Abstract
She2p is an RNA-binding protein that recognizes a zipcode on specific mRNAs necessary for the assembly of a protein complex that localizes them to the yeast bud tip. In this issue of Genes & Development, Shen and colleagues (pp. 1914–1926) demonstrate that She2p associates with RNAPII globally, but then recognizes the nascent chain only if it contains a zipcode. This demonstrates yet another case where the mRNA's cytoplasmic fate is determined by the RNAPII complex.
Keywords: mRNA, localization, yeast, She2p, Spt4p, Spt5p, cotranscriptional
The functional outcome of an mRNA is determined in the cytoplasm through binding proteins that specify its localization, translation, and degradation. It is a misperception that the fate of a localized mRNA is determined after nuclear export. In fact, evidence is accumulating to the contrary (Farina and Singer 2002). In this issue of Genes & Development, Shen et al. (2010) further this evidence by describing the nuclear component of localization in budding yeast, and demonstrating that localization of several mRNAs to the bud tip is determined cotranscriptionally. They show that the RNA-binding protein She2p, which is responsible for assembling a complex in the cytoplasm that contains myosin for RNA transport (Long et al. 1997, 2000), recognizes its target nascent mRNA in the nucleus. Importantly, Shen et al. (2010) show mechanistically that She2p interacts first with RNA polymerase II (RNAPII) throughout the entire elongation process via the association with the transcription factor Spt4–Spt5p/DSIF (DRB [5,6-dichloro-1-b-d-ribofuranosylbenzimidazole] sensitivity-inducing factor), and then “hops” onto the nascent mRNA in a sequence-dependent manner. Interestingly, the initial interaction with RNAPII is general, but the move to the RNA presumably requires the presence of the She2p-binding sites on the RNA. These results, taken together with examples published over recent years and described below, shift our attention back into the nucleus, demonstrating that the ultimate function of an mRNA can be determined in an RNAPII-dependent process.
Cotranscriptional regulation of mRNA localization
In budding yeast, the She2 protein binds to specific RNA sequences called “zipcodes” (Kislauskis and Singer 1992) within the localizing transcript (Olivier et al. 2005), and directs localization of several mRNAs to the bud tip. When deleted, cells no longer properly localize their transcripts (Long et al. 1997). She2p recruits a type V myosin, Myo4p, through the She3p adaptor protein, thereby promoting the formation of the “locasome” (Long et al. 2000) needed for proper mRNA localization. The best understood example of a localized mRNA in budding yeast is ASH1, which encodes a transcriptional repressor of the HO locus and is a regulator of mating type switching in yeast. It is expressed in late mitosis and selectively represses mating type switching in the daughter cell so that only mothers can switch mating types after each cell division. She2p is a shuttling protein, and possesses a nonclassical nuclear localization sequence (NLS). When mutated, She2p became excluded from the nucleus, leading to mislocalization of ASH1 mRNA (Shen et al. 2009), suggesting the possibility that She2p could bind localizing mRNAs in the nucleus.
Different regulators of mRNA localization have been shown to bind their targets in the nucleus in both Saccharomyces cerevisiae and in higher eukaryotes (Holt and Bullock 2009; Martin and Ephrussi 2009). For example, ZBP1 and ZBP2 are two RNA-binding proteins that regulate localization of β-actin mRNA to the leading edge of chicken embryo fibroblasts and growth cones in developing neurons. This localization is important for maintenance of cell polarity and directed cell motion (Shestakova et al. 2001; Farina et al. 2003). Using immunofluorescence and fluorescent in situ hybridization (FISH), ZBP1, ZBP2, and β-actin mRNA were found colocalized in nuclear foci identified as active β-actin transcription sites (Oleynikov and Singer 2003; Pan et al. 2007). However, it is unclear if RNAPII orchestrates ZBP1 and ZBP2 binding to their target mRNAs, or whether they bind to the β-actin mRNA independently.
Shen et al. (2010) offer the first account that provides a mechanistic interpretation of how this association is achieved, and how it is coupled with the transcription machinery. They show that nuclear association of She2p with the nascent mRNA occurred cotranscriptionally through the interaction with the RNAPII-bound Spt4–5p transcriptional elongators. This interaction was necessary for proper localization of ASH1 mRNA to the bud tip and subsequent localization of Ash1p in the daughter nucleus to inhibit mating type switching (Jansen et al. 1996). Consequently, mutation of either Spt4 or Spt5 strongly reduced the efficiency of ASH1 localization to the bud tip, phenocopying the She2p deletion. Remarkably, chromatin immunoprecipitation (ChIP) experiments showed that She2p associated with the actively transcribing RNAPII of nonlocalizing transcripts ACT1, FBA1, PMA1, ASC1, or PGK1 as well. However, the ChIP signal was greatly enriched when the localizing mRNAs ASH1, IST2, or EAR1 were transcribed, because She2p could apparently “hop” from an RNAPII-bound Spt4p–5/DSIF directly onto the zipcode sequence in the nascent mRNA. Enrichment of the She2p signal was RNase-sensitive, while RNase treatment had no effect on the level of She2p associated with the nonlocalizing ACT1, FBA1, PMA1, ASC1, and PGK1 genes. Finally, ChIP experiments also revealed that the presence of a zipcode inserted into the nonlocalizing LacZ mRNA resulted in the RNA-dependent interaction of She2p with the LacZ gene.
The “hopping” of the She2p regulator from the Spt4/5p-associated RNAPII onto the mRNA in a sequence-dependent manner is a conceptual teaser. When the zipcode sequence that forms a secondary structure on the mRNA (there are four of them in ASH1: three in the ORF, and one in the 3′ untranslated region [UTR]) (Chartrand et al. 1999) first emerges from the elongating RNAPII, the affinity of She2p for the RNA relative to Spt4p–Spt5p has to be greater than for the RNAPII in order to switch. This jump is specific for the zipcode, since it does not occur when a nonlocalizing mRNA is transcribed. Here, Spt4p and Spt5p could act as a binding adjuvant to increase the likelihood that each zipcode will be occupied. How the specific mRNA sequence is recognized and how the transfer of the RNAPII-bound She2p onto the nascent chain is achieved remain to be addressed. Does the reading of the mRNA sequence occur concomitantly with the elongation process, or does the RNAPII pause every so often, giving RNAPII-bound She2p adequate time to find its target? In case of the latter, this mechanism would recall the capping reaction of the newly transcribed mRNA, and, possibly, this mechanism could serve as a model for She2p interaction with transcribing zipcode RNA.
The capping process begins soon after transcription initiation, when RNAPII is paused ∼20–30 nucleotides (nt) into the pre-mRNA synthesis by the joint action of DSIF (an Spt4p–Spt5p complex) and NELF (a four-protein complex) (Hartzog et al. 1998; Wu et al. 2003). This promoter-proximal pausing provides a temporal window for capping execution as soon as the nascent transcript emerges from the RNAPII (Chiu et al. 2001; Pei and Shuman 2002). After the relief of the pause, RNAPII engages in a productive elongation, and Spt4–5/DSIF remains associated with it throughout this process (Andrulis et al. 2000).
It is possible that, similar to the capping reaction, She2p could be transferred onto the nascent mRNA in an RNAPII-induced transcriptional pause. Such a scenario could occur farther downstream into the coding sequence because elongating RNAPII is known to pause away from the transcription start site (Darzacq et al. 2007). The pause sites are usually found in mammalian cells and are less well characterized in budding yeast but are not unlikely. With the help of Spt4p and Spt5p, a stalled RNAPII would have adequate time to search for a zipcode sequence to allow She2p to “hop” onto it.
For instance, the RNAPII–Spt4/5p–She2p complex could recognize its target mRNA if the RNAPII would proceed with transcription more slowly on genes coding for localizing mRNAs. ASH1 zipcodes, for example, are long, 118- to 250-nt sequences that form strong secondary structures (Chartrand et al. 1999); the ones located in the ORF could slow down the elongating polymerase. A slower (or paused) RNAPII could assure that each and every zipcode will be identified as a binding site, and enable She2p to disassociate from the RNAPII and bind to the mRNA in a timely fashion. This dissociation step may alternatively provide a resistance to polymerase elongation. In the case of nonlocalizing mRNAs that do not have a zipcode, the RNAPII with the bound She2p would therefore move through the gene more rapidly. This mechanism is reminiscent of alternative splicing, where the recognition of the alternative splice site depends on the velocity of the elongating RNAPII (Muñoz et al. 2009). In this case, it was described that the transcribing RNAPII will recognize the weaker splice site only if it traverses through the coding sequence slowly enough for the splicing machinery to assemble on the newly synthesized transcript. If the velocity of the RNAPII is too high, spliceosomal assembly occurs only on stronger splice sites.
She2p is only one of the components of the locasome structure (Long et al. 2000) that is responsible for bud-specific transport of mRNAs. Other components, like She1p–5p, are essential for proper localization of ASH1 mRNA and Ash1p sorting (Long et al. 1997). Interestingly, when the She2p NLS is mutated so that it can no longer enter the nucleus, it can still bind to the target mRNA on its own in the cytoplasm (Du et al. 2008), but with a diffuse RNA phenotype rather than bud tip-anchored mRNA localization (Shen et al. 2009). Furthermore, when the N terminus of She3p, an adapter of She2 interaction with Myo4p, was fused with She2p to prevent its nuclear import, ASH1 mRNAs could localize to the bud tip as in wild-type cells, but, remarkably, failed to redistribute Ash1p asymmetrically (Du et al. 2008). While in the nucleus, She2p also recruits the Loc1p and Puf6p proteins that are involved in the translational repression of the ASH1 transcript (Deng et al. 2008; Shen et al. 2009). Alhough cytoplasmic association of ASH1 mRNA with She2p does occur (and these mRNAs can localize to the bud tip with a reduced efficiency), nuclear mRNA processing mediated by the activity of nuclear She2p appears to be critical for coordination of cellular localization and appropriate Ash1p translation. This demonstrates that the order of binding of regulatory proteins to the mRNA involved in both processes is paramount to achieve a highly efficient localization and translation, and She2p appears to orchestrate these events. However, it is unclear whether an entire locasome assembles on the nascent mRNA chain (plausibly also in the Spt4p/5p-dependent manner), or if some of the components of the locasome interact in the cytoplasm. While association of some components, like Loc1p and Puf6p, could occur cotranscriptionally together with She2p, others, like Myo4p, appear to bind in the cytoplasm. Of course, there could be other factors that are mRNA-specific or are found to be involved in locasome formation only during specific stages of the cell cycle. Controlling which proteins would participate in mRNA localization would provide a cell with greater control over the formation of an mRNP and its cytoplasmic fate. It is tempting to hypothesize that such regulation could begin cotranscriptionally by controlling which locasome components would be recruited to the RNAPII and later bind to the target mRNA in response to a cellular hierarchy. A cell could synthesize mRNPs of different compositions from a specific gene, depending on the phase of the cell cycle or its stage during development or differentiation, and this could increase the number of localizing patterns and, potentially, other mRNA functions. Such regulatory mechanisms would become more important in higher eukaryotes, where temporal and spatial regulation of localization occurs during differentiation and development.
In the Drosophila oocyte, for example, oskar mRNA travels to the posterior pole of an oocyte during oocyte development. This process is regulated through nuclear splicing, rather than recognition of a specific zipcode sequence in the mRNA itself (Hachet and Ephrussi 2004). In this case, the nuclear shuttling proteins Y14/Tsunagi and Mago nashi, which are also components of the exon–junction complex (EJC), are required for oskar mRNA localization. As with other zipcode-binding proteins, these travel with the localizing mRNA, and are found to colocalize with oskar mRNA at the posterior pole of an oocyte. Intronless oskar mRNA not only mislocalized, but was also poorly translated relative to the wild-type oskar mRNA. Therefore, splicing in the nucleus not only regulated localization of oskar mRNA, but also the efficiency of its translation in the cytoplasm. Hence, alternative splicing could give rise to different mRNA–protein complexes with diverse cytoplasmic localization patterns that could participate in regulating embryo development. During Drosophila embryogenesis, 71% out of 3370 mRNAs investigated were found to be localized, and several dozen new localization patterns were identified (Lécuyer et al. 2007), indicating that different mRNAs interact with different protein regulators to assure variability. As with oskar mRNA, the locasome assembly could begin cotranscriptionally in a regulated manner to maximize regulation of localization patterning.
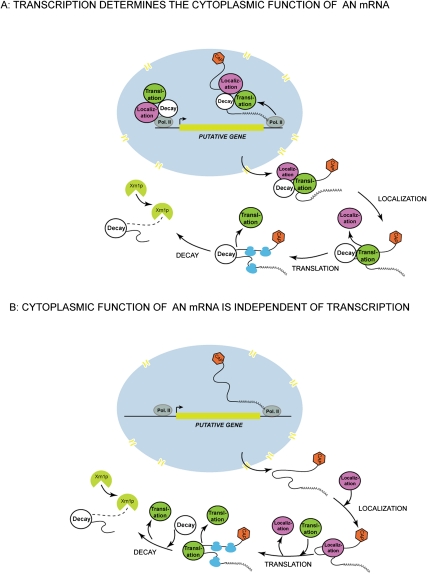
Cytoplasmic processes as diverse as mRNA decay and cellular localization were shown to be regulated on a transcriptional level in both yeast and higher eukaryotes (Goler-Baron et al. 2008; Holt and Bullock 2009; Martin and Ephrussi 2009). To date, such examples are rare, and therefore the regulatory path that cotranscriptionally defines the composition of the cytoplasmic mRNP is still an exception rather than a rule. However, these cases will likely become more prominent in the future. Experimental evidence presented by Shen et al. (2010) and bolstered by the similar findings reported for other genes suggests a scenario as shown in Figure 1. An mRNA becomes equipped with all of the critical regulatory proteins cotranscriptionally, and then progressively sheds them in the cytoplasm with the completion of each successive step in the mRNA life cycle (Fig. 1A). This cotranscriptional view of the mRNP assembly is markedly different from the one that is generally assumed to exist for a “typical” mRNA, where mRNP assembly is regarded as a step-by-step process that depends on the successive, diffusion-driven association and disassociation of individual regulatory proteins in the cytoplasm (Fig. 1B).
Figure 1.
(A) An mRNP is fully configured in the nucleus cotranscriptionally. It exits the nucleus equipped with proteins that define its localization, translation, and decay patterns. After each completed step, individual regulatory proteins are shed away in a step-by-step fashion. (B) An mRNP structure changes dynamically throughout the lifetime of an mRNA in the cytoplasm through the diffusion-driven association and disassociation process. Transcriptional events in the nucleus are separated physically and temporally from those that occur in the cytoplasm, and have little effect on the formation of a functional mRNP.
Perspective
Transcription of nascent mRNA molecules is a highly dynamic and regulated process, and many processes that affect mRNP configuration occur cotranscriptionally and are independent of the cytoplasmic association of protein regulators with the mRNA. This brings the RNAPII enzymatic complex into the center of mRNP regulation, where it can be viewed as both a catalyst of mRNA synthesis and a scaffold onto which regulatory proteins assemble. According to the Saccharomyces genome database, Rpb1p, the large RNAPII subunit, interacts physically with 129 other proteins, and each one of these proteins further interacts with a number of other proteins. For example, the TFIID subunit Tef14p interacts with Rpb1p directly and with 88 other proteins, illustrating that RNAPII holds a huge capacity to engage in a variety of unique complexes, all of which can ultimately determine the functional outcome of a cytoplasmic mRNA. She2p, Spt4p, and Spt5p are fairly abundant proteins, and were reported to be present in ∼4070, 4490, and 2340 protein molecules per cell, respectively (Ghaemmaghami et al. 2003). The majority of their protein is concentrated in the nucleus, and they can potentially interact with the RNAPII. It would be interesting to know whether every RNAPII associates with the Spt4p–Spt5p–She2 complex, or if there are RNA polymerases that do not. If so, are there zipcodes that are not occupied by the She2 protein, and how would this affect localization (and translation) efficiency of an mRNA such as ASH1? Varying the scope of these interactions during the synthesis of a single gene could vary the way a single mRNA species is localized, translated, and decayed, by carrying with it into the cytoplasm a regulatory mechanism that could play an important role during differentiation and development. Looking forward, it will be exciting to see whether this type of regulation becomes accepted as a general mechanism of mRNP assembly.
Acknowledgments
We thank Jonathan R. Warner for critical reading of the manuscript. This work was supported by NIH GM57071 to R.H.S.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1972810.
References
- Andrulis ED, Guzmán E, Döring P, Werner J, Lis JT 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes Dev 14: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH, Long RM 1999. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 9: 333–336 [DOI] [PubMed] [Google Scholar]
- Chiu YL, Coronel E, Ho CK, Shuman S, Rana TM 2001. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J Biol Chem 276: 12959–12966 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH 2007. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14: 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Singer RH, Gu W 2008. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev 22: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du TG, Jellbauer S, Müller M, Schmid M, Niessing D, Jansen RP 2008. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep 9: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Singer RH 2002. The nuclear connection in RNA transport and localization. Trends Cell Biol 12: 466–472 [DOI] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M 2008. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev 22: 2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A 2004. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428: 959–963 [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev 12: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bullock SL 2009. Subcellular mRNA localization in animal cells and why it matters. Science 326: 1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K 1996. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell 84: 687–697 [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Singer RH 1992. Determinants of mRNA localization. Curr Opin Cell Biol 4: 975–978 [DOI] [PubMed] [Google Scholar]
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277: 383–387 [DOI] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P 2000. She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J 19: 6592–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A 2009. mRNA localization: Gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz MJ, Pérez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. 2009. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 137: 708–720 [DOI] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH 2003. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol 13: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P 2005. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol 25: 4752–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Hüttelmaier S, Singer RH, Gu W 2007. ZBP2 facilitates binding of ZBP1 to β-actin mRNA during transcription. Mol Cell Biol 27: 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Shuman S 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem 277: 19639–19648 [DOI] [PubMed] [Google Scholar]
- Shen Z, Paquin N, Forget A, Chartrand P 2009. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell 20: 2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, St-Denis A, Chartrand P 2010. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4–Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev (this issue). doi: 10.1101/gad.1937510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova EA, Singer RH, Condeelis J 2001. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci 98: 7045–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev 17: 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]