Abstract
Tea catechin is one of the compounds that are closely related to obesity and insulin sensitivity. In order to determine the effect of catechin on adipocyte differentiation, we treated 3T3-L1 preadipocytes with different kinds of catechins. Our results showed that catechins, especially epigallocatechin gallate (EGCG), significantly reduced intracellular lipid accumulation and repressed the activity of glycerol-3-phosphate dehydrogenase, an enzyme involved in lipid synthesis. Furthermore, glucose and fatty acid transport were also suppressed by catechin. We then analyzed the activity of transcription factors—forkhead transcription factor class O1 (FoxO1) and sterol regulatory element-binding protein-1c (SREBP1c)—which are involved in adipocyte differentiation and lipid synthesis, respectively. The transcriptional activities of both these factors significantly decreased by EGCG. Western blot analysis revealed that EGCG induced the insulin signal-mediated phosphorylation of FoxO1 (Thr24, Ser256). These results suggest that EGCG suppresses the differentiation of adipocytes through the inactivation of FoxO1 and SREBP1c.
Keywords: Catechin, 3T3-L1, FoxO1, SREBP1c, Insulin signaling
Introduction
The recent increase in the number of patients with lifestyle-related diseases such as high blood pressure, hyperlipemia, cancer, heart disease, diabetes due to obesity, and metabolic syndromes related to eating habits has become a problem in developing countries (Deitel 2003). Obesity is a complex disorder caused by both genetic and environmental factors. Although the correlation between obesity and insulin sensitivity should be well understood, so far, little is known. Hormones such as insulin and insulin-like growth factor 1 (IGF-1) conjunct with the IGF-1 receptor and activate the insulin signaling cascade; this in turn induces phosphatidylinositol-3 kinase (PI3 K) to activate Akt (protein kinase B) via insulin receptor substrates (IRSs) (Longo and Finch 2003). Forkhead transcription factor class O (FoxO) proteins have highly conserved phosphorylation sites (Thr24, Ser256, and Ser-319 in human FoxO1) (Nakae et al. 1999) that are targets of phosphorylation by Akt. Phosphorylated FoxO is transported from the nucleus to the cytosol, resulting in its eventual inactivation (Muslin and Xing 2000). The FoxO family is composed of the subtypes FoxO1, FoxO3, and FoxO4 (Ogg et al. 1997). In a recent study, Nakae et al. reported that FoxO1 specially inhibits adipocyte differentiation via insulin signaling (Nakae et al. 2003), whereas we revealed that knockdown of FoxO1 mRNA expression markedly suppressed adipocyte differentiation (Munekata and Sakamoto 2009). Those results suggest that FoxO1 plays an essential role in adipocyte differentiation. Moreover, it is known that reactive oxygen species (ROSs) interfere with insulin signaling and causes apoptosis via FoxO activation (Hansen et al. 1999).
Sterol regulatory element-binding proteins (SREBPs) regulate the expression of genes that control fatty acid and cholesterol synthesis. SREBPs are composed of 3 subtypes: SREBP1a (Christenson et al. 2001), SREBP1c (Tontonoz et al. 1993), and SREBP2 (Hua et al. 1993). SREBP1c prevents fatty acid synthesis, SREBP2 prevents cholesterol formation, and SREBP1a prevents both cholesterol formation and fatty acid synthesis. SREBP1c, in particular, is expressed remarkably in the liver and adipocytes and is regulated by nutritional state. Generally, the SREBP1c-SCAP complex is transported to the Golgi complex in response to stimulation by insulin or glucose (Nohtufft et al. 1998). In addition, activated SREBP also promotes the expression of glucose and fatty acid synthesis enzymes. We also revealed the fact that ROSs accelerate SREBP1c transcriptional activity (Sekiya et al. 2008).
In general, ROSs are involved in a variety of physiological cell processes. Low levels of ROSs induce the expression of ROS scavenger enzymes (catalase, glutathione peroxidase, or super oxide dismutase) as a cellular antioxidant protective mechanism (Wang et al. 2004). High levels of ROSs cause cancer, aging, and neurodegenerative disorders by damaging cells and DNA (Hussain et al. 2003; Martindale and Holbrook 2002). Moreover, ROSs act as second messengers that regulate the function of the target proteins through phosphorylation and oxidation of cysteine residues (Cross and Templeton 2006). In addition, it is known that ROSs mediate dephosphorylation of Akt, causing apoptosis (Cao et al. 2009). Further, catechins are polyphenols that possess antioxidant activities. EGCG is the strongest antioxidant in green tea polyphenols and prevents neurodegenerative diseases (Mendel and Youdim 2004), streptozotocin-induced diabetes (Song et al. 1999), cancer (Ahmad and Mukahtar 1999; Liao et al. 2001; Lin et al. 1999; Mitcher et al. 1997; Yang and Wang 1993), and collagen-induced arthritis (Haqqi et al. 1999) that are caused by oxidative stress. In addition, EGCG can reduce body weight and body fat. Injection of EGCG into rats lowers both blood glucose and insulin levels (Kao et al. 2000). EGCG may also participate in insulin signaling because superoxide scavengers such as N-acetylcysteine (NAC) induce the activation of PI3K (Waltner-Law et al. 2002).
Generally, adipocytes play a central role in maintaining energy balance by storing triacylglyceroles (TGs) and releasing free fatty acids (FFAs) (Fruhbeck et al. 2001). The mouse preadipocyte cell line, 3T3-L1, has the capability to differentiate into adipocytes when treated with dexamethasone, 3-iso-butyl-1-methylxanthine, and insulin (DMI) (Rubin et al. 1978). It is easy to determine the molecular mechanism of catechin by analyzing the mechanism of differentiation in detail.
In this report, we aimed to determine the physiological function of EGCG and elucidate its molecular mechanism underlying adipocyte differentiation via the insulin signaling cascade. In particular, we focused on the transcription factors FoxO and SREBP1c that participate in adipocyte differentiation and lipid synthesis.
Materials and methods
Cathechins
(−) Epigallocatechin gallate (EGCG), (−) Epicatechin gallate (ECG), (−) Epigallocatechin (EGC), (−) Catechin gallate (CG) were provided from Mitsui Norin (Minato, Tokyo, Japan). The catechins used in this study were dissolved in methanol and stored at −20 °C.
Cell culture
The 3T3-L1 cells (Health Science Research Resources Bank, Sennan, Osaka, Japan) were cultured at 37 °C in Dulbecco’s Modified Eagle’s Medium (DMEM High-glucose) supplemented with 10% FBS (Sanko Junyaku, Chiyoda, Tokyo, Japan). After the confluence, cells were cultured for 2 days with 0.25 μM Dexamethason (Sigma–Aldrich), 0.5 mM 3-iso-butyl-1-methylxanthine (Sigma–Aldrich) and 10 μg/mL insulin (Wako, Chuo, Osaka, Japan) (DMI induction). Then cells were cultured in medium containing 5 μg/mL insulin for 2 days, and in standard culture medium for 4 days (Rubin et al. 1978).
Oil red O staining
Cells were cultured in medium containing DMI and catechin (EGCG, ECG) (0, 50, 100, 200 μM) for 2 days and in standard culture medium for 6 days. Cells were fixed with 4% paraformaldehyde for 1 h, and then stained with 3 mg/mL Oil red O (in 60% isopropanol) for 10 min. After washing, cells were observed under microscope [DMIRBE M2FLIII (Leica Microsystems Inc., Bannockburn, IL, USA)]. In addition to the aforementioned, the dye was eluted for 10 min with 100% isopropanol. The concentration of the eluted dye was determined from measurements of absorbance (OD 420 nm).
Triglyceride assay
Cells were lysed in lysis buffer (20 mM HEPES [pH 7.6], 420 mM NaCl, 1% Triton X-100, 0.1% SDS) and total fat was extracted by Bligh and Dyer method (Bligh and Dyer 1959). The cell extract (600 μL) was incubated in 2 mL methanol and 1 mL chloroform for 1 h, and then 1 mL chloroform and 1 mL of sterile water were added, centrifuged briefly to collect the chloroform phase. This extract was dried for overnight, and was dissolved in 10% trition-isopropanol solution. According to manual of triglyceride E-test Wako (Wako Chuo, Osaka, Japan), the quantity of triglyceride was measured. The quantity of triglycerides (μg/μL) was normalized by each protein contents.
GPDH assay
Cells were treated with supersonicator (UP-50H, B. Braun Biotech International GmbH, Melsungen, Germany) for 2 min, and the cell extract was collected after centrifugation (4 °C, 21,206g, 10 min, MX-100, Tomy, Tokyo, Japan). The protein concentration was quantified using the BCA protein assay kit (Pierce, Rockford, IL, USA). The protein (100 μg) was added to TAE solution (0.5 M Tris [pH 8.0], 10 mM EDTA, 10 mM β-mercaptoethanol), 5 mM dihydroxyacetone phosphate, 0.5 mM NADH, dDW at 37 °C, 20 min, 65 °C, 5 min and analyzed by measuring OD 340 nm.
MTT assay
Cells cultured in DMEM medium were treated with catechin (EGCG, ECG) (0, 10, 50, 100, 200 μM) for 2 days and then treated by 5 mg/mL MTT (3-(4, 5-Dimetyl-2-thiazolyl)-2, 5-diphenyltetrazoliumbromide) solution (Sigma) for 3 h. After cells were dissolved in 0.04 N HCl (in isopropanol), formazane level was analyzed by measuring OD 570 nm (against OD 630 nm).
Glucose transport
Cells were cultured in medium containing DMI induction and catechin (EGCG, ECG 100 μM) for 2 days. After 6 days culture, cells were treated with KRP-H buffer (131.2 mM NaCl, 4.7 mM KCl, 2.5 mM NaH2PO4, 2.5 mM CaCl2, 1 mM MgSO2, 1 mM HEPES [pH 7.4] containing 10 μM 2-NBDG (2-(N-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl) amino)-2-deoxyglucose) (Invitrogen Life Technologies, Carlsbad, CA, USA) at 37 °C for 5 min. After washing the cells with KRP-H buffer, cells were observed under a LEITZ DMRXA HC RXA-6 fluorescence microscope (Leica).
Fatty acid transport
Cells were cultured in a medium containing DMI and catechin (EGCG, ECG 100 μM) for 2 days. After 6 days culture, cells were treated with 20 μM BODIPY3823 (Invitrogen) and 20 μM BSA (fatty acid free) at 37 °C for 2 min. After 20 μM BSA treatment, cells were observed under a LEITZ DMRXA HC RXA-6 fluorescence microscope (Leica).
Luciferase assay
COS7 cells were transfected with FoxO1 expressing plasmid (pcDNA3-FoxO1, 0.25 μg/dish), luciferase reporter plasmid with a consensus sequence of FoxO1 binding site (IRS-Luc, 0.5 μg/dish) and β-galactosidase expressing plasmid (PCMV-β-gal, 0.25 μg/dish). After 45 h incubation, cells were treated with EGCG (0, 10, 20, 50, 100 μM) for 3 h. We prepared the DNA fragments which carry the genomic sequence (Catalase promoter region; Catalase) of FoxO binding site, and the consensus sequence (SREBP-1c responsive element; SRE) of SREBP-1c binding site. The luciferase reporter plasmids with a FoxO DNA binding site (Catalase-Luc) and with a SREBP-1c DNA binding site (SRE-Luc) were constructed. The stable 3T3-L1 cells which carry Catalase-Luc or SRE-Luc were prepared (3T3-L1/Catalase-Luc, 3T3-L1/SRE-Luc). These cells were treated with catechin (100 μM of EGCG, ECG, EGC), resveratrol (100 μM) and N-acetylcystein (10 mM) for 2 days. After, brief centrifugation (15,800g, 2 min, MX-100, Tomy, Tokyo, Japan) of the cell lysate, supernatant was collected and mixed with Luciferase Assay Reagent (Promega). Luciferase activity was measured with a Luminometer Microlumat LB69p (Berthold Technology, Germany).
Real-time PCR
Quantitative PCR analysis was performed with an Gene Amp 5700 Sequence Detection System (Applied Biosystems, CA, USA) and Thunder Birds SYBR qPCR mix reagent (TOYOBO, Kita, Osaka, Japan). PCR (95 °C for 15 s, 60 °C for 1 min, for 40 cycles) was performed using the specific primers (5′-AAACTCTGGGAGATTCTCCT-3′ and 5′-TGGCATCTCTGTGTCAAC-3′) for PPARγ, (5′-GCCAAACTGAGACTCTTC-3′ and 5′-GGAAGCCTAAGTCTTAGC-3′) for C/EBPα, and (5′-CTGTGCTGCTCACCGAGG-3′ and 5′-AGCCTGGATGGCTACGTA-3′) for β-actin. β-actin was used as an internal standard for correction of the error determined between each sample.
RT-PCR
The cDNA was synthesized using M-MLV Reverse Transcriptase (Takara Bio, Otsu, Shiga, Japan), and PCR (95 °C for 5 min; 25–40 cycles of 95 °C for 30 s, 57 °C for 30 s, 72 °C for 1.5 min; 72 °C for 7 min) was performed using the specific primers (5′-GATCTACGAGTGGATGGT-3′ and 5′-CAGCGTAGACGCCATCTT-3′) for FoxO1, (5′-TTGTACCACTGGTAGAGC-3′ and 5′-CTGTGGCCTCATGTAGGAAT-3′) for SREBP1c, (5′-GACCCCTTCATTGACCT-3′ and 5′-CCACCACCCTGTTGCTGT-3′) for GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Western blotting
Cells were suspended in a sample buffer (50 mM Tris–HCl [pH 6.8], 2% SDS, 6% β-mercaptoethanol, 10% glycerol) containing protease inhibitors (1 mM AEBSF, 130 μM Bestatin, 14 μM E-64, 1 mM EDTA, 1 μM pepstatin A). Then cells were treated with supersonicator (UP-50H, B. Braun Biotech International GmbH, Melsungen, Germany) for 2 min, and the cell extract was collected after centrifugation (4 °C, 21,206g, 10 min, MX-100, Tomy, Tokyo, Japan). The protein concentration was quantified using the BCA protein assay kit (Pierce, Rockford, IL, USA). SDS–PAGE was performed with 80 μg of protein and the separated proteins were transferred to a PVDF membrane (Schleicher & Schuell BioScience, Keene, NH, USA). Non-specific binding was blocked by soaking the membrane in 5% skim milk. The membrane was then incubated with the primary antibody overnight followed by incubation with the secondary antibody for 1 h and detection of specific binding using the Lumi GLO reagent (Cell Signaling, Danvers, MA, USA). Finally, each protein band was detected by chemiluminescence (Las 1000, Fuji FILM, Minato, Tokyo, Japan), and analyzed with Image Gauge Software (FUJI-FILM). Anti-FoxO1 (Cell signaling), anti-phospho-FoxO1 (Thr24) (Cell signaling), anti-phospho-FoxO1 (Ser256) (Cell signaling) and anti-β-actin (Sigma) were used as primary antibodies.
Results
Effect of catechin on fat accumulation
3T3-L1 cells were treated with DMI and catechin (EGCG, ECG) (0, 50, 100, 200 μM) for 2 days. After 6 days of culturing, cells were dyed and observed. We showed that the differentiation of 3T3-L1 cells was suppressed by catechin in a dose dependent manner (Fig. 1a), and ECG was found to exert a stronger suppressive effect than EGCG. In addition, we extracted the dye from the cells and measured its absorbance [optical density (OD. 420 nm)] (Fig. 1b). As observed in Fig. 1a, several kinds of catechin suppressed adipocyte differentiation, and the suppressive effect of ECG was more remarkable than EGCG. Under the same conditions, we also examined the accumulation of triglyceride and found that catechin treatment reduced its accumulation (Fig. 1c). Treatment with NAC also resulted in reduced triglyceride accumulation. Because intracellular level of reactive oxygen species (ROS) generated during adipocyte differentiation was reduced by either EGCG and NAC treatment (data not shown), EGCG possibly prevented fat accumulation in 3T3-L1 mainly through the antioxidant activity of catechin.
Fig. 1.
Effect of catechin on adipocyte differentiation. a Adipose accumulation was observed in the cells treated with EGCG or ECG (0, 50, 100, 200 μM). Cells were treated by EGCG or ECG with DMI for 2 days. Then cells were cultured for 2 days with insulin (5 μg/mL) and 4 days in standard medium, cells were fixed for Oil Red O staining. b The Oil Red O stained cells (100 μM of EGCG, ECG, EGC, CG) were treated with isopropanol to extract the dye, and the concentration of the eluted dye was determined by measuring the absorbance at OD 420 nm. An errorbar shows standard deviation of the mean, n = 3; ** P < 0.05. c Cells were treated by EGCG (100 μM), ECG (100 μM) or NAC (10 mM) with DMI for 2 days. Then cells were cultured for 2 days with insulin (5 μg/mL) and 4 days in standard medium. Lipid was then extracted by using the Bligh and Dyer methods (Bligh and Dyer 1959). Triglyceride level was corrected by total protein level. An errorbar shows standard deviation of the mean, n = 3; ** P < 0.05
To elucidate the role of catechin in the prevention of adipocyte differentiation, we analyzed GPDH activity, a parameter of fat synthesis in adipose tissues and adipocytes. As observed in Fig. 1, catechin also reduced GPDH activity (Fig. 2). We used EGCG and ECG (0, 10, 50, 100, 200 μM); the latter exerted the highest suppressive effect on 3T3-L1 cell differentiation. Cell viability was determined by the MTT assay. EGCG and ECG treatments did not change cell viability (data not shown).
Fig. 2.
Effect of catechin on GPDH activity. Cells were treated by catechin (100 μM of EGCG, ECG, EGC and CG) with DMI for 2 days. Then cells were cultured for 2 days with insulin (5 μg/mL) and 4 days in standard medium, and GPDH activity was analyzed. An errorbar shows standard deviation of the mean, n = 3; ** P < 0.05
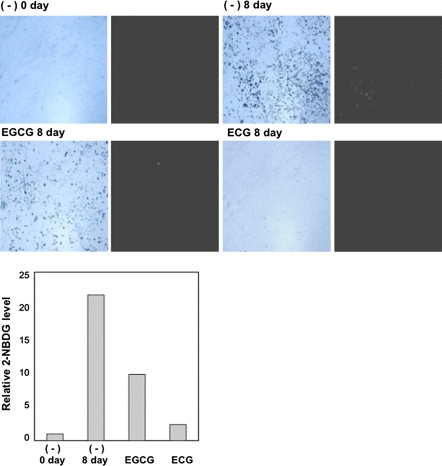
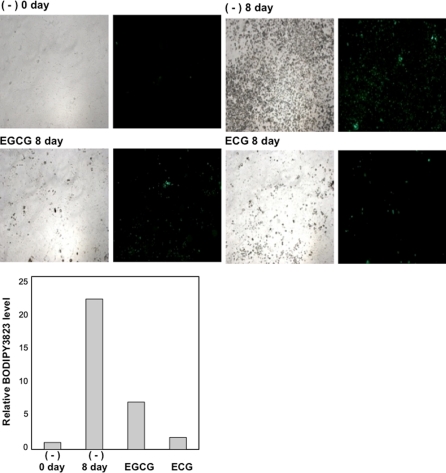
Next, we investigated glucose and fatty acid incorporation to determine the suppressive effect of catechin on adipocyte differentiation. We exposed differentiated 3T3-L1 cells to fluorescent-labeled glucose (2-NBDG) and observed them under a fluorescence microscope. We observed that catechin inhibited glucose incorporation (Fig. 3). Simultaneously, differentiated 3T3-L1 cells were exposed to the fluorescent-labeled fatty acid analog BODIPY3823 instead of 2-NBDG. As observed in Fig. 3, catechin also remarkably reduced fatty acid incorporation (Fig. 4).
Fig. 3.
Effect of catechin on glucose uptake. Cells were treated by EGCG (100 μM) or ECG (100 μM) with DMI for 2 days. Then cells were cultured for 2 days with insulin (5 μg/mL) and 4 days in standard medium: Glucose uptake was analyzed by treating cells with fluorescent-labeled glucose 2-NBDG (10 μM) for 5 min. The bright field (left) and fluorescent field (right) are shown. Fluorescence densities were quantified with the image J, and the relative fluorescence levels are shown in graph as relative value against control ((−) 0 day)
Fig. 4.
Effect of catechin on fatty acid uptake. Cells were treated by EGCG (100 μM) or ECG (100 μM) with DMI for 2 days. Then cells were cultured for 2 days with insulin (5 μg/mL) and 4 days in standard medium: Fatty acid metabolism was analyzed by measuring fatty acid incorporation in cells treated with the fluorescent-labeled fatty acid analog BODIPY3823 (20 μM) for 2 min. The bright field (left) and fluorescent field (right) are shown. Fluorescence densities were quantified with the image J, and the relative fluorescence levels are shown in graph as relative value against control ((−) 0 day)
Effect of catechin on the transcriptional activity of FoxO1 and SREBP1c
To analyze the effect of EGCG on the activity of FoxO1, COS7 cells were treated with varying concentrations of EGCG (0, 10, 20, 50, 100 μM) for 3 h and analyzed with the luciferase assay. EGCG reduced the transcriptional activity of transiently introduced-FoxO1 in COS7 cells at the concentration of 100 μM (Fig. 5a). We also prepared 3T3-L1 cells that carry luciferase reporter plasmids with a FoxO1 binding site of catalase promoter (3T3-L1/Catalase-Luc) and with a consensus sequence of SREBP1c binding site (3T3-L1/SRE-Luc). The effects of catechin on the transcriptional activity of these cells were determined. We treated cells with catechin (EGCG, ECG, EGC), resveratrol, and NAC for 2 days, then analyzed them with the luciferase assay. Catechin, resveratrol, and NAC reduced the transcriptional activity of FoxO1 (Fig. 5b). Although ECG more strongly suppressed lipid accumulation as compared to EGCG (Fig. 1), it did not significantly decrease FoxO1 transcriptional activity. As shown in Fig. 5b, EGCG reduced SREBP1c transcriptional activity (Fig. 5c). These results indicate that EGCG suppressed the differentiation of the adipocytes by reducing the transcriptional activity of FoxO1 and SREBP1c through its antioxidant effect.
Fig. 5.
Effect of catechin on transcriptional activity of FoxO1 and SREBP1c. a Luciferase activity in the cells treated with EGCG is indicated. Cells were transfected with FoxO1 expressing plasmid (pcDNA3-FoxO1, 0.25 μg/dish), luciferase reporter plasmid with a consensus sequence of FoxO1 binding site (IRS-Luc, 0.5 μg/dish) and β-galactosidase expressing plasmid (PCMV-β-gal, 0.25 μg/dish). (−) indicates the cell transfected with these plasmids except FoxO1 expressing plasmid. After 45 h incubation, cells were treated with EGCG (0, 10, 20, 50, 100 μM) for 3 h and analyzed for luciferase assay. Luciferase activity was corrected by β-galactosidase activity. An errorbar shows standard deviation of the mean, n = 6; ** P < 0.05. b The luciferase activity of catechin-treated cells is shown. 3T3-L1 cells which stably carry a luciferase reporter plasmid with a FoxO1 binding site (3T3-L1/Catalase-Luc) were treated with catechin (100 μM of EGCG, ECG, EGC), resveratrol (RES, 100 μM), and NAC (10 mM) for 2 days. An errorbar shows the standard deviation of the mean, n = 3; ** P < 0.05. c The luciferase activity of catechin-treated cells is shown. 3T3-L1 cells which stably carry a luciferase reporter plasmid with a SREBP1c binding site (3T3-L1/SRE-Luc) were treated with catechin (100 μM of EGCG, ECG, EGC), resveratrol (RES, 100 μM), and NAC (10 mM) for 2 days. An errorbar shows standard deviation of the mean, n = 3; ** P < 0.05
Effect of catechin on mRNA and protein expression of FoxO1 and SREBP1c
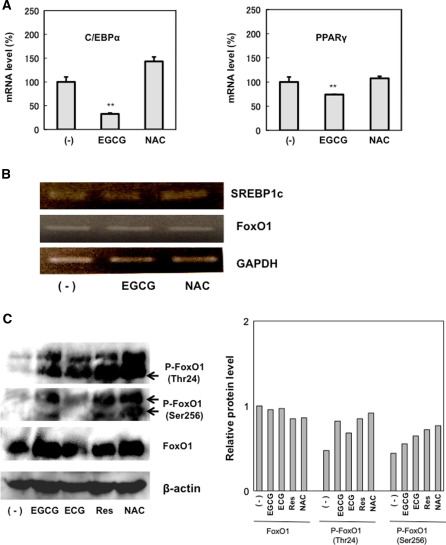
To elucidate the signaling through which EGCG acts on differentiation of 3T3-L1, real-time PCR was performed to analyze the endogenous mRNA expression of clonal expansion-related genes, including PPARγ and C/EBPα. As observed in Fig. 6a, EGCG remarkably reduced mRNA expression of PPARγ and C/EBPα. These results suggest that EGCG arrest the differentiation by reducing mRNA expression of PPARγ and C/EBPα in 3T3-L1. Then, we used RT-PCR to analyze the mRNA expression of FoxO1 and SREBP1c. EGCG treatment did not affect FoxO1 and SREBP1c mRNA expression (Fig. 6b). Figure 5 shows that EGCG remarkably reduced FoxO1 transcriptional activity; therefore, we analyzed Akt-dependent FoxO1 phosphorylation because its activity is regulated by insulin signaling. Although FoxO1 protein expression was not affected by catechin (EGCG, ECG) treatment (as compared to β-actin), there was increased phosphorylation at residues Thr24 and Ser256, which are targets of Akt (Fig. 6c). In addition, NAC treatment resulted in increased FoxO1 phosphorylation. These results suggest that the antioxidant effect of catechin suppressed insulin signaling.
Fig. 6.
Effect of catechin on mRNA expression and phosphorylation. a The mRNA level of each gene was analyzed by real-time PCR. Cells were treated with EGCG (100 μM) or NAC (10 mM) with DMI for 2 days. After 8 days culture, RNA was extracted for real-time PCR assay. b The mRNA level of each gene was analyzed by RT-PCR. Cells were treated by EGCG (100 μM) or NAC (10 mM) with DMI for 2 days, and RNA was extracted for RT-PCR assay. GAPDH was used as an internal RNA control. c The protein levels of FoxO1 and phosphorylated FoxO1 (Thr24, Ser256) were analyzed by western blot assay. Cells were treated by EGCG (100 μM), ECG (100 μM) or NAC (10 mM) with DMI for 2 days, and proteins analyzed by western blot analysis were treated with antibodies against FoxO1 (78–82 kDa), phospho-FoxO1 (Thr24, 78–82 kDa), and phospho-FoxO1 (Ser256, 82 kDa). β-actin was used as a loading control. Band densities were quantified with the image J, and protein levels are shown in graph as relative value against control ((−) FoxO1)
Discussion
Our results clearly revealed that catechin suppressed the differentiation of preadipocyte 3T3-L1 cells. Resveratrol and NAC, which have antioxidant activities, similarly suppressed 3T3-L1 differentiation. These results suggest that the antioxidant activity of catechin inhibits adipocyte differentiation. Actually, catechin decreased the accumulation of ROS generated during the differentiation process of adipocyte (data not shown). Since it is reported that catechin decreases blood insulin levels (Kao et al. 2000), it is possible that the ability of catechin to inhibit differentiation is associated with insulin signaling.
In this experiment, we suspected that catechin may inhibit adipocyte differentiation through its regulation of a transient cell proliferation stage known as clonal expansion since the 3T3-L1 cells were treated with DMI and catechin for only the first 2 days. It was recently reported that the FoxO family inhibits the clonal expansion stage (Nakae et al. 2003). Nakae et al. suggested that FoxO1 participates in differentiation, whereas we clarified the fact that the differentiation of 3T3-L1 preadipocytes infected with an adenovirus expressing FoxO1-siRNA was markedly inhibited (Munekata and Sakamoto 2009). It is known that FoxO proteins have highly conserved phosphorylation sites (Thr24, Ser256, Ser319 in human FoxO1), which are phosphorylated by Akt (Longo and Finch 2003). Phosphorylated FoxO is transported from the nucleus to the cytosol, resulting in its inactivation (Muslin and Xing 2000). In this study, EGCG reduced FoxO1 transcriptional activity in COS7 cells (Fig. 5a) and 3T3-L1 cells (Fig. 5b). Therefore, we think that the inhibition of the adipocyte differentiation by catechin is at least partially caused by the inactivation of FoxO. To study the modification of the FoxO protein in insulin signaling, we analyzed the Akt-dependent phosphorylation of FoxO and observed that catechin increased FoxO1 phosphorylation (at sites Thr24, Ser256) (Fig. 6c). Additionally, since ROSs result in the dephosphorylation of Akt (Cao et al. 2009), we think that the antioxidant effect of EGCG may promote the phosphorylation of Akt, resulting in the phosphorylation and inactivation of FoxO1.
The transcriptional activity of SREBP1c, which regulates the expression of enzymes such as fatty acid synthase, was reduced by EGCG (Fig. 5c). It is known that SREBP is activated by Akt, and it regulates the expression of cholesterol and fatty acid synthetic enzymes (Hua et al. 1993). We also showed that ROSs induce SREBP1c transcriptional activity (Sekiya et al. 2008). These findings indicate that the antioxidant effect of EGCG possibly reduces the transcriptional activity of SREBP1c. In addition, we propose that the transcription of FoxO and SREBP1c is controlled by the same signaling pathway since catechin reduces the transcriptional activity of both FoxO and SREBP1c.
The findings of this study indicate that the antioxidant effect of EGCG suppresses 3T3-L1 preadipocyte differentiation by reducing the transcriptional activity of FoxO1 and SREBP1c via the insulin signaling pathway. Future studies on the inhibitory mechanism of green tea catechin on insulin signaling and FoxO may aid in the prevention of diabetes and obesity.
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan. We wish to thank Mitsui Norin Co. Ltd. for providing catechin compounds.
Abbreviation
- Akt
Protein kinase B
- CG
(−) Catechin gallate
- DMEM
Dulbecco’s Modified Eagle’s Medium
- EGCG
(−) Epigallocatechin gallate
- ECG
(−) Epicatechin gallate
- EGC
(−) Epigallocatechin
- FoxO
Forkhead transcription factor class O
- GPDH
Glycerol-3-phosphate
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- IGF-1
Insulin-like growth factor1
- IRS
Insulin receptor substrates
- MTT
3-(4, 5-dimetyl-2-thiazolyl)-2, 5-diphenyltetrazolium bromide
- 2-NBDG
2-(n-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl) amino)-2-deoxyglucose
- PI3 K
Phosphatidyl inositol 3 kinase
- ROS
Reactive oxygen species
- SREBPs
Sterol regulatory element binding protein
References
- Ahmad N, Mukahtar H. Green tea polyphenols and cancer: biologic mechanisms and practical implications. Nutr Rev. 1999;57:78–83. doi: 10.1111/j.1753-4887.1999.tb06927.x. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Cao J, Xu D, Wang D, Wu R, Zhang L, Zhu H, He Q, Yang BO. ROS-driven Akt dephosphorylation at Ser-473 is involved in 4-HPR-mediated apoptosis in NB4 cells. Free Radic Biol Med. 2009;5:536–547. doi: 10.1016/j.freeradbiomed.2009.05.024. [DOI] [PubMed] [Google Scholar]
- Christenson KL, Osborne FT, McAllister MJ, Strauss JF., 3rd Conditional response of the human steroidogenic acute regulatory protein gene promoter to sterol regulatory element binding protein-1a. Endocrinology. 2001;142:28–36. doi: 10.1210/en.142.1.28. [DOI] [PubMed] [Google Scholar]
- Cross JV, Templeton DJ. Regulation of signal transduction through protein cystein oxidation. Antioxid Redox Signal. 2006;8:1819–1827. doi: 10.1089/ars.2006.8.1819. [DOI] [PubMed] [Google Scholar]
- Deitel M. Overweight and obesity worldwide now estimated to involve 1.7 billion people. Obes Surg. 2003;13:329–330. doi: 10.1381/096089203765887598. [DOI] [PubMed] [Google Scholar]
- Fruhbeck G, Gomez-Ambrosi J, Muruzabal FJ, Burrell MA. The adipocyte: a model for integration of endocrine and metabolic signaling in energy metabolism regulation. J Physiol Endocrinol Metab. 2001;280:827–847. doi: 10.1152/ajpendo.2001.280.6.E827. [DOI] [PubMed] [Google Scholar]
- Hansen LL, Ikeda Y, Olsen GS, Busch Ak, Mosthaf L. Insulin signaling is inhibited by micromolar concentrations of H2O2. Evidence for a role of H2O2 in tumor necrosis factor alpha-mediated insulin resistance. J Biol Chem. 1999;274:25078–25084. doi: 10.1074/jbc.274.35.25078. [DOI] [PubMed] [Google Scholar]
- Haqqi TM, Anthony DD, Gupta S, Ahmad N, Lee MS, Kumar GK, Mukhtar H. Prevention of collagen- induced arthritis in mice by a polyphenolic fraction from green tea. Proc Natl Acad Sci USA. 1999;96:4524–4529. doi: 10.1073/pnas.96.8.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, Wang X. SREBP-2a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- Kao YH, Hiipakka RA, Liao S. Modulation of endocrine systems and food intake by green tea epigallocatechin gallate. Epigallocatechin, a constituent of green tea, represses hepatic glucose production. Endocrinology. 2000;141:980–987. doi: 10.1210/en.141.3.980. [DOI] [PubMed] [Google Scholar]
- Liao S, Kao YH, Hiipakka RA. Green tea: biochemical and biological basis for health benefits. Vitam Horm. 2001;62:1–94. doi: 10.1016/S0083-6729(01)62001-6. [DOI] [PubMed] [Google Scholar]
- Lin JK, Liang YC, Lin SY. Cancer chemoprevention by tea polyphenols through mitotic signal transduction blockade. Biochem Pharmacol. 1999;58:911–915. doi: 10.1016/S0006-2952(99)00112-4. [DOI] [PubMed] [Google Scholar]
- Longo VD, Finch CE. Evolutionary medicine: from dwarf model systems to healthy centenarians. Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- Martindale JL, Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- Mendel S, Youdim MB. Catechin polyphenols; neurodegeneration and neuroprotection in neurodegenerative disease. Free Radic Biol Med. 2004;37:304–317. doi: 10.1016/j.freeradbiomed.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Mitcher LA, Jung M, Shankel D, Dou JH, Steele L, Pillai SP. Chemoprotection: a review of the potential therapeutic antioxidant properties of green tea and certain of its constituents. Med Res Rev. 1997;17:327–365. doi: 10.1002/(SICI)1098-1128(199707)17:4<327::AID-MED2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Munekata K, Sakamoto K. Forkhead transcription factor FOXO1 is essential for adipocyte differentiation. In vitro Cell Dev Biol Anim. 2009;45:642–651. doi: 10.1007/s11626-009-9230-5. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Xing H. 14–3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/S0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- Nakae J, Park BC, Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- Nakae J, Kitamura T, Kitamura Y, Biggs WH, III, Arden KC, Accili D. The Forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–129. doi: 10.1016/S1534-5807(02)00401-X. [DOI] [PubMed] [Google Scholar]
- Nohtufft A, Brown H, Goldstein LJ. Topology of SREVP cleavage- activating protein, a polytopic membrane protein with a sterol-sensing domain. J Biol Chem. 1998;273:17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterspm GL, Lee L, Tissembaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Rubin CS, Hirsch A, Fung C, Rosen OM. Development of hormone receptors and hormonal responsivness in vitro. Insulin receptor and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3–L1 cells. J Biol Chem. 1978;253:7570–7578. [PubMed] [Google Scholar]
- Sekiya M, Hiraishi A, Touyama M, Sakamoto K. Oxidative stress induced lipid accumulation via SREBP1c activation in HepG2 cells. Biochem Biophys Res Commun. 2008;375:602–607. doi: 10.1016/j.bbrc.2008.08.068. [DOI] [PubMed] [Google Scholar]
- Song EK, Hur H, Han MK. Epigallocatechin gallate prevents autoimmune diabetes induced by multiple low doses of streptozotocin in mice. Arch Pharm Res. 1999;26:559–563. doi: 10.1007/BF02976881. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Kim JB, Graves RA, Spiegelman BM. A novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltner-Law ME, Wang XL, Law BK, Hall RK, Nawano M, Ganner DK. Epigallocatechin, a constituent of green tea, represses hepatic glucose production. J Biol Chem. 2002;277:34933–34940. doi: 10.1074/jbc.M204672200. [DOI] [PubMed] [Google Scholar]
- Wang JF, Zhang X, Groopman JE. Activation of vascular endothelial growth factor receptor-3 and its downstream signaling promote cell survival under oxidative stress. J Biol Chem. 2004;279:27088–27097. doi: 10.1074/jbc.M314015200. [DOI] [PubMed] [Google Scholar]
- Yang CS, Wang ZY. Tea and cancer. J Natl Cancer Inst. 1993;85:1038–1049. doi: 10.1093/jnci/85.13.1038. [DOI] [PubMed] [Google Scholar]