Abstract
Objective
The goal of this study was to guide bone marrow-derived human mesenchymal stem cells (hMSC) into a cardiac progenitor phenotype, and assess therapeutic benefit in chronic myocardial infarction.
Background
Adult stem cells, delivered in their naïve state, demonstrate a limited benefit in patients with ischemic heart disease. Preemptive lineage pre-specification may optimize therapeutic outcome.
Methods
hMSC were harvested from a coronary artery disease patient cohort. A recombinant cocktail consisting of TGFβ1, BMP-4, Activin-A, retinoic acid, IGF-1, FGF-2, α-thrombin and IL-6 was formulated to engage hMSC into cardiopoiesis. Derived hMSC were injected into the myocardium of a nude infarcted murine model, and followed over 1-year for functional and structural end-points.
Results
While the majority of patient-derived hMSC in their native state demonstrated limited effect on ejection fraction, stem cells from rare individuals harbored a spontaneous capacity to improve contractile performance. This reparative cytotype was characterized by high expression of Nkx2.5, Tbx5, Mesp-1 and Mef2C, markers of cardiopoiesis. Recombinant cardiogenic cocktail guidance secured the cardiopoietic phenotype across the patient cohort. Compared to unguided counterparts, cardiopoietic hMSC delivered into infarcted myocardium achieved superior functional and structural benefit without adverse side effects. Engraftment into murine hearts was associated with increased human-specific nuclear, sarcomeric and gap junction content along with induction of myocardial cell cycle activity.
Conclusions
Guided cardiopoiesis thus enhances the therapeutic benefit of bone marrow-derived human mesenchymal stem cells in chronic ischemic cardiomyopathy.
Keywords: Cell therapy, Ischemic cardiomyopathy, Heart failure, Patient derived, Transplantation
Cell-based reparative approaches are increasingly considered in the management of ischemic heart disease. Stem cell capacity for multi-lineage tissue specification provides the opportunity to extend treatment options for ischemic cardiomyopathy beyond current practices, and achieve heart repair (1–3).
Emerging experience has focused on adult stem cells delivered unaltered, in their native state (4,5). Of increased interest are mesenchymal stem cells with a documented aptitude for multipotent differentiation, cytoprotection and myocardial restoration demonstrated when derived from allogeneic healthy donors (6–8). However, in autologous therapy, patient-related modifiers of regenerative capacity, including age, sex and co-morbidities, translate into variability in repair compromising efficacy (9–11). Development of technologies that would enhance stem cell benefit is thus warranted (1–13).
We here tracked inter-patient variation in stem cell effectiveness, and identified that expression of cardiac markers in hMSC biases repair. Accordingly, non-reparative hMSC were guided to acquire regenerative potency through cardiogenic priming. Derived cardiopoietic hMSC demonstrated improved efficacy compared to their naïve source, establishing an adult stem cell-derived progenitor with enhanced benefit in the treatment of chronic ischemic cardiomyopathy.
Methods
Study protocols were approved by pertinent Institutional Ethics Committee, Institutional Review Board and Institutional Animal Care and Use Committee.
Ischemic heart disease patient cohort
Patients undergoing coronary artery bypass surgery provided informed consent. Information concerning age, gender, history, medications, perioperative laboratory evaluation and coronary assessment is presented as Supplemental Tables 1–3.
Stem cell culture
Human bone marrow aspirates, obtained during coronary artery bypass surgery following sternotomy, yielded plastic-adherent hMSC with identity confirmed by Fluorescence-Activated Cell Sorting (FACS) using the CD34−/CD45−/CD105+/CD133+ panel (Supplemental Figure 1). Cells were cultured at 37°C in advanced-MEM supplemented with 5% human platelet lysate (14), and stimulated with TGF-β1 (2.5 ng/ml), BMP-4 (5 ng/ml), Activin-A (5 ng/ml) (15,16), FGF-2 (10 ng/ml) (6), IL-6 (100 ng/ml) (17), Factor IIa (hα-thrombin, 1 U/ml) (18), IGF-1 (50 ng/ml) (6) and/or retinoic acid (1 μM) (19). Phospho-AKT inhibition was achieved with SR13668 (50 nM). Differentiation beyond the cardiopoietic state was carried-out with 1% platelet lysate in the presence of FGF-2 and hα-thrombin for 10 days. To scale-up production under GMP compliance, cell culture procedures were site-certified by independent regulatory review as meeting mandatory environmental and quality assurance criteria required to ensure traceability, purity, homogeneity and sterility for batch testing and release (14).
RNA profiling
Real-time quantitative polymerase chain reaction (qRT-PCR) was performed using a TaqMan PCR kit (Applied Biosystems) in triplicate. Threshold cycle (CT) values were determined using the 2−ΔΔCT method, normalized to human specific GAPDH (P/N 435,2662-0506003). Genes representative of cardiac transcriptional activity included Nkx2.5 (Hs00231763_m1), GATA4 (Hs00171403_m1), MEF2C (Hs00231149_m1), Tbx5 (Hs00361155_m1), FOG1 (Hs00542350_m1), MESP1 (Hs00251489_m1), GATA6 (Hs00232018_m1), and Flk-1 (Hs00911699_m1).
Protein and DNA probing
Immunostaining for cardiac transcription factors Mef2C (1:400, Cell Signaling Technologies), Nkx2.5 (1:150, Santa Cruz Biotechnology), Gata-4 (1:150, Santa Cruz Biotechnology), Tbx5 (1:5000, Abcam), Mesp-1 (1:250, Novus Bio), Fog-2 (1:100, Santa Cruz Biotechnology), along with Phospho-AKTSer473 (1:100, Cell Signaling Technologies), human-specific Troponin-I (1:100, Abcam), sarcomeric α-actinin (1:500, Sigma-Aldrich), MLC2v (1:500, Synaptic Systems), Sca-1 (1:100, R&D Systems), CD-31/PECAM-1 (1:500; Beckman Coulter), α-smooth muscle actin (1:500; Sigma), human Lamin A/C (1:50, Novacastra), Connexin-43 (1:100, Zymed Laboratories) and Ki67 (1:500, NeoMarkers) was performed following fixation in 3% paraformaldehyde and permeabilization with 1% Triton X-100. Human ALU-DNA probing (Biogenex) required hybridization (85°C, 10 min) and incubation at 37°C overnight followed by anti-Fluorescein GFP-labeled secondary detection. For genomic probing, human and murine DNA were labeled in Vysis® Spectrum Green or Orange d-UTP via nick translation followed by hybridization and denaturation (Abbott Molecular). Frozen tissue was fixed (3:1 methanol:acetic acid), and pretreated in 2xSSC for 2 min (73°C), placed 5 min in 1 M Tris, 0.5 M EDTA (90°C), 2 min in 4 mg Pepsin/L, 0.9% NaCl (37°C), and 30 s in 70% ETOH. Slides were denatured in 70% formamide (73°C), followed by a 70%/85%/100% ETOH series. Denatured genomic probes were applied to sections, hybridized overnight (37°C), and washed in 2xSSC/0.1% NP40 solution (73°C). Slides were counterstained with DAPI-containing mounting medium.
Cell imaging
Images were obtained with LSM 510/LSM 700 laser scanning confocal or Apotome structured illumination microscopes (Carl Zeiss). Quantification of cytosolic and nuclear induction of Nkx2.5, Mef2C or GATA-4 was performed using Zeiss Axioplan and Metamorph software (Molecular Devices). Cell cycle activity was quantified using a hematocytometer. Cell structure was resolved by transmission electron microscopy (20). Calcium transients were monitored in cells loaded with 5 μM of the calcium-selective probe fluo-4-acetoxymethyl ester (Molecular Probes) using a temperature controlled Zeiss LSM 510 or Apotome microscope with line-scan or phase images acquired during 1 Hz stimulation (15).
Cell therapy
Myocardial infarction was performed in 8–12 weeks old nude, immunocompromised mice (Harlan) (21). Following intubation and ventilation under isoflurane anesthesia, proximal left anterior descending artery was ligated with 9-0 suture. Injury induced ST-changes on electrocardiography (MP150; Biopac) and akinetic regions on echocardiography (Sequoia 512, Siemens; Vevo2100, VisualSonics). Following a blinded design, 1-month post-infarction a total of 600,000 hMSC, suspended in 12.5 μl phosphate buffered saline, was injected under microscopic visualization in five epicardial sites (120,000 cells per site) in the anterior wall of the left ventricle. Saline-treated group underwent identical procedure. Left ventricular function and structure were serially followed by echocardiography at pre-infarction, 1-month after infarction (pre-cell therapy), 1-, 2-, 6-, 12-, and 20-month post-cell therapy. Ejection fraction (%) was calculated as [(LVVd-LVVs)/LVVd]x100, where LVVd is left ventricular end-diastolic volume (μl), and LVVs is left ventricular end-systolic volume (μl).
Statistics
Data are presented as mean±SD unless otherwise indicated (JMP 8; SAS Institute). Paired group analysis was performed using Student’s t-test for each pair of samples without correction for type I error, and non-parametrically validated using Wilcoxon signed rank test (JMP). Kaplan-Meier analysis with log-rank testing was applied for survival evaluation. A p value <0.05 was considered significant.
Results
Reparative profile extracted from patient stem cells demonstrating innate efficacy
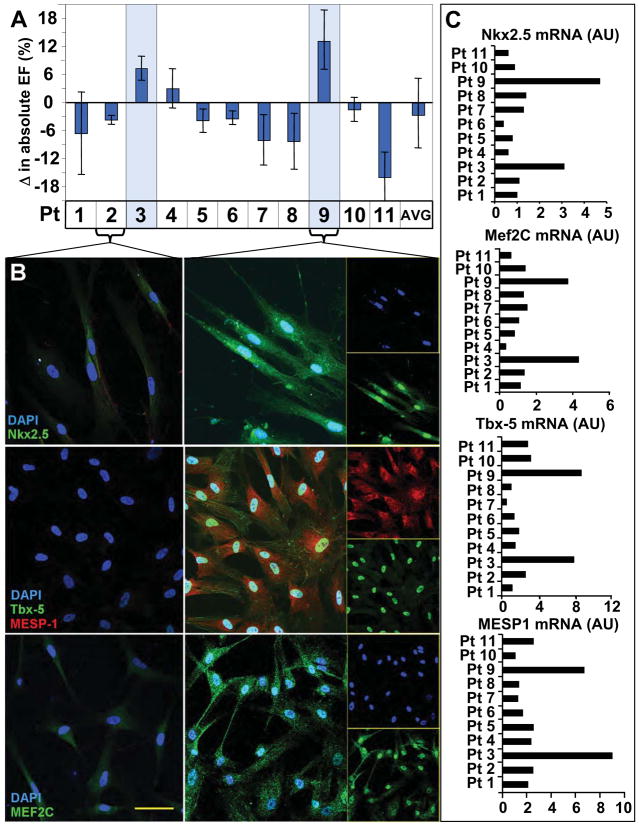
Bone marrow-derived human mesenchymal stem cells (hMSC) obtained from patients undergoing coronary artery bypass revealed individuals with an innate capacity for heart repair. Specifically, hMSC derived from two out of eleven individuals significantly improved ejection fraction following myocardial injection into a nude murine model of chronic ischemic cardiomyopathy (Figure 1A). No clinical features separating these patients from the cohort were apparent (Supplemental Tables 1–3). Reparative stem cells were distinguished from non-effective hMSC counterparts by robust expression of early (homeobox transcription factor Nkx2.5 (22), T-Box Transcription Factor Tbx-5 (23) and the helix–loop–helix transcription factor MESP1) (24) and late (myocyte enhancer factor MEF2C) (25) cardiac transcription factors (Figure 1B). Distinctive protein abundance was corroborated on qPCR with a 3 to 6-fold upregulation in Tbx-5 and MESP1 and a 2 to 3-fold upregulation of MEF2C mRNA in effective versus non-effective hMSC, respectively (Figure 1C). Upregulated cardiac transcription factors in hMSC thereby provided a molecular signature consistent with ejection fraction improvement.
Figure 1. Repair efficacy in hMSC correlates with cardiac transcription factor expression.
A, Patient hMSC demonstrated variable repair after injection into an infarction model. Only rare stem cells (patients #3 and #9 highlighted) demonstrated benefit. B, Low expression of cardiac transcription factors in non-reparative hMSC (patient #2 illustrated). Robust Nkx2.5 (83±5% and 86±2% of hMSC in patients #3 and #9), MEF2C, Tbx-5, and MESP-1 expression in hMSC with reparative capacity (patient #9 shown, with single channels provided in inset). Bar, 20 μm. C, Reparative pattern validated with upregulated Nkx2.5, Mef2c, Tbx-5 and MESP1 mRNA in reparative versus non-reparative hMSC.
Induction of cardiac program in hMSC
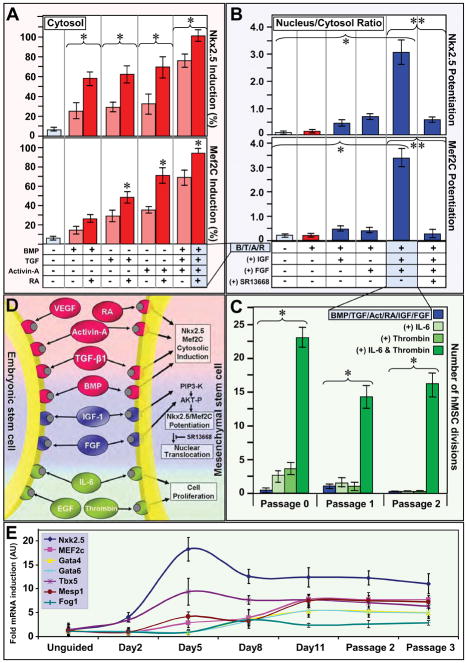
To impose expression and potentiation of cardiac transcription factors, while maintaining proliferative status, endoderm-based pathways (26,27) contributing to embryonic cardiogenesis (15,16) were here tested on the adult hMSC phenotype (Figure 2). TGFβ superfamily members (TGFβ1, BMP-4 and Activin-A), which induce Nkx2.5 expression in embryonic stem cells (16), here individually promoted cytosolic Nkx2.5 and Mef2C expression at protein (Figure 2A and Supplemental Figure 2A) and mRNA (Supplemental Figure 2B) levels. Stimulation of hMSC with a combination of TGFβ1, BMP-4 and Activin-A resulted in augmented Nkx2.5 and Mef2C cytosolic expression, further enhanced by retinoic acid (Figure 2A and Supplemental Figure 3). TGFβ members and retinoic acid did not achieve nuclear translocation of cardiac transcription factors, maintaining nuclear to cytosol ratios equivalent to untreated hMSC (Figure 2B, 1st–2nd column). Potentiation of cardiac transcription factors was achieved with IGF-1 and FGF-2 whose combination triggered nuclear translocation of Nkx2.5 and Mef2C (Figure 2B, 5th column and Supplemental Figure 4), correlating with AKT activation (Supplemental Figure 5) as nuclear expression was abolished by the AKT inhibitor, SR13668 (Figure 2B, 6th column and Supplemental Figure 6). Stimulation of hMSC with TGFβ1, BMP4, Activin-A, retinoic acid, FGF-2, and IGF-1 (Figure 2C, Passage 0) compromised cell proliferation, rescued by α-thrombin and IL-6 (Figure 2C). Applied as a human recombinant cardiogenic cocktail, within a 5% platelet lysate at the onset of a 5 day incubation period, TGFβ1, BMP4, Activin-A, retinoic acid, FGF-2, IGF-1, α-thrombin and IL-6 induced cardiac transcription factor expression, potentiated nuclear translocation, and maintained cell cycle progression (Figure 2D). Induction of pro-cardiogenic transcription factors was chronicled with mRNA profiling during and after cocktail exposure (Figure 2E), and the derived cardiopoietic profile stabilized within 5 days was maintained for 7 passages.
Figure 2. Derivation of cardiogenic cocktail that induces and potentiates cardiac transcription factor expression, and maintains cell cycle activity.
A, BMP-4, TGFβ1, Activin-A induced cytosolic expression of Nkx2.5 and Mef2C (light red columns). Addition of retinoic acid (RA) boosted cytosolic induction (red columns). B, BMP-4, TGFβ1, Activin-A and RA (B/T/A/R, red) potentiated nuclear translocation of Nkx2.5 and Mef2C when combined with IGF-1 and FGF-2. AKT inhibitor (SR13668) inhibited nuclear translocation. C, Low hMSC cell cycle activity following BMP-4, TGFβ1, Activin-A, RA, IGF-1, and FGF-2 treatment (blue) rescued by IL-6 and human α-thrombin. Brackets indicate pared analysis. Star and double star, p<0.05. D, Cardiogenic cocktail, applied as a combined regimen, induced and potentiated nuclear translocation of Nkx2.5 and Mef2C while maintaining cell cycle activity. E, Nkx2.5, Tbx5, MEF2C, GATA4, GATA6, FOG1 and MESP1 hMSC mRNA in response to cocktail treatment. Panels A-C and E evaluate non-reparative patient hMSC.
Cardiogenic cocktail guides cardiopoiesis to yield functional cardiomyocytes
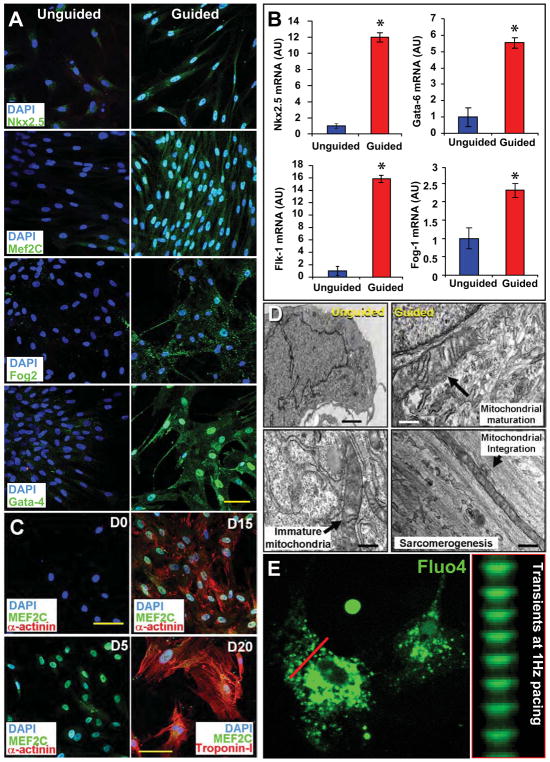
Guidance with the cardiogenic cocktail induced nuclear translocation of cardiac transcription factors, documented for Nkx2.5 (86±10% of cells), Mef2c (84±8%), Fog2 (78±15%), Tbx5 (76±7%), MESP1 (83±11%) and GATA4 (81±3%) (5,15,25,28), augmenting baseline expression limited to the cytosol in unguided hMSC (Figure 3A). In this way, hMSC from each patient (n=12) were guided to derive a cardiopoietic phenotype with nuclear upregulation of cardiac transcription factors in the absence of sarcomere protein expression (Supplemental Figure 7A). This lineage engagement was further documented with mRNA analysis revealing a 12, 16, 6, and 2-fold increase in Nkx2.5, Flk-1, Gata-6, and Fog-1, respectively in treated versus untreated hMSC (Figure 3B). Thereby, hMSC from patients with ischemic heart disease were converted in vitro from a naïve state of sequestered plasticity to active tissue specification adopting a phenotype consistent with enhanced repair aptitude.
Figure 3. Cardiogenic cocktail guides cardiogenesis to yield functional cardiomyocytes.
A, Nuclear translocation of Nkx2.5, Mef2C, Fog-2, and Gata-4 in cocktail-guided hMSC. B, Guidance of hMSC, from 12 coronary artery disease patients, increased mRNA expression of Nkx2.5, Flk-1, Gata-6, and Fog-1. Star, p<0.01. C, Cocktail-based cardiogenic conversion of naïve hMSC (D0) resulted in Mef2C activation at day 5 (D5), and sarcomerogenesis (α-Actinin) achieved by day 15 (D15) to 20 (D20, Troponin I expression). Bar, 20 μm. D, Compared to naïve hMSC with abundant nuclei (Left), transmission electron microscopy of cocktail-guided hMSC at day 15 revealed mitochondrial maturation and integration with organizing myofibrils. Bar, 1 μm (upper left); 250 nm (other panels). E, Calcium transients elicited in response to 1-Hz electrical stimulation. See also Supplemental Movie depicting calcium transients in Fluo4AM-loaded and paced cardiopoietic hMSC at 20 days following 1% platelet lysate differentiation.
Differentiation beyond the cardiopoietic state, in 1% human platelet lysate medium, ultimately achieved sarcomerogenesis tracked by expression of α-actinin (48±10% of cells at day 10 and 75±8% at day 15) and Troponin-I (75±6% of cells at day 10 and 86±5% at day 15), which demonstrated immature organization at 20 days post-cocktail stimulation (Figure 3C and Supplemental Figure 7B). Smooth muscle actin stained myocytes were rarely observed (1±1% of cells) at day 20 of differentiation (Supplemental Figure 7B). Nascent cardiomyocytes derived from guided hMSC contained ~80% of mitochondria with structured cristae and aligned with forming sarcomeres, in contrast to unguided hMSC characterized by prominent nuclei and immature mitochondria (Figure 3D and Supplemental Figure 7C). At day 20 (15 days upon end of cocktail stimulation), functional excitation-contraction coupling was documented by rhythmic calcium transients in Fluo-4 loaded cells seen in response to external electrical stimulation (Figure 3E and Supplemental Movie). Thus, the cardiopoietic phenotype derived from hMSC can yield sarcomere-containing myocytes capable of electro-mechanical response.
Cardiopoietic guidance enhances therapeutic efficacy of hMSC
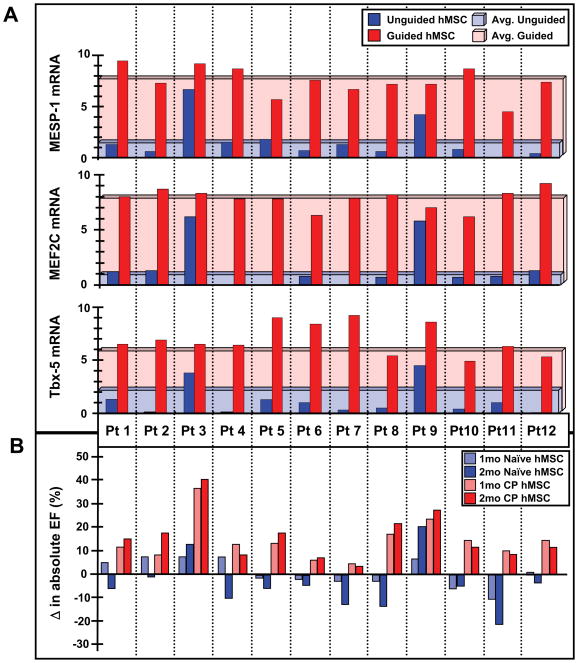
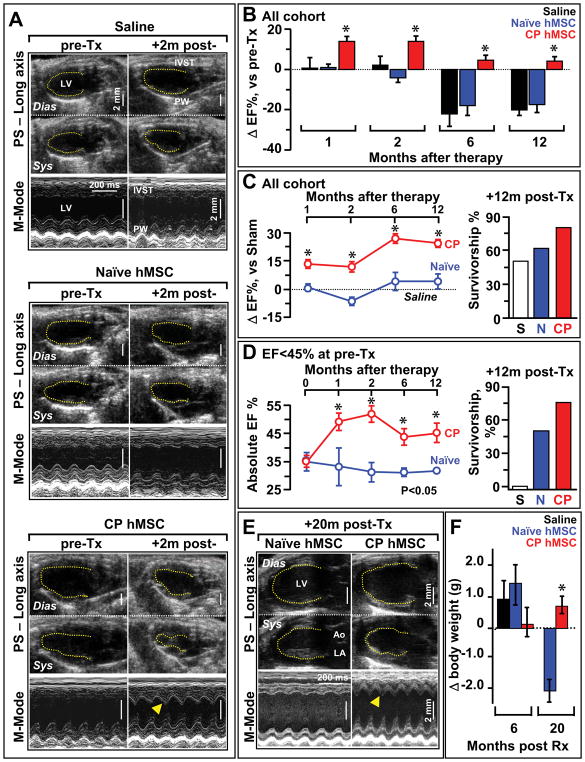
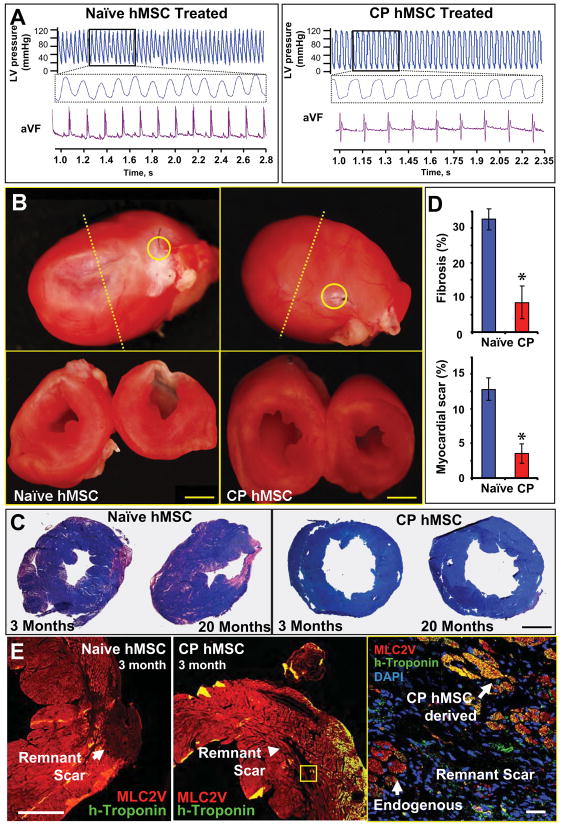
Guided hMSC (n=12 patients) demonstrated on average a 3-fold increase in Tbx-5, and 8-fold increase in Mesp-1 and Mef2C (Figure 4A). Induction of the cardiopoietic phenotype was verified with RT-qPCR prior to transplantation into nude mice 1-month after coronary ligation. Permanent ligation of the left anterior descending artery induced akinesis and dyskinesis in the anterior wall (ejection fraction 78.8±2.1% and 38.2±1.2%, pre versus post-infarction, p<0.05) with an 84.6% survivorship at 1-month (n=52). Surviving infarcted mice were randomized into saline-, naïve hMSC- and cardiopoietic hMSC-treatment groups. Direct myocardial delivery of cardiopoietic hMSC, demonstrating nuclear translocation of cardiac transcription factors in the absence of sarcomeric proteins, was utilized to eliminate variability introduced by reliance on distal homing. Despite equivalent akinesis (saline 31.3±2.6%, naïve hMSC 33.1±2.6%, cardiopoietic hMSC 35.3±2.3%), guided cardiopoietic hMSC improved ejection fraction over patient-matched naïve hMSC at 1 and 2 months post-transplantation (Figure 4B). Echocardiography of cardiopoietic hMSC treated chronically infarcted mice (n=14, typically 1 mouse per patient) revealed progressive reanimation of the anterior left ventricular wall on 1-year follow-up (Figure 5A and Supplemental Movies), compared to marginal change recorded with patient-matched naïve hMSC (n=17 mice; Figure 5A, left panels) or saline (n=10 mice). Across cohorts, cardiopoietic hMSC were superior throughout 12 months preventing heart failure progression (Figure 5B). Relative to saline-treated hearts, cardiopoietic hMSC demonstrated sustained benefit superior to naïve therapy (Figure 5C) resulting in a 1-year survival of 78% and 61%, respectively compared to 48% for the saline-treated cohort (Figure 5C inset). The benefit of cardiopoietic hMSC was particularly prominent in the subgroup with ejection fraction <45% (Figure 5D) resulting in 1-year survival of 75% in contrast to 48% for naïve and 0% for saline-treated counterparts. Naïve hMSC demonstrated limited capacity for recovery (Figure 5D) with myocardial dysfunction (Figure 5F) and cachexia observed at 20-month follow-up, typically absent from the cardiopoietic hMSC-treated group (Figure 5G). Distinct outcomes were corroborated with cardiac catheterization that revealed abnormal end-diastolic pressure increase in the naïve hMSC-treated group, absent from cardiopoietic hMSC-treated mice despite equivalent afterload pressure profiles (Figure 6A). Histopathology revealed progressive scar formation in naïve hMSC-treated hearts, attenuated with cardiopoietic hMSC treatment over 3 to 20 months (Figure 6B and 6C, Supplemental Figure 8A). Global myocardial fibrosis and scar size were 32±4% and 12±2% in naïve hMSC treated hearts versus 8±6% and 3±2% in cardiopoietic hMSC treated hearts (Figure 6D, n=10 for each group, p<0.01), respectively. In contrast to persistent scar and low human-troponin staining in the naïve hMSC group, the anterior wall of cardiopoietic hMSC-treated hearts revealed human-specific troponin positive cardiomyocytes (Figure 6E; Supplemental Figure 8B and 8C). Thus, treatment with cardiopoietic hMSC resulted in stable functional and structural benefit in chronically infarcted hearts.
Figure 4. Cardiopoietic hMSC phenotype associated with ejection fraction improvement.
A, Induction of cardiac transcription factors Tbx-5, MEF2c, and MESP-1 mRNA documented for each patient. B, Echocardiography demonstrated ejection fraction improvement in chronically infarcted mice treated with cardiopoietic (CP) over patient-matched naïve hMSC at 1–2 months. Experiment in B distinct from Figure 1A.
Figure 5. Cardiopoietic (CP) hMSC provide benefit following transplantation.
A, Echocardiography of infarcted hearts 4 weeks after coronary ligation, prior to therapy (pre-Tx) revealed an akinetic anterior wall. Saline and naïve hMSC treated hearts maintained akinesis in the anterior wall, in contrast to re-animation in CP treated (+2m post-Tx, arrowhead). See Supplemental Movies depicting anterior wall reanimation in the long and short axis on echocardiography from cardiopoietic but not naïve hMSC-treated cohorts at 1-year post-therapy. B, Cardiopoietic transplantation (CP; n=14) was associated with ejection fraction improvement acutely and limited progression towards failure chronically, not observed in saline (S; n=10) or naïve groups (N; n=17). C, Sustained benefit of CP hMSC therapy over 1-year follow-up, compared to naïve hMSC treatment, was associated with survival benefit (right). Ejection fraction expressed relative to sham (dotted line). D, Subgroup analysis of mice with heart failure (pre-Tx ejection fraction <45%), CP hMSC had superior benefit versus naïve, with maintained survival (right). Saline treated mice in this subgroup did not survive beyond 2 months. E-F, CP hMSC treatment prevented heart failure and blunted weight loss, compared to naïve (n=5) at 20 months. In E, LV, left ventricle; Ao, aorta; LA, left atrium. Dias, diastole; Sys, systole. In E, arrowhead indicates reanimated anterior wall. Error bars represent SEM. Star, p<0.01 in B, C and D; p<0.05 in F.
Figure 6. Echocardiographic improvement validated with pressure catheter and histopathological evaluation.
A, Catheterization revealed no afterload discrepancy between cohorts, yet elevated end-diastolic pressures were noted in naïve versus cardiopoietic (CP) hMSC groups. B, Pathological evaluation demonstrated diminished scar downstream of left anterior descending (LAD) artery ligation (yellow circle) with re-muscularization and diminished remodeling in CP (right) in contrast to naïve (left) treated hearts at 3-months (yellow circle). C, Diffuse fibrosis and larger scar area at 3 and 20 months in naïve compared to CP hMSC treated hearts. Bar, 2 mm for B and C. D, Scar size and total myocardial fibrosis (n=10 per condition). Star, p<0.01. E, Immunohistological evaluation of scar reveals limited contribution of human-troponin cells in naïve (left) in contrast to cardiopoietic treated hearts (middle, right). Bar, 500 μm - left and 20 μm - right.
Delivery of cardiopoietic hMSC triggers safe repair
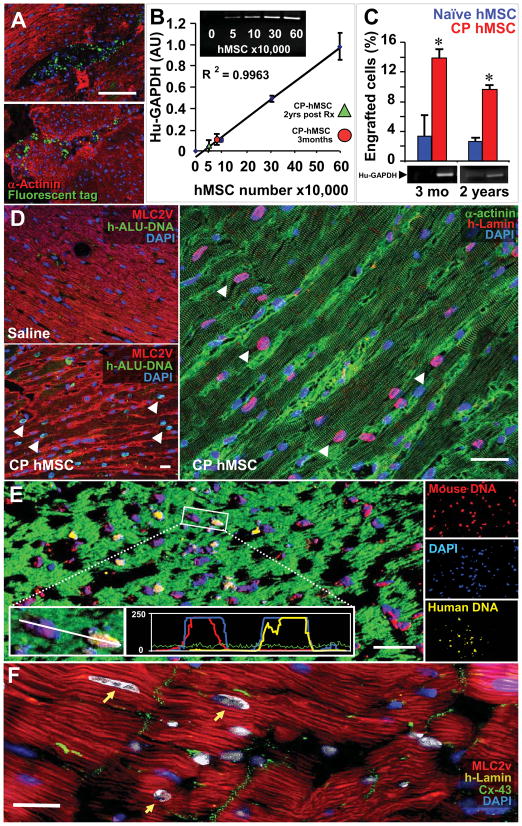
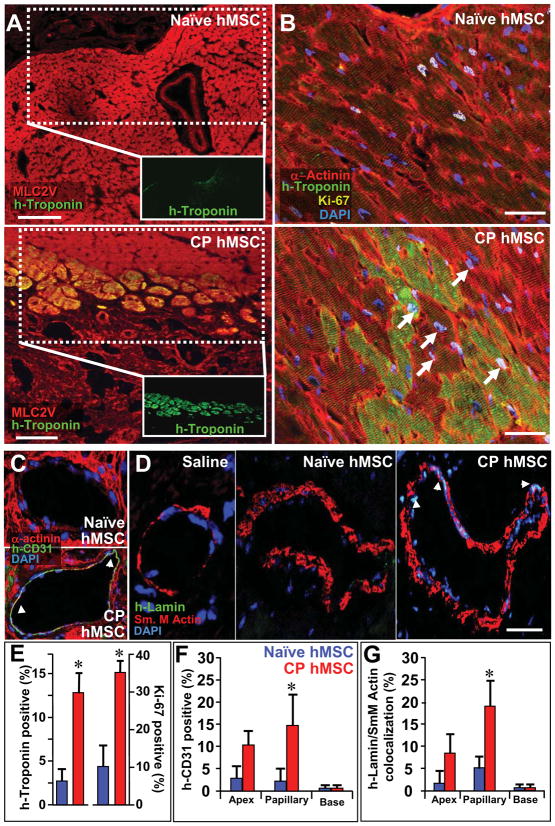
Histological evaluation of murine hearts 48 h after delivery of fluorochrome-loaded cardiopoietic hMSC validated epicardial delivery demonstrating cellular implantation (Figure 7A). hMSC engraftment within treated myocardium, quantified utilizing a human-specific GAPDH standard curve derived from murine hearts treated with escalating doses of hMSC (Figure 7B), was 3.3±2.8% and 3.4±0.7% of cells injected for naïve, and 13.8±2.4% and 9.6±1.2% of cells injected for cardiopoeitic hMSC at 3 months and 2 years, respectively (Figure 7C). Cardiopoietic hMSC-treated murine myocardium revealed widespread presence of human-derived cells with positive staining for ALU DNA sequences specific to the human species, absent in saline-treated infarcted controls (Figure 7D). Quantification of human lamin within the infarcted anterior wall, validated with ALU-DNA staining, revealed 3±2% (n=5) versus 25±5% (n=5) of nuclei in naïve and cardiopoietic hMSC-treated hearts, respectively (p<0.01). Typically, nuclear fusion was absent in cardiopoietic hMSC-treated hearts as determined by species-specific DNA hybridization, albeit cytosolic fusion could not be ruled out (Figure 7E; see rare event in Supplemental Figure 9A). Cardiopoietic hMSC derived human lamin positive myocytes integrated with host tissue as demonstrated by co-localization with connexin-43, a gap junction protein (Figure 7F). Cardiopoietic hMSC transplantation was thus associated with long-term cellular retention and non-fusogenic integration within the myocardium. Engrafted cardiopoietic hMSC displayed a ventricular phenotype as counter-staining of human-troponin positive cells co-localized with ventricular myosin light chain (MLC2v) in the resolving scar (Figure 8A and Supplemental Figure 8B). Within the anterior wall of cardiopoietic hMSC-treated hearts, co-localization of human cardiac troponin-I with α-actinin (Figure 8B and Supplemental Figure 8C) accounted for 12±3% of cardiomyocytes, in contrast to 2.5±2% of naïve hMSC-treated hearts (Figures 8B and 8F). Furthermore, human-troponin positive cardiomyocytes and adjacent murine myocardium in cardiopoietic hMSC-treated hearts had a significantly higher expression of Ki-67 (35±3%) compared to naïve hMSC-treated heart (10±5%), indicative of cell cycle engagement of endogenous and hMSC-derived cardiomyocytes up to 3 months after delivery (Figures 8B and 8F). Specifically in the cardiopoietic hMSC treated myocardium, 10±3% of human troponin and 3±2% of non-human α-actinin/MLC2v positive cardiomyocytes expressed Ki-67. In addition, non-human Sca-1 cells were induced from 5.1±3 to 18.6±5.7 cells per field in naïve versus cardiopoietic hMSC-treated hearts (Supplemental Figure 9B and 9C) (29). Cardiopoietic hMSC-treated myocardium demonstrated vasculogenic potential, observed distal to the occluded coronary vessel with expression of human PECAM-1 (CD-31; Figure 8D and 8G) and co-localization of human lamin with α-smooth muscle actin (Figure 8E and 8H). As such, transplanted cardiopoietic hMSC contributed to the myocardial pool, upregulated cell cycle activity, induced endogenous Sca-1-positive stem cell proliferation and contributed to neovasculogenesis distal to the site of ligation. Long-term safety of cardiopoietic hMSC was documented by absence of myocardial tumorigenesis (Supplemental Figure 10A) or electrical ectopy on electrocardiography (Supplemental Figure 10B). Beyond the heart, at 1-year follow-up no evidence of abnormal cell proliferation or neoplastic transformation was found on gross or histological examination of other well perfused organs (Supplemental Figure 10C). Thus, transplantation of cardiopoietic hMSC achieved lineage specification for safe repair in the setting of ischemic cardiomyopathy.
Figure 7. Cardiopoietic (CP) hMSC demonstrate long-term integration.
A, Fluorochrome loaded CP hMSC detected within murine myocardium 48 h after delivery. B, mRNA standard curve for human-specific GAPDH with escalating doses of hMSC within murine myocardium. Quantified hMSC engraftment (3-month, red circle; 2-years, green triangle). C, Higher CP hMSC engraftment compared to naïve. Star, p<0.01. D, Compared to saline (left), CP hMSC within treated hearts confirmed by human h-ALU DNA (green, middle) and immuno-probing for human-specific lamin (red, right). E, Concurrent human (yellow) and mouse (red) DNA probing demonstrated negligible fusion through species-specific nuclear staining on confocal histogram and single channel analysis (insets). Bar, 20 μm for A, D, E. F, Human lamin (yellow, arrows) expressing myocytes demonstrated connexin-43 (green) expression. Bar, 10 μm.
Figure 8. Cardiopoietic (CP) hMSC demonstrate cardiogenic and vasculogenic potential within host myocardium.
A, Human-specific troponin-I (green) in the anterior wall of naive- versus CP-treated hearts, respectively, co-localized with ventricular myosin light chain (MLC2v, red). Bar, 100 μm. B, Human troponin-I staining of naïve versus CP hMSC treated hearts, counterstained with α-Actinin (red), demonstrated engraftment of human cells. Cell cycle activation, documented by Ki-67 expression (yellow, arrows), noted in human troponin positive and endogenous cardiomyocytes. C, Confocal evaluation of collateral vessels from CP hMSC treated hearts demonstrated human-specific CD-31 (PECAM-1) staining. D, Human lamin staining (arrows) co-localized with nuclei of smooth muscle in vessels from CP hMSC treated but not saline or naïve treated hearts. Bar, 20 μm for B-D. E, Percent of human troponin and Ki-67 positive cells in myocardial wall, upregulated in CP versus naïve hMSC treated hearts (n=10). F-G, Quantification of human CD-31 and co-localization between human-lamin and smooth muscle actin reveals significant contribution at the level of the papillary muscle from CP hMSC compared to naïve (n=10 in F and G). Star, p<0.05.
Scaled-up GMP production of cardiopoietic hMSC
Following safety and efficacy testing, the feasibility of scaled-up cardiopoietic hMSC production under Good Manufacturing Practice (GMP) criteria was evaluated (7,14). With regulatory approval, cardiopoietic guidance was carried out utilizing Standard Operating Procedures (SOP) with each cocktail component adhering to GMP guidelines. In this way, ≥600 million cardiopoietic hMSC were derived within 6-weeks of patient bone marrow harvest meeting sterility, purity and homogeneity release criteria. GMP-grade cardiopoietic hMSC were successfully derived from 10 patients, and released following demonstration of cardiac transcription factor upregulation consistent with an aptitude for repair. Thus, guided cardiopoiesis under GMP stringency demonstrated the capacity for scalable manufacturing of a stem cell biologics with imposed cardioreparative traits.
Discussion
Heterogenous outcome among patients treated with autologous stem cells is recognized as an impediment to broader clinical acceptance of cell-based therapy in cardiovascular medicine (5,9,11,30,31). The present study identified individuals that harbored hMSC with an innate capacity for myocardial repair. Reparative stem cells were characterized by upregulation of cardiac transcription factors, suggesting that preemptive cardiogenic specification could ensure enhanced therapeutic efficacy of hMSC.
Indeed, hMSC are a desirable source for organ repair due to accessibility for harvest, propensity to propagate in culture, and favorable biological profile (32,33). Discovered as a byproduct of hematopoietic stem cell isolation, derivation of hMSC is however confounded by phenotype heterogeneity among patients compromising therapeutic efficacy (34). Here, evaluation of adult stem cells with innate reparative potential identified traits associated with regenerative efficacy, consistent with expression of early and late cardiac transcription factors (15,25,35). Although expression of cardiac transcription factors is also seen in embryonic stem cells and is pivotal in their response to a natural sequence and duration of cardiogenic stimuli, the pluripotent phenotype has the distinctive capacity to readily engage into a program of lineage specification with orchestrated and targeted signaling driving cardiac differentiation (15,26,36,37). In contrast, adult stem cells demonstrate a latent plasticity, unmasked within strongly conducive in vivo environments to yield tissues emanating from all germinal layers (38–40). In vitro, adult stem cells demonstrate a limited capacity for differentiation, requiring protracted and continuous growth factor exposure to trigger cell fate specification (30,38). In this study, a combinatorial application of a set of recombinant factors was formulated to induce and potentiate the expression of cardiac transcription factors in patient-derived hMSC while maintaining the progenitor profile of cellular proliferation at an intermediate stage that preceded induction of sarcomerogenesis. Although components of embryonic cardiogenesis were represented, the cardiogenic cocktail regimen applied here in the hMSC model system was distinct from the complex kinetics and morphogen gradients of developmental biology. Yet, this cocktail was sufficient to secure across the studied patient cohort the reparative cardiopoietic phenotype. Furthermore, there is limited evidence for the capacity of adult hMSC to execute cardiac fate despite an aptitude for in vivo transdifferentiation (12,13,15,36,41). Here, the cocktail-guided cardiopoietic phenotype demonstrated an acquired capacity for extended differentiation yielding adult stem cell-derived sarcomerogenesis and electromechanical coupling.
The present proof-of-concept study thus establishes a platform to enhance the benefit of patient-derived stem cells in treating chronic ischemic cardiomyopathy. Identifying the molecular properties of reparative hMSC in an ischemic heart disease patient cohort established criteria consistent with an aptitude for cardiac repair. Application of a recombinant inductive milieu proved sufficient to guide hMSC isolated from a coronary artery disease cohort into cardiopoiesis, and thereby optimize efficacy prior to transplantation. Cardiogenic cocktail-induced preemptive cell fate specification ensured safe and lasting functional and structural benefit following transplantation in the failing ischemic heart. Within the healing murine heart, human-specific nuclear, sarcomeric and gap junction proteins documented an active contribution to infarct repair. Moreover, upregulation of endogenous stem cells and promotion of cell cycle activity within the host myocardium indicated a concomitant paracrine role in repair (5,34,42,43). Scaled-up GMP production of cardiopoietic hMSC indicated translational feasibility. Ultimately, the value of this technology to the patient will require controlled clinical trials.
Supplementary Material
Acknowledgments
Supported by NIH (HL83439, HL85208), AHA, Marriott Heart Disease Research Program, Cardio3 BioSciences, Ted Nash Long Life Foundation, Ralph Wilson Medical Research Foundation, Mayo Clinic General Mills Clinician-Investigator Fellowship, Mayo Graduate School, Mayo Clinic FUTR Career Development Award, and Mayo Clinic Discovery Translation Program.
We thank Dr. Allan B. Dietz for contribution of platelet lysate.
Non-standard Abbreviations and Acronyms
- CP
cardiopoietic stem cell
- GMP
good manufacturing practice
- hMSC
human mesenchymal stem cell
- SOP
standard operating procedure
Footnotes
Conflict of Interest:
A.B., J.B., and A.T. serve on the research advisory board of Cardio3 BioSciences.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dimmeler S, Zeiher AM, Schneider MD. Unchain my heart: the scientific foundations of cardiac repair. J Clin Invest. 2005;115:572–83. doi: 10.1172/JCI24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srivastava D, Ivey KN. Potential of stem-cell-based therapies for heart disease. Nature. 2006;441:1097–9. doi: 10.1038/nature04961. [DOI] [PubMed] [Google Scholar]
- 3.Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–42. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 4.Abdel-Latif A, Bolli R, Tleyjeh IM, et al. Adult bone marrow-derived cells for cardiac repair: a systematic review and meta-analysis. Arch Intern Med. 2007;167:989–97. doi: 10.1001/archinte.167.10.989. [DOI] [PubMed] [Google Scholar]
- 5.Passier R, van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–9. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 6.Hahn JY, Cho HJ, Kang HJ, et al. Pre-treatment of mesenchymal stem cells with a combination of growth factors enhances gap junction formation, cytoprotective effect on cardiomyocytes, and therapeutic efficacy for myocardial infarction. J Am Coll Cardiol. 2008;51:933–43. doi: 10.1016/j.jacc.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 7.Hare JM, Traverse JH, Henry TD, et al. A randomized, double-blind, placebo-controlled, dose-escalation study of intravenous adult human mesenchymal stem cells (prochymal) after acute myocardial infarction. J Am Coll Cardiol. 2009;54:2277–86. doi: 10.1016/j.jacc.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quevedo HC, Hatzistergos KE, Oskouei BN, et al. Allogeneic mesenchymal stem cells restore cardiac function in chronic ischemic cardiomyopathy via trilineage differentiating capacity. Proc Natl Acad Sci USA. 2009;106:14022–7. doi: 10.1073/pnas.0903201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenzweig A. Cardiac cell therapy--mixed results from mixed cells. N Engl J Med. 2006;355:1274–7. doi: 10.1056/NEJMe068172. [DOI] [PubMed] [Google Scholar]
- 10.Chavakis E, Urbich C, Dimmeler S. Homing and engraftment of progenitor cells: a prerequisite for cell therapy. J Mol Cell Cardiol. 2008;45:514–22. doi: 10.1016/j.yjmcc.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Dimmeler S, Leri A. Aging and disease as modifiers of efficacy of cell therapy. Circ Res. 2008;102:1319–30. doi: 10.1161/CIRCRESAHA.108.175943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston PV, Sasano T, Mills K, et al. Engraftment, differentiation, and functional benefits of autologous cardiosphere-derived cells in porcine ischemic cardiomyopathy. Circulation. 2009;120:1075–83. doi: 10.1161/CIRCULATIONAHA.108.816058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith RR, Barile L, Cho HC, et al. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- 14.Dietz AB, Padley DJ, Gastineau DA. Infrastructure development for human cell therapy translation. Clin Pharmacol Ther. 2007;82:320–4. doi: 10.1038/sj.clpt.6100288. [DOI] [PubMed] [Google Scholar]
- 15.Behfar A, Perez-Terzic C, Faustino RS, et al. Cardiopoietic programming of embryonic stem cells for tumor-free heart repair. J Exp Med. 2007;204:405–20. doi: 10.1084/jem.20061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behfar A, Zingman LV, Hodgson DM, et al. Stem cell differentiation requires a paracrine pathway in the heart. FASEB J. 2002;16:1558–66. doi: 10.1096/fj.02-0072com. [DOI] [PubMed] [Google Scholar]
- 17.Plenz G, Eschert H, Erren M, et al. The interleukin-6/interleukin-6-receptor system is activated in donor hearts. J Am Coll Cardiol. 2002;39:1508–12. doi: 10.1016/s0735-1097(02)01791-6. [DOI] [PubMed] [Google Scholar]
- 18.Messina E, De Angelis L, Frati G, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004;95:911–21. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 19.Wobus AM, Kaomei G, Shan J, et al. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J Mol Cell Cardiol. 1997;29:1525–39. doi: 10.1006/jmcc.1997.0433. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Terzic C, Behfar A, Mery A, van Deursen JM, Terzic A, Puceat M. Structural adaptation of the nuclear pore complex in stem cell-derived cardiomyocytes. Circ Res. 2003;92:444–52. doi: 10.1161/01.RES.0000059415.25070.54. [DOI] [PubMed] [Google Scholar]
- 21.Yamada S, Nelson TJ, Behfar A, Crespo-Diaz RJ, Fraidenraich D, Terzic A. Stem cell transplant into preimplantation embryo yields myocardial infarction-resistant adult phenotype. Stem Cells. 2009;27:1697–705. doi: 10.1002/stem.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou B, Ma Q, Rajagopal S, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–13. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruneau BG, Nemer G, Schmitt JP, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–21. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 24.David R, Brenner C, Stieber J, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–45. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- 25.Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes Dev. 2003;17:1937–56. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- 26.Arrell DK, Niederlander NJ, Faustino RS, Behfar A, Terzic A. Cardioinductive network guiding stem cell differentiation revealed by proteomic cartography of tumor necrosis factor alpha-primed endodermal secretome. Stem Cells. 2008;26:387–400. doi: 10.1634/stemcells.2007-0599. [DOI] [PubMed] [Google Scholar]
- 27.Mummery C, Ward-van Oostwaard D, Doevendans P, et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–40. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 28.Grepin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development. 1997;124:2387–95. doi: 10.1242/dev.124.12.2387. [DOI] [PubMed] [Google Scholar]
- 29.Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–76. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 30.Clavel C, Verfaillie CM. Bone-marrow-derived cells and heart repair. Curr Opin Organ Transplant. 2008;13:36–43. doi: 10.1097/MOT.0b013e3282f428d1. [DOI] [PubMed] [Google Scholar]
- 31.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726–36. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 32.Amado LC, Schuleri KH, Saliaris AP, et al. Multimodality noninvasive imaging demonstrates in vivo cardiac regeneration after mesenchymal stem cell therapy. J Am Coll Cardiol. 2006;48:2116–24. doi: 10.1016/j.jacc.2006.06.073. [DOI] [PubMed] [Google Scholar]
- 33.Sotiropoulou PA, Perez SA, Salagianni M, Baxevanis CN, Papamichail M. Characterization of the optimal culture conditions for clinical scale production of human mesenchymal stem cells. Stem Cells. 2006;24:462–71. doi: 10.1634/stemcells.2004-0331. [DOI] [PubMed] [Google Scholar]
- 34.Psaltis PJ, Zannettino AC, Worthley SG, Gronthos S. Concise review: mesenchymal stromal cells: potential for cardiovascular repair. Stem Cells. 2008;26:2201–10. doi: 10.1634/stemcells.2008-0428. [DOI] [PubMed] [Google Scholar]
- 35.Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132:537–43. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Behfar A, Faustino RS, Arrell DK, Dzeja PP, Perez-Terzic C, Terzic A. Guided stem cell cardiopoiesis: discovery and translation. J Mol Cell Cardiol. 2008;45:523–9. doi: 10.1016/j.yjmcc.2008.09.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faustino RS, Behfar A, Perez-Terzic C, Terzic A. Genomic chart guiding embryonic stem cell cardiopoiesis. Genome Biol. 2008;9:R6.1–16. doi: 10.1186/gb-2008-9-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, Jahagirdar BN, Reinhardt RL, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–9. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 39.Liechty KW, MacKenzie TC, Shaaban AF, et al. Human mesenchymal stem cells engraft and demonstrate site-specific differentiation after in utero transplantation in sheep. Nat Med. 2000;6:1282–6. doi: 10.1038/81395. [DOI] [PubMed] [Google Scholar]
- 40.Orlic D, Kajstura J, Chimenti S, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410:701–5. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 41.Bird SD, Doevendans PA, van Rooijen MA, et al. The human adult cardiomyocyte phenotype. Cardiovasc Res. 2003;58:423–34. doi: 10.1016/s0008-6363(03)00253-0. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez A, Rota M, Nurzynska D, et al. Activation of cardiac progenitor cells reverses the failing heart senescent phenotype and prolongs lifespan. Circ Res. 2008;102:597–606. doi: 10.1161/CIRCRESAHA.107.165464. [DOI] [PubMed] [Google Scholar]
- 43.Deuse T, Peter C, Fedak PW, et al. Hepatocyte growth factor or vascular endothelial growth factor gene transfer maximizes mesenchymal stem cell-based myocardial salvage after acute myocardial infarction. Circulation. 2009;120:S247–54. doi: 10.1161/CIRCULATIONAHA.108.843680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.