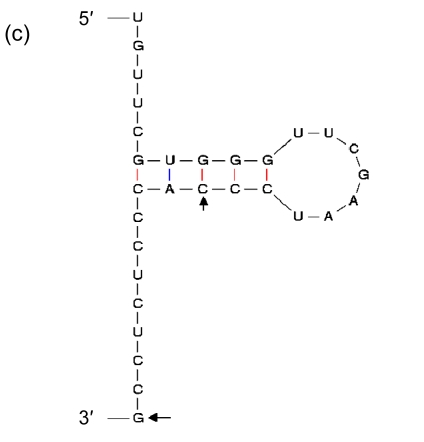
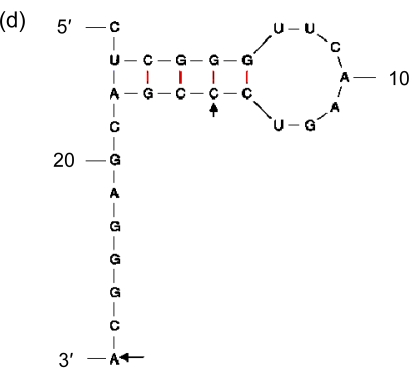
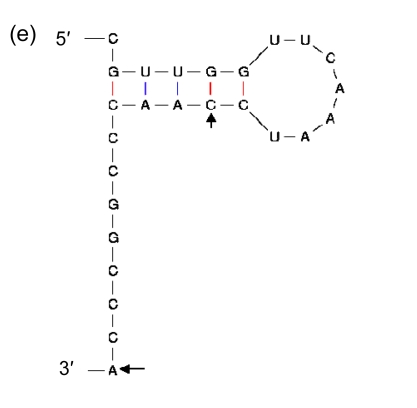
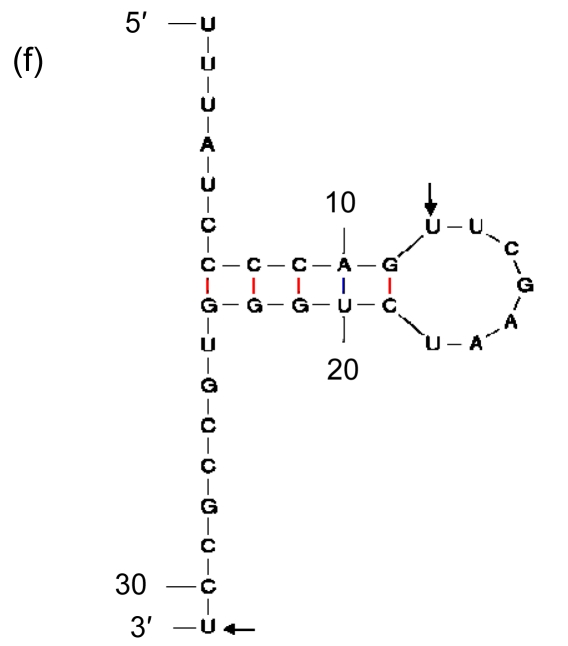
Being the duplications of a 3′ end of tRNA, direct repeats are not preferred in insertions. It suggests that there should be more elements than high GC contents that affect the process. Therefore, we included in our analysis the sequences immediately in front of the segments of the 3′ ends that are repeated elsewhere in GIs, and found indeed the second general characteristic: they have palindromic regions (Fig. 8). As the sequence of high GC content can form a stable DNA-DNA hybridization in recombination, and the palindromic structure can bind an integrase, we presume that the palindromic regions, whose ends are adjacent to the sequences of high GC contents, are the real insertion sites of GIs in Prochlorococcus.
Fig. 8.
Secondary structures of the 3′ ends of tRNA
(a) Asn-AAC; (b) Pro-CCA; (c) Ser-TCG; (d) Thr-ACC; (e) Tyr-TAC; (f) Cys-TGC. The letters in front of a hyphen stand for the amino acid carried by tRNA, and the letters behind a hyphen stand for the codon corresponding to tRNA. The segment between two arrows is identical with the direct repeat
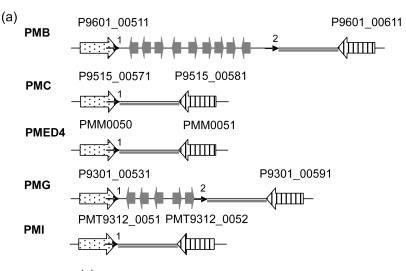
To confirm our supposition, we analyzed the GIs that are not flanked by tRNA, including the region between P9601_00511 and P9601_00611 in PMB, and its counterparts in PMC, PMED4, PMG, and PMI (Fig. 9a). In P9601_00511 and its orthorlogs P9515_00571, PMM0050, P9301_00531, and PMT9312_0051, we found a palindromic structure at each of their 3′ ends (Fig. 9b), but only the 3′ ends of P9601_00511 and P9301_00531 are adjacent to an intergeneic sequence of high GC content (“CCCA” and “TCCCA” respectively) (Fig. 9c). It is in these two genes that the insertion of foreign genetic materials happens. Having both high GC content and palindromic structure is a necessary condition for a real insertion site. As we all know, there is a great tendency of mutation from base “C” into “T”. If a sequence of high GC content lies within a gene such as tRNA, it is not likely to mutate from “G or C” to “A or T”, and therefore can undergo repeated insertion. On the contrary, an intergentic sequence, due to its high mutation rate, can hardly be inserted.
Fig. 9.
GIs between the homologs of P9601_00511 and P9601_00611 in five Prochlorococcus strains
(a) Schematic representation of the GIs between the homologs of P9601_00511 and P9601_00611 in five Prochlorococcus strains. The location and orientation of the genes flanking the GIs are indicated by spotted and striped thick arrows, and the corresponding orthologs are indicated by the same pattern. The genes within the GIs are indicated by gray thick arrows. Direct repeats are indicated by thin arrows. “1” indicates the position of the repeated sequences of the 3′ ends of genes, and the other numbers indicate the positions of the direct repeats. Gray lines indicate homologous sequences; (b) The secondary structure of the 3′ end of P9601_00511. The segment between two arrows is identical with the direct repeat; (c) The alignment of the 3′ end of P9601_00511 and its orthologs in PMC, PMED4, PMG, and PMI and their direct repeats. The 3′ ends and the direct repeats are in the box. The regions in the right of the box are intergenic sequence
4. Conclusions
GIs that confer fitness on an organism to occupy a particular ecological niche are horizontally transferred sequences. The tRNA loci usually serve as the target site for GI integration. Evidence shows that four different hypotheses have been proposed to elucidate the mechanism of the insertion of GIs into tRNA, thoroughly but insufficiently. We consequently propose that the real insertion site of GIs prefers the region characterized by a palindromic structure adjacent to a sequence of high GC content, and as the 3′ end of a conserved tRNA gene can maintain this property, it can be inserted repeatedly.
Acknowledgments
We thank Prof. Jeff ELHAI in Virginia Commonwealth University, Richmond, USA, for helpful comments and insights, and Ms. Xin XU in Chengdu University of Information Technology, Chengdu, China, and Dr. Guo-bo CHEN in Zhejiang University, Hangzhou, China, for their helps with the manuscript.
Appendix
Table A1.
Characteristics of organisms used in this study
| Organism | Abbreviation | Light adaptation | Size (Mb) | Gene number | Source |
| Prochlorococcus MED4 | PMED4 | HL(I) | 1.66 | 1757 | GI:33772317 |
| Prochlorococcus MIT9515 | PMC | HL(I) | 1.7 | 1948 | GI:123965234 |
| Prochlorococcus AS9601 | PMB | HL(II) | 1.67 | 1964 | GI:123967536 |
| Prochlorococcus MIT9301 | PMG | HL(II) | 1.64 | 1949 | GI:126695337 |
| Prochlorococcus MIT9312 | PMI | HL(II) | 1.71 | 1855 | GI:78778385 |
The source of genome sequence is the National Center for Biotechnology Information (NCBI), with the given accession number. The other source is Joint Genome Institute (JGI). The HL represents high-light-adapted ecotypes
Table A2.
GIs and the tRNA genes associated with them in PMB, PMC, PMED4, PMG, and PMI
| GI | tRNA* |
| PMB | |
| P9601_04001-P9601_04371 | Thr-ACC, Tyr-TAC |
| P9601_12291-P9601_12581 | Pro-CCA |
| P9601_13391-P9601_13511 | Ser-TCG |
| P9601_13891-P9601_14601 | Arg-AGA, Ala-GCC |
| P9601_17001 | Thr-ACG |
| P9601_17311-P9601_17341 | Ser-AGC |
| P9601_17951 | Cys-TGC |
| PMC | |
| P9515_01401-P9515_01541 | Asn-AAC |
| P9515_04071-P9515_04481 | Thr-ACC, Tyr-TAC |
| P9515_06341 | Leu-TTA |
| P9515_07221-P9515_07291 | Ser-TCA |
| P9515_08881-P9515_08951 | Met-ATG |
| P9515_12091 | Lys-AAA |
| P9515_12161-P9515_12401 | Pro-CCA |
| P9515_13301 | Ser-TCG |
| P9515_13681-P9515_14201 | Arg-AGA, Ala-GCC |
| P9515_17061-P9515_17081 | Ser-AGC |
| P9515_18311-P9515_18321 | Gly-GGC |
| PMED4 | |
| PMM0126 | Asn-AAC |
| PMM0362-PMM0386 | Thr-ACC, Tyr-TAC |
| PMM0657-PMM0659 | Ser-TCA |
| PMM0813-PMM0820 | Met-ATG |
| PMM0859-PMM0861 | Thr-ACG, Met-ATG |
| PMM1123-PMM1127 | Pro-CCA |
| PMM1162-PMM1163 | Ser-TCG |
| PMM1197-PMM1261 | Arg-AGA, Ala-GCC |
| PMG | |
| P9301_01431-P9301_01451 | Asn-AAC |
| P9301_03991-P9301_04061 | Thr-ACC, Tyr-TAC |
| P9301_06841-P9301_07091 | Ser-TCA |
| P9301_12301-P9301_12651 | Pro-CCA |
| P9301_13541-P9301_13601 | Ser-TCG |
| P9301_14341-P9301_14431 | Ala-GCC |
| PMI | |
| PMT9312_0312-PMT9312_0315 | Phe-TTC |
| PMT9312_0366-PMT9312_0382 | Thr-ACC, Tyr-TAC |
| PMT9312_0657-PMT9312_0659 | Ser-TCA |
| PMT9312_1132-PMT9312_1205 | Pro-CCA |
| PMT9312_1317-PMT9312_1355 | Ala-GCC |
| PMT9312_1619 | Ser-AGC |
The letters in front of a hyphen stand for the amino acid carried by tRNA, and the letters behind a hyphen stand for the codon corresponding to tRNA
Footnotes
Project (No. 2006AA10A102) supported by the National High-Tech R & D Program (863) of China
References
- 1.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baar C, Eppinger M, Raddatz G, Simon J, Lanz C, Klimmer O, Nandakumar R, Cross R, Rosinus A, Keller H, et al. Complete genome sequence and analysis of Wolinella succinogenes . PNAS. 2003;100(20):11690–11695. doi: 10.1073/pnas.1932838100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boto L. Horizontal gene transfer in evolution: facts and challenges. Proc R Soc B. 2010;277(1683):819–827. doi: 10.1098/rspb.2009.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheetham BF, Katz ME. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18(2):201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 5.Choi IG, Kim SH. Global extent of horizontal gene transfer. PNAS. 2007;104(11):4489–4494. doi: 10.1073/pnas.0611557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF, Chisholm SW. Genomic islands and the ecology and evolution of Prochlorococcus . Science. 2006;311(5768):1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 7.Elhai J, Taton A, Massar JP, Myers JK, Travers M, Casey J, Slupesky M, Shrager J. BioBIKE:a Web-based, programmable, integrated biological knowledge base. Nucleic Acids Res. 2009;37(web server):W28–W32. doi: 10.1093/nar/gkp354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Vallvé VS, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 2000;10(11):1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Rep. 2001;2(5):376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hess WR, Rocap G, Ting CS, Larimer F, Stilwagen S, Lamerdin J, Chisholm SW. The photosynthetic apparatus of Prochlorococcus: insights through comparative genomics. Photosynth Res. 2001;70(1):53–71. doi: 10.1023/A:1013835924610. [DOI] [PubMed] [Google Scholar]
- 11.Hou YM. Transfer RNAs and pathogenicity islands. Trends Biochem Sci. 1999;24(8):295–298. doi: 10.1016/S0968-0004(99)01428-0. [DOI] [PubMed] [Google Scholar]
- 12.Hsiao W, Wan I, Jones SJ, Brinkman FSL. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics. 2003;19(3):418–420. doi: 10.1093/bioinformatics/btg004. [DOI] [PubMed] [Google Scholar]
- 13.Kettler GC, Martiny AC, Huang K, Zuker J, Coleman ML, Rodrigue S, Chen F, Lapidus A, Ferriera S, Johnson J, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus . PLoS Genet. 2007;3(12):e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin EV. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009;37(4):1011–1034. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55(1):709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu HL, Zhu J. Identification of genomic islands in the genomes of five Prochlorococcus strains by multiple genomic comparison. J Zhejiang Univ (Agric & Life Sci) 2010;36(5):473–484. [Google Scholar]
- 17.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36(7):760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 18.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritime . Nature. 1999;399(6734):323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 19.Parreira VR, Gyles CL. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect Immun. 2003;71(9):5087–5096. doi: 10.1128/IAI.71.9.5087-5096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63(1):106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiter WD, Palm P, Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989;17(5):1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritter A, Blum G, Emody L, Kerenyi M, Bock A, Neuhieri B, Rabsch W, Scheutz F, Hacker J. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic Escherichia coli . Mol Microbiol. 1995;l7(1):109–121. doi: 10.1111/j.1365-2958.1995.mmi_17010109.x. [DOI] [PubMed] [Google Scholar]
- 23.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424(6952):1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 24.Semsey S, Blaha B, Koles K, Orosz L, Papp PP. Site-specific integrative elements of rhizobiophage 16-3 can integrate into proline tRNA (CGG) genes in different bacterial genera. J Bacteriol. 2002;184(1):177–182. doi: 10.1128/JB.184.1.177-182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Syvanen M. Horizontal gene transfer: evidence and possible consequences. Annu Rev Genet. 1994;28(1):237–261. doi: 10.1146/annurev.ge.28.120194.001321. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 27.Tian YJ, Yang H, Wu XJ, Li DT. Molecular analysis of microbial community in a groundwater sample polluted by landfill leachate and seawater. J Zhejiang Univ-Sci B. 2005;6(3):165–170. doi: 10.1631/jzus.2005.B0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanum V, Brettin TS, Nierman WC, Peacock SJ, et al. Genomic islands from five strains of Burkholderia pseudomallei . BMC Genomics. 2008;9(1):566. doi: 10.1186/1471-2164-9-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Aartsen JJ. The Klebsiella pheV tRNA locus: a hotspot for integration of alien genomic islands. Biosci Horiz. 2008;1(1):51–60. doi: 10.1093/biohorizons/hzn006. [DOI] [Google Scholar]
- 30.Wang XS, Zhu J, Mansueto L, Bruskiewich R. Identification of candidate genes for drought strees tolerance in rice by the integration of a genetic (QTL) map with the rice genome physical map. J Zhejiang Univ-Sci B. 2005;6(5):382–388. doi: 10.1631/jzus.2005.B0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams KP. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30(4):866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu XZ, Liu QB, Fan LJ, Cui XF, Zhou XP. Analysis of synonymous codon usage and evolution of begomoviruses. J Zhejiang Univ-Sci B. 2008;9(9):667–674. doi: 10.1631/jzus.B0820005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhaxybayeva O, Gogarten JP, Charlebois RL, Doolittle WF, Papke RT. Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events. Genome Res. 2006;16(9):1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zinser ER, Johnson ZI, Coe A, Karaca E, Veneziano D, Chisholm SW. Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr. 2007;52(5):2205–2220. [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]