Abstract
tRNA biology has come of age, revealing an unprecedented level of understanding and many unexpected discoveries along the way. This review highlights new findings on the diverse pathways of tRNA maturation, and on the formation and function of a number of modifications. Topics of special focus include the regulation of tRNA biosynthesis, quality control tRNA turnover mechanisms, widespread tRNA cleavage pathways activated in response to stress and other growth conditions, emerging evidence of signaling pathways involving tRNA and cleavage fragments, and the sophisticated intracellular tRNA trafficking that occurs during and after biosynthesis.
Keywords: Cleavage, modifications, processing, splicing, tRNA, trafficking, turnover
tRNA biogenesis involves the synthesis of the initial transcript, followed by processing to remove the 5′ leader, trim the 3′ trailer, add CCA, splice introns that may be present, modify multiple nucleoside residues (Fig. 1), and, for eukaryotes, export the tRNA to the cytoplasm, before its use in translation (Hopper and Phizicky 2003). Although superficially a simple process, results in the last several years have highlighted an unexpected complexity and breadth in tRNA processing and trafficking pathways, and have uncovered multiple levels of regulation of tRNA biosynthesis and function. For the first time, there is a nearly complete working knowledge of the essential toolkit of genes required for tRNA biogenesis in the yeast Saccharomyces cerevisiae (Table 1), and the pace of discovery in bacteria, archaea, and other eukaryotes is accelerating rapidly. Exciting stories have unfolded that describe unexpected layers of regulation in tRNA gene transcription, new insights in end maturation and splicing, and newly defined roles of tRNA modifications in translation and tRNA quality control. Intriguing new results demonstrate the widespread existence of tRNA cleavage pathways activated by stress and other growth conditions, the unexpected signaling roles of tRNA fragments and tRNA molecules, and the surprisingly intricate pathways of tRNA trafficking within the cell. In this review, we discuss some of the highlights of these findings in tRNA biology.
Figure 1.
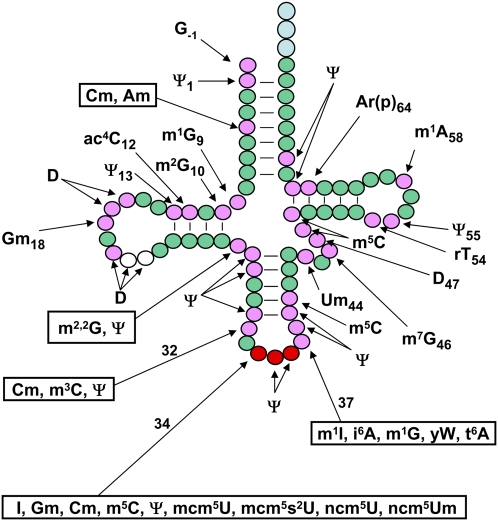
A schematic of modifications found in cytoplasmic tRNA in S. cerevisiae. tRNA is shown in its usual secondary structure form, with circles representing nucleotides and lines representing base pairs. (Green circles) Residues that are unmodified in all yeast tRNA species; (pink circles) residues that are modified in some or all tRNA species; (white circles) additional residues (20a and 20b) that are present in some, but not all, tRNAs and are sometimes modified; (red circles) anticodon residues, which are modified in some tRNAs; (light-blue circles) the CCA end. Conventional abbreviations are used; see the Modomics database (http://modomics.genesilico.pl). (Ψ) Pseudouridine; (Am) 2′-O-methyladenosine; (Cm) 2′-O-methylcytidine; (m1G) 1-methylguanosine; (m2G) 2-methylguanosine; (ac4C) 4-acetylcytidine; (D) dihydrouridine; (Gm) 2′-O-methylguanosine; (m2,2G) N2,N2-dimethylguanosine; (m3C) 3-methylcytidine; (I) inosine; (m5C) 5-methylcytidine; (mcm5U) 5-methoxycarbonylmethyluridine; (mcm5s2U) 5-methoxycarbonylmethyl-2-thiouridine; (ncm5U) 5-carbamoylmethyluridine; (ncm5Um) 5-carbamoylmethyl-2′-O-methyluridine; (m1I) 1-methylinosine; (i6A) N6-isopentenyl adenosine; (yW) wybutosine; (t6A) N6-threonylcarbamoyladenosine; (Um) 2′-O-methyluridine; (m7G) 7-methylguanosine; (rT) ribothymidine; [Ar(p)] 2′-O-ribosyladenosine (phosphate). The pictured molecule starts at position −1 and is numbered consecutively from the next base (+1) to 76 (with the insertion of two residues [20a and 20b]). Several tRNA species have a longer variable arm starting after residue 44, and some tRNAs have different numbers of residues in the D-loop and the variable arm, but the anticodon is always numbered residues 34, 35, and 36, and the CCA end is always numbered residues 74, 75, and 76.
Table 1.
S. cerevisiae genes implicated in tRNA processing and tRNA trafficking
aFor complex modifications, the underlined portion indicates the part of the modification due to the corresponding gene(s).
Multiple layers of regulation of tRNA transcription
tRNA and rRNA genes are highly transcribed, leading to the production in yeast of ∼3 million tRNAs per generation and 300,000 ribosomes (Waldron and Lacroute 1975), compared with about 60,000 mRNAs (Ares et al. 1999). Because of the energy devoted to tRNA and rRNA transcription, and because of the required coordination of tRNA and ribosome function, tRNA transcription via RNA polymerase III (Pol III) and rRNA transcription via Pol I need to be coordinated and regulated in response to cellular nutrient availability and other environmental information. The consequences of inappropriate regulation of tRNA transcription have been underscored by the results of Marshall et al. (2008) showing that elevated tRNAiMet transcription can promote cell proliferation and immortalization as well as tumors in mice. Within the past decade, there has been much progress delineating mechanisms by which Pol III transcription is regulated and coordinated with environmental signals (for review, see Willis and Moir 2007; Ciesla and Boguta 2008).
Pol III is negatively regulated by a single protein, Maf1, first discovered in yeast by its effects on tRNA-mediated nonsense suppression (Murawski et al. 1994; Moir et al. 2006). Maf1 is conserved throughout eukaryotes, although mammalian Maf1 negatively regulates Pol I and Pol II transcription in addition to Pol III transcription (Pluta et al. 2001; Reina et al. 2006; Johnson et al. 2007). Yeast and mammalian Maf1 interacts directly with Pol III subunits (Pluta et al. 2001; Gavin et al. 2006; Oficjalska-Pham et al. 2006; Reina et al. 2006) and components of the TFIIIB transcription factor (Upadhya et al. 2002; Desai et al. 2005; Reina et al. 2006; Rollins et al. 2007; for review, see Ciesla and Boguta 2008).
Substantial evidence suggests that the PKA and TOR pathways regulate Maf1. Under favorable growth conditions, Maf1 is phosphorylated by both PKA and the TOR-dependent kinase Sch9 (Huber et al. 2009; J Lee et al. 2009; Wei et al. 2009; for review, see Boguta 2009). Maf1 activity is also regulated by TOR via a Sch9-independent mechanism (J Lee et al. 2009; Wei and Zheng 2009). Phosphorylation of Maf1 prevents its negative regulation of Pol III transcription.
Maf1 is dephosphorylated in response to conditions that slow growth—nutrient deprivation, shift from fermentation to respiration carbon sources, DNA damage, and various other environmental stresses (Boisnard et al. 2009; for review, see Willis and Moir 2007; Ciesla and Boguta 2008)—and unphosphorylated Maf1 is able to negatively regulate Pol III transcription. Earlier studies had implicated Tpd3, a regulatory subunit of the TOR-dependent protein phosphatase PP2A, in tRNA transcription (van Zyl et al. 1992), and it has since been learned that Maf1 remains phosphorylated in PP2A mutants (Boisnard et al. 2009), showing that PP2A is a phosphatase acting on Maf1.
In some yeast strains, phosphorylated Maf1 is located in the cytoplasm, and is therefore unable to access and repress Pol III transcription. Maf1's cytoplasmic location is mediated by two mechanisms: phosphorylation-dependent inactivation of the Maf1 nuclear location signals (NLSs) (Moir et al. 2006), and nuclear export of phosphorylated Maf1 by the exportin Msn5 (Towpik et al. 2008). Conversely, in nutrient-deprived and stress conditions, dephosphorylated Maf1 is located in the nucleus, and is thereby able to access Pol III and down-regulate tRNA transcription.
Although the different distribution of Maf1 between the nucleus and the cytoplasm could, in principle, account for Maf1's regulation of Poll III transcription, two lines of evidence suggest that the nuclear–cytoplasmic dynamics of Maf1 may instead be a mechanism to fine-tune regulation. First, Maf1 remains able to appropriately regulate Pol III transcription in cells lacking Msn5, which is required to export phosphorylated Maf1 from the nucleus (Towpik et al. 2008). Second, although Maf1 is constitutively located in the nucleus in the common W303 yeast strain background, Maf1 regulation of Pol III is nevertheless responsive to environmental signaling (Wei et al. 2009). As phosphorylation of Maf1 also prevents the interaction of Maf1 with Pol III (Oficjalska-Pham et al. 2006), it would appear that this interaction may be the primary level of negative regulation of Pol III transcription by Maf1.
tRNA end-processing and splicing reveal new insights in biology and evolution
Surprises in 5′ processing
The study of RNase P catalysis has revealed a series of fascinating discoveries about the nature of catalysis and the universal requirement for the reaction to occur. The accepted dogma of tRNA processing has always involved synthesis of a precursor tRNA containing a 5′ leader sequence, which was subsequently removed by the endonuclease RNase P (Walker and Engelke 2006). It has also been accepted dogma since the early 1980s that RNase P is a ribonucleoprotein (RNP), and that RNA is the catalytic component in bacteria (Guerrier-Takada et al. 1983; Torres-Larios et al. 2005), which have one protein component, and in archaea (Pannucci et al. 1999), which have five protein components (Cho et al. 2010). However, the source of RNase P catalysis was unknown in eukaryotes such as yeast or humans, which have nine (Table 1) or 10 protein subunits, respectively, in addition to their RNA components (Walker and Engelke 2006), particularly since their RNA components have substantial differences in regions important for stability and catalysis (Marquez et al. 2005, 2006). Nonetheless, results from Kirsebom and coworkers (Kikovska et al. 2007) have demonstrated that the RNA component of eukaryotic RNase P of humans or of Giardia lamblia is, in fact, catalytic, albeit with very poor kinetics, likely due in part to the absence in eukaryotes of helices P15–P17 (Marquez et al. 2005, 2006). This low level of catalytic activity supports the idea of a prominent role of one or more of the eukaryotic protein components in stimulating catalysis by the RNA component (Marquez et al. 2006). Indeed, Gopalan and coworkers (Tsai et al. 2006) have shown with reconstituted archaeal RNase P holoenzyme that the protein subunits have a 4000-fold stimulatory effect on catalytic efficiency.
In light of this evidence that the RNA component of RNase P is always the catalytic component, it was a distinct surprise to find that human mitochondrial RNase P has no RNA components at all (Holzmann et al. 2008). Although there had been prior reports hinting that RNase P activity in chloroplasts (Wang et al. 1988) and human mitochondria (Rossmanith and Karwan 1998) did not require an RNA component, the chloroplast data had not been confirmed in >20 years and the human mitochondrial data had been challenged (Puranam and Attardi 2001), leaving many convinced that active RNase P invariably required RNA. However, Rossmanith and colleagues (Holzmann et al. 2008) have shown convincingly that human mitochondrial RNase P is instead comprised of only three seemingly unrelated polypeptides: a protein related to the tRNA m1G9 methyltransferase, a member of the short chain dehydrogenase/reductase complex, and an uncharacterized ORF. Since active mitochondrial RNase P was reconstituted after expression and purification of these three protein subunits in Escherichia coli, it is certain that the enzyme does not require RNA.
New results have also identified an exception to the apparent universal requirement for RNase P activity during tRNA biogenesis. Computational analysis suggested that RNase P RNA was missing from Nanoarchaeum equitans, Pyrobaculum aerophylum, and Aquifex aeolicus (Li and Altman 2004). Subsequently, Söll and coworkers (Randau et al. 2008) provided experimental evidence that N. equitans has tRNA genes that are transcribed without a leader sequence, and that are apparently functional without further processing. These tRNAs have triphosphorylated 5′ ends, and, in several cases, also have an additional purine base at the −1 position, and are functional in vitro for charging with these unusual 5′ ends (Randau et al. 2008).
RNase P is also now known to have other substrates in bacteria, yeast, plants, and vertebrate cells (for review, see Kirsebom 2007). In yeast, genome-wide studies to identify authentic physiologically relevant RNase P substrates revealed a role for RNase P in the processing of box C/D small nucleolar RNAs (snoRNAs) (Coughlin et al. 2008) and in the processing of the HRA1 noncoding RNA (ncRNA) (Samanta et al. 2006; Yang and Altman 2007), whereas, in vertebrates, RNase P has been shown to process both the MALAT RNA and the Men β transcript (Wilusz et al. 2008; Sunwoo et al. 2009). It remains to be determined how many other RNAs are processed by RNase P, and whether, and to what extent, there is a biological connection linking these activities with tRNA processing.
3′ Processing and the rise of RNase Z
New data over the last several years have substantially improved understanding of the process by which the 3′ end of tRNAs is processed in different organisms. Maturation of the 3′ end of tRNA always requires removal of the 3′ trailer from the original transcript, and also often requires the subsequent addition of CCA after N73, although, in some bacteria and archaea, some or all tRNA genes have encoded CCA (Vogel et al. 2005; Hartmann et al. 2009). Earlier biochemical and in vivo work in E. coli had established an important role for the endonuclease RNase E in initial cleavage of most 3′ trailers (Li and Deutscher 2002; Ow and Kushner 2002), and for subsequent trimming by 3′ exonucleases, primarily by RNase PH and RNase T, but also by RNase II, PNPase, and RNase BN (Li and Deutscher 1994, 1996). Similarly, prior biochemical work in eukaryotes had implicated an endonuclease activity (Castano et al. 1985; Furter et al. 1992) and an exonuclease activity (Garber and Altman 1979; Engelke et al. 1985), and fractionation in yeast suggested more than one source of each activity (Papadimitriou and Gross 1996). Subsequently, Wolin and colleagues (Yoo and Wolin 1997) demonstrated that the 3′ endonuclease activity in yeast required the yeast La protein (Lhp1), and provided evidence that, in the absence of La protein, a number of tRNAs were processed by an alternative pathway involving exonuclease activity.
A body of emerging data demonstrates that the endonuclease RNase Z (also called tRNase Z) (Schiffer et al. 2002) plays a major role in removal of the 3′ trailer from tRNAs in several organisms (for review, see Vogel et al. 2005). Most RNase Z enzymes specifically cleave tRNAs immediately after the discriminator base (N73) to remove the 3′ trailer, prior to the addition of CCA, and available evidence suggests that RNase Z has this same role in vivo. Thus, Bacillus subtilis strains depleted for RNase Z accumulate a large number of tRNAs containing 3′ extensions from among the set of tRNAs lacking an encoded CCA sequence (Pellegrini et al. 2003), and RNAi treatment of Drosophila S2 cells to knock down RNase Z expression results in the accumulation of both nuclear and mitochondrial tRNAs that retain their 3′ trailers (Dubrovsky et al. 2004).
One remarkable biochemical feature of many RNase Z enzymes is their lack of cleavage of tRNA species with a mature CCA end (Mohan et al. 1999). Thus, for the B. subtilis RNase Z, substrates with a CCA sequence at the beginning of the 3′ trailer have a KM that is 2.5-fold higher and a Vmax that is 0.4 % of that of substrates with a UAA sequence at this position, with most of the effect due to the first C at position N74 (Pellegrini et al. 2003). This dramatic anti-determinant property of the CCA end prevents mature tRNA from being subject to cleavage, and a resulting futile cycle. This anti-determinant property also means that other mechanisms must exist for 3′ end formation of tRNAs with encoded CCA sequences in organisms such as B. subtilis, which have tRNAs both with and without encoded CCAs. In this organism, RNase PH and another RNase T-like exonuclease are implicated in maturation of tRNAs with encoded CCA sequences (Wen et al. 2005; Redko et al. 2007).
Understanding of the molecular basis for RNase Z binding and activity and CCA inhibition has been propelled by extensive structural data (de la Sierra-Gallay et al. 2005; Ishii et al. 2005, 2007; Kostelecky et al. 2006; Li de la Sierra-Gallay et al. 2006), as well as by biochemical data. From the cocrystal structure (Li de la Sierra-Gallay et al. 2006), it is evident that the tRNA substrate binds one subunit and is cleaved by the other subunit, that the protein binds tRNA through numerous contacts in the tRNA backbone, and that the D-stem and anticodon stem are largely devoid of contacts, which is consistent with biochemical data that these regions are not important for activity (Nashimoto et al. 1999a). From modeling and biochemical data, it is likely that the 3′ trailer binds through an exit tunnel that precludes base-pairing (Nashimoto et al. 1999b; de la Sierra-Gallay et al. 2005; Redko et al. 2007), and that the anti-determinant function of the CCA end of RNase Z is due to the loop between strands β1 and β2 and to the flexible arm, also called the exosite (de la Sierra-Gallay et al. 2005; Redko et al. 2007; Minagawa et al. 2008).
The CCA end does not behave universally as an anti-determinant (Schiffer et al. 2003; Minagawa et al. 2004). For example, the RNase Z enzyme from Thermotoga maritima chooses its cleavage site based on the presence or absence of a CCA sequence; the 45 tRNAs with an encoded CCA sequence are cleaved after A76, whereas the lone tRNA without an encoded CCA sequence is cleaved at N75 (Minagawa et al. 2004).
There are several additional fascinating aspects of RNase Z function in vivo. While its prominent role in tRNA 3′ end maturation is well documented, it is still unclear how the gene is linked to increasing the risk of prostate cancer (Tavtigian et al. 2001). It is also unclear which other substrates RNase Z has in vivo, since the enzyme can act in the 5′ processing of 5S RNA in vitro in Halferax volcanii (Holzle et al. 2008) and has been implicated in the turnover of several mRNAs in vivo in E. coli (Perwez and Kushner 2006), consistent with the known biochemical activity of the E. coli RNase Z enzyme on unstructured RNAs (HS Shibata et al. 2006). Multiple different substrates might partially explain the presence of four Arabidopsis RNase Z homologs—including one found in both the nucleus and mitochondria, one found solely in the mitochondria, one found in the cytoplasm—and an essential homolog found in the chloroplast (Canino et al. 2009).
Splicing endonuclease structure and connection to disease
Several results in the last several years have cast new light on the components and mechanisms of tRNA splicing, the medical importance of tRNA splicing, and other uses and interactions of the tRNA splicing components. It remains clear from analysis of multiple sequenced genomes that, although introns occur in only a minority of tRNA genes, they are found in all sequenced eukaryotes and archaea, and splicing is essential (or nearly essential) in all of these organisms, based on the presence of at least one family of tRNA genes in each organism in which all or almost all of the tRNA genes have introns (Genomic tRNA Database, http://gtrnadb.ucsc.edu). While eukaryotic tRNA introns are invariably found between nucleotides 37 and 38 of tRNAs, and are most often found at that position in archaea, archaeal tRNA introns are also found at 14 other positions around the tRNA molecule (Marck and Grosjean 2003), all of which feature versions of the bulge–helix–bulge motif originally identified some years ago, comprised of a 4-base-pair (bp) helix surrounded on each side by 3-nucleotide (nt) bulges and a following helix (Thompson and Daniels 1990; Marck and Grosjean 2003).
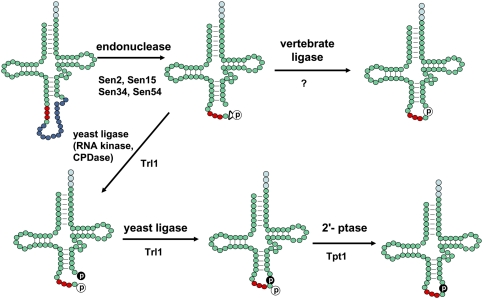
Splicing of tRNA is superficially much simpler than the more common spliceosome-mediated mRNA splicing, because it involves only a limited number of proteins, each carrying out a defined reaction (Fig. 2). In all organisms, tRNA splicing is initiated by an endonuclease that excises the intron, leaving a 5′ tRNA half-molecule ending in a 2′–3′ cyclic phosphate, and a 3′ tRNA half-molecule beginning with a 5′-OH group (Peebles et al. 1983). In yeast, the endonuclease has four different subunits (Table 1), including two related subunits (Sen2 and Sen34) with catalytic activity (Trotta et al. 1997), whereas, in archaea, the endonuclease is often comprised of fewer subunits, with α2, α2β2, or α4 configurations (see Xue et al. 2006).
Figure 2.
tRNA splicing and ligation pathways. tRNA is shown in its usual secondary structure, with the antidocodon indicated by red circles, and the intron after residue 37 indicated by blue circles. The endonuclease (comprised of Sen2, Sen15, Sen34, and Sen54 in yeast) excises the intron by cleaving the pre-tRNA at each exon/intron border, leaving tRNA half-molecules with a 2′–3′ cyclic phosphate (indicated by a triangle with a white circle containing the phosphate) and a 5′-OH group at their ends. In yeast and plants, the ligase (Trl1 in yeast) RNA 5′ kinase activity phosphorylates the 5′-OH end of the 3′ half-molecule (black circle), and the ligase cyclic phosphodiesterase activity opens the 2′–3′ cyclic phosphate to a 2′ phosphate. Then ligase joins the half-molecules (after activation of the 5′ phosphate, which is not shown), using the 5′ phosphate (black circle) as the junction phosphate, and leaving the 2′ phosphate at the splice junction (white circle). This 2′ phosphate is subsequently transferred to NAD by the 2′ phosphotransferase (Tpt1 in yeast). The yeast-like ligation pathway is also found in vertebrates, but, in vertebrates and some archaea, the predominant vertebrate ligase directly joins the phosphate of the 2′–3′ cyclic phosphate (white circle) to the 3′ half-molecule.
New structural information has provided a clear image of a homodimeric endonuclease and its active site. The structure of the endonuclease in complex with a model substrate (Xue et al. 2006) demonstrates that all 3 nt of each bulge of the RNA substrate are flipped out from their stacking positions. Furthermore, each bulge of the RNA is bound by amino acid residues of the adjacent endonuclease subunit, as well as by R280 and R302 of the other subunit, as part of a composite active site (Xue et al. 2006). The composite nature of the active site was also cleverly documented biochemically with the yeast enzyme (Trotta et al. 2006). The structure also clearly shows a catalytic triad, with Y246 poised for attack of the 2′-OH, H257 available to protonate the 5′-O− leaving group, and K287 stabilizing the negative charge of the transition state, although none of these residues appears to be absolutely required for activity (Calvin et al. 2008).
Intriguingly, recent results demonstrate that mutations in specific subunits of the tRNA splicing endonuclease subunits are associated with two of the subtypes of the neurodegenerative disease pontocerebellar hypoplasia. In many afflicted individuals, the Sen54 endonuclease subunit is mutated in a region that is conserved in mammals (but not lower eukaryotes). In two other individuals, there are alterations in either a strictly conserved amino acid of the Sen2 subunit or a position in the Sen34 subunit that is highly conserved in mammals (Budde et al. 2008). Although the finding that pontocerebellar hypoplasia is associated with mutations in any of three different endonuclease subunits virtually proves a direct connection between the endonuclease complex and the disease, it remains to be determined exactly how these mutations exert their effect on endonuclease function.
Partial resolution of the ligation pathway in vertebrates
One of the long-standing unsolved problems in tRNA splicing has been the nature and identity of the ligase that catalyzes the second step of tRNA splicing in vertebrates (Fig. 2). In yeast and plants, ligation requires a cyclic phosphodiesterase to hydrolyze the 2′–3′ cyclic phosphate of the 5′ half-molecule and generate a 2′ phosphate, an RNA kinase to phosphorylate the 5′-OH of the 3′ half-molecule, and a ligase to join the half-molecules after adenylylation of the 5′-P end (Greer et al. 1983). A single yeast protein, Trl1 (Phizicky et al. 1986), which is essential for tRNA splicing in vivo (Phizicky et al. 1992), catalyzes all four of these activities. Trl1 also catalyzes the ligation step of the nonconventional tRNA-like splicing of yeast HAC1 mRNA, which is required for the unfolded protein response, and is initiated by excision of the HAC1 intron by the endonuclease Ire1 (Sidrauski et al. 1996; Sidrauski and Walter 1997). The resulting ligated RNA from either tRNA splicing or HAC1 mRNA splicing bears a splice junction 2′ phosphate that, for tRNA, is subsequently transferred to NAD by the 2′ phosphotransferase Tpt1 to form ADP-ribose 1″–2″ cyclic phosphate (Culver et al. 1993; Spinelli et al. 1997).
In contrast to yeast and plants, vertebrates appear to have two tRNA ligation pathways. The classical vertebrate ligation pathway involves a “vertebrate” ligase activity that directly joins the tRNA half-molecules in vitro, using the phosphate of the 2′–3′ cyclic phosphate as the junction phosphate (Nishikura and De Robertis 1981; Filipowicz and Shatkin 1983; Laski et al. 1983). In addition, vertebrates have a second yeast-like ligase/2′ phosphotransferase pathway, since both of these activities have been detected in HeLa extracts (Zillmann et al. 1991, 1992), and the mammalian 2′ phosphotransferase is known to be functional in yeast (Spinelli et al. 1998). Moreover, mammals have retained the nonconventional tRNA-like splicing of their HAC1 mRNA ortholog (XBP1) during the unfolded protein response (Yoshida et al. 2001; Calfon et al. 2002). Therefore, it has been unclear which ligation pathway is involved in which reaction.
Some recent results have favored the involvement of the yeast-like ligation pathway in vertebrate tRNA splicing. Thus, human Clp1 protein was shown to have an RNA 5′ kinase activity, and siRNA experiments in mammalian cells implicated Clp1 in tRNA splicing (Weitzer and Martinez 2007). This finding is consistent with the yeast-like ligase/2′ phosphotransferase pathway, since this pathway requires an RNA 5′ kinase activity. In addition, prior results had shown that Clp1 copurifies with the human tRNA splicing endonuclease (Paushkin et al. 2004), although it is known to be involved in cleavage and polyadenylation of mRNA. Thus, it seemed likely that splicing in vertebrate cells required the yeast-like ligation pathway.
However, it now seems more likely that the yeast-like ligation pathway is not required in mice for either tRNA splicing or the unfolded response, since a mouse strain lacking the 2′ phosphotransferase gene is fully functional for translation of mRNAs requiring decoding by spliced tRNA, and is normal for the unfolded protein response (Harding et al. 2008). Thus, it seems likely that the classical vertebrate ligation pathway is used for tRNA and XBP1 mRNA splicing in vertebrates, similar to the pathway used for tRNA splicing in archaea (Zofallova et al. 2000; Salia et al. 2003). The identity of the ligase awaits discovery.
Baroque tRNA gene organization and tRNA processing
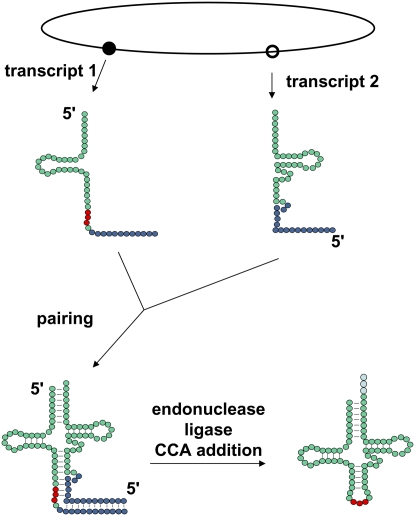
An additional surprise is the discovery of two highly unusual arrangements of tRNA genes that appear to use tRNA processing enzymes in unexpected ways. In the first example, Söll and coworkers (Randau et al. 2005b) have found that N. equitans generates functional tRNAs by combining separate tRNA halves, which are located in different parts of the chromosome and are individually transcribed (Fig. 3). The 5′ tRNA half-molecule transcripts terminate in the anticodon loop, and are followed by an additional sequence of ∼12 nt, which pairs exactly with a corresponding additional sequence at the 5′ end of the 3′ tRNA half-molecule transcripts (Randau et al. 2005b). These paired halves are then processed by the N. equitans splicing endonuclease and, presumably, by a ligase (Randau et al. 2005a). In the second example, Sekine and coworkers (Soma et al. 2007) found that the red alga Cyanidioschyzon merolae had circularly permuted tRNA genes in which the 5′ end of the transcript began in the D-loop, the anticodon loop, the T-stem, or the T-loop. The resulting transcripts were subsequently processed to remove linking sequences, presumably by a combination of the splicing machinery, RNase P, and RNase Z (Soma et al. 2007).
Figure 3.
Formation of mature tRNA from split tRNA genes. In N. equitans, split tRNA genes are transcribed from distant chromsomal loci (indicated by filled and open circles) as half-molecules, each containing additional nucleotides at their 5′ or 3′ end (blue circles). The additional sequences at the 3′ end of the 5′ half-molecule occur after nucleotide 37, the usual position of an intron, and pair with the additional 5′ sequences of the 3′ half-molecule, forming a hybrid intron. Splicing by the endonuclease, followed by ligation and CCA addition, results in formation of the mature tRNA.
These unexpected mechanisms for engineering functional RNAs from unusual use of tRNA processing enzymes are as intriguing as the conserved use of the tRNA ligation machinery for splicing of HAC1 (XBP1) mRNA in the unfolded protein response (Sidrauski et al. 1996; Sidrauski and Walter 1997; Yoshida et al. 2001; Calfon et al. 2002), and the use of RNase P to generate the 3′ end of MALAT1 RNA in humans (Wilusz et al. 2008). Another recent example of the flexibility of tRNA processing machinery is the demonstration in yeast that a designed STE2 mRNA containing an embedded intron-containing pre-tRNA can be spliced by the tRNA splicing machinery and RNase P to yield both a functional Ste2 protein and a functional tRNA (Di Segni et al. 2008).
Modification biology takes its place at the translation table
One of the striking features of tRNA from all organisms is their large number of post-transcriptional modifications. A total of 92 different tRNA modifications are listed in the RNA Modification Database (http://biochem.ncsu.edu/RNAmods), and a survey of 561 sequenced tRNAs from several different organisms (Sprinzl and Vassilenko 2005) shows that modifications are found on 11.9% of the residues, with a median of eight modifications per tRNA species (Phizicky and Alfonzo 2010). In the yeast S. cerevisiae, 25 different modifications are found among the 34 sequenced cytoplasmic tRNA species (Fig. 1). These modifications occur at 36 different positions, with an average of 12.6 modifications per species.
The last decade has witnessed great progress in the identification of the genes and corresponding enzymes that are required for modification, as well as in our understanding of their biochemistry and their biological roles. Thus, in S. cerevisiae, the vast majority of the genes that are responsible for modifications have now been identified (Table 1), and a large number of modification genes and enzymes have been identified in other organisms (see the Modomics database, http://modomics.genesilico.pl).
Many of the functions of these modifications have begun to be elucidated, and conform to three general rules. First, many modifications in or around the anticodon loop affect translation or growth. For example, yeast strains lacking I34 are inviable (Gerber and Keller 1999); strains lacking m1G37 or t6A are extremely sick (Bjork et al. 2001; El Yacoubi et al. 2009); strains lacking Nm32 and Nm34, or Ψ38 and Ψ39, grow poorly (Lecointe et al. 1998; Pintard et al. 2002); and strains lacking i6A37 are mildly defective for translation (Laten et al. 1978; Dihanich et al. 1987). Other specific examples of the effects of modifications around the anticodon loop are discussed below. Second, many modifications in the main body of the tRNA affect tRNA folding or stability. Thus, the lack of T54 lowers the Tm of tRNA by 2°C–6°C in tRNAPhe and tRNAfMet (Davanloo et al. 1979; Sengupta et al. 2000), the lack of m1A9 in human tRNALys leads to formation of an alternative structure (Helm et al. 1999), the lack of 2′-O-methylated nucleotides can destabilize the 3′ endo form of RNA (Kawai et al. 1992), and the lack of Ψ can destabilize helices (Durant and Davis 1999; Newby and Greenbaum 2001). Although the specific biological consequences of the lack of such body tRNA modifications are not as apparent, emerging evidence elaborated later in this review suggests that the lack of a number of different body modifications can elicit tRNA degradation by two different tRNA turnover pathways. Third, some modifications at various positions specifically affect tRNA identity. Thus, G−1 of tRNAHis is a positive determinant for HisRS (Rudinger et al. 1994; Nameki et al. 1995), and Ar(p) at position 64 is an identity element for tRNAiMet in yeast (Astrom and Bystrom 1994), whereas m1G37 of tRNAAsp prevents misacylation by ArgRS in E. coli (Putz et al. 1994), and lysidine at position 34 prevents misacylation of tRNAIle by E. coli MetRS (Muramatsu et al. 1988).
The Elp complex is revealed as a modification enzyme
One major highlight has been the discovery that the central role of the Elp complex in cell function is due to its activity in modifying tRNA. The Elp complex had been characterized previously as a complex that copurifies with active elongating hyperphosphorylated Pol II (Otero et al. 1999), acetylates histones H3 and H4 (Wittschieben et al. 1999; Winkler et al. 2002), and interacts with Sec2 to regulate exocytosis (Rahl et al. 2005). This complex is comprised of a subcomplex containing Elp1, Elp2, and Elp3, and another subcomplex containing Elp4, Elp5, and Elp6 (Krogan and Greenblatt 2001; Winkler et al. 2001), and is associated with Kti11 and Kti12 (Fichtner et al. 2002, 2003).
Remarkably, available evidence suggests that all of the phenotypes associated with the lack of the Elp complex derive from the lack of formation of mcm5s2U at position 34 of certain substrate tRNAs. First, it was found that all six subunits of the Elp complex, as well as Kti11, Kti12, and Kti13, were required for formation of mcm5U, mcm5s2U, and ncm5U at position 34 (Huang et al. 2005). Then it was shown that the Kluyveromyces lactis γ-toxin acts by cleaving tRNAGlu(UUC), tRNALys(UUU), and tRNAGln(UUG) after U34 (Lu et al. 2005), and that γ-toxin targets these tRNAs because of their mcm5s2U modification, since γ-toxin resistance is conferred by mutation of the Elp complex, KTI11, KTI12, or KTI13 (Jablonowski et al. 2001; Lu et al. 2005). Finally, it was shown that overproduction of tRNAGln(UUG) and tRNALys(UUU) suppresses all of the known Elp complex phenotypes associated with transcription or exocytosis, and that mutation of NCS2, which results in the lack of s2U modification of these same tRNAs, results in the same phenotypes ascribed to the lack of the Elp complex (Esberg et al. 2006). Thus, it appears that the effects on transcription and on exocytosis are secondary consequences of the lack of the mcm5s2U modifications on these tRNAs (Esberg et al. 2006). It remains to be determined if the requirement of the Elp complex for filamentous growth signaling is also directly due to effects on modified tRNAs (Abdullah and Cullen 2009). It also remains to be determined why the activity of the Elp complex is defective in mutants lacking Sit4 phosphatase (critical for cell cycle progression and modification of Pkc1 function) (Sutton et al. 1991; Torres et al. 2002), in mutants lacking both of the Sit4-associated proteins (Sap185 and Sap190), and in mutants lacking the Kti14 protein kinase (Mehlgarten and Schaffrath 2003; Jablonowski et al. 2004; Huang et al. 2008).
The function of the Elp complex in tRNA modification and the importance of the mcm5s2U modification are conserved in metazoans. For example, in Caenorhabditis elegans, the ELP1 ortholog is also required for formation of the mcm5 moiety of mcm5s2U and ncm5U (Chen et al. 2009), and the TUC1 ortholog (Bjork et al. 2007) is required for formation of the s2U moiety of mcm5s2U (Chen et al. 2009). Furthermore, the metazoan Elp complex is consistently associated with defects in neurological function. Mutation of the human ELP1 ortholog is associated with the human neurodegenerative disease familial dysautonomia (Anderson et al. 2001; Slaugenhaupt et al. 2001). Similarly, mutation of the C. elegans ELP1 or ELP3 orthologs is associated with a defect in salt chemotaxis learning, and mutation of both the ELP1 (or ELP3) and TUC1 orthologs results in developmental defects (Chen et al. 2009).
The decoding function of the mcm5s2U, mcm5U, and ncm5U modifications has recently been investigated in detail by examination of a series of multiple deletion strains lacking specific tRNA genes and/or the genes responsible for synthesis of the mcm5 or s2 moiety (Johansson et al. 2008). These studies reveal that the mcm5U modification is important for reading G in the wobble position (wobble G) for both tRNAs with this modification [tRNAArg(UCU) and tRNAGly(UCC)], based on growth of the corresponding strain lacking tRNA with C34. They also reveal that the mcm5s2U modification is important for reading a wobble A and, in the context of tRNAGln(UUG), also helps to read a wobble G (but not well enough to allow growth of a knockout of the corresponding tRNA with C34). Similar experiments show that the ncm5 moiety is important for reading a wobble G residue for tRNAVal(UAC), tRNASer(UGA) , and tRNAThr(UGU), and that tRNAPro(UGG) can decode codons ending in all four residues with or without its ncm5U moiety (Johansson et al. 2008), a clear statement that this property does not require an unmodified U.
A connection between TRM9 or its mcm5U product and translation of DNA repair genes
Recent experiments have also revealed a striking translation role for Trm9 (Begley et al. 2007), which catalyzes conversion of cm5U to mcm5U at position 34 of substrate tRNAs (Kalhor and Clarke 2003). trm9-Δ mutants are paromomycin-sensitive, implying some translation defect that is related to ribosome A site function (Kalhor and Clarke 2003). Expression analysis (Begley et al. 2007) suggests that trm9 mutants are defective in reading arginine AGA codons as well as glutamate GAA codons, which are read by tRNAs with the mcm5 moiety, as well as arginine AGG codons, which are read both by tRNA with an exact match anticodon and tRNA with the mcm5 moiety (Johansson et al. 2008). Since trm9 mutants have reduced amounts of Rnr1, Rnr3, and Yef3 proteins, each of which has an overrepresentation of GAA and AGA codons, and the sensitivity of trm9 mutants to DNA-damaging treatments can be overcome by overexpression of RNR1 or RNR3, these results suggest that lack of Trm9 and/or its mcm5U product causes these phenotypes (Begley et al. 2007). Consistent with this explanation, recent results show that depletion of the human Trm9 homolog ABH8 reduces the amount of mcm5U in tRNA, and leads to increased sensitivity to DNA-damaging treatments (Fu et al. 2010). Since Trm9 does not appear to have a major effect on the growth of strains requiring tRNAArg(UCU) to read AGG codons, or tRNAGly(UCC) to read GGG codons, and each of these tRNAs has an mcm5U moiety (Johansson et al. 2008), the precise effects of Trm9 on translation may be specific for certain codons or tRNAs. It is intriguing to note also that silencing of human ABH8 also leads to apoptosis of urothelial carcinoma lines and down-regulation of NOX-1-dependent reactive oxygen species, as well as suppression of angiogenesis and invasion (Shimada et al. 2009). Since ABH8 does not have demethylase activity (Fu et al. 2010), these effects may also be due to the lack of the mcm5U moiety of tRNAs.
Formation and function of s2U revealed
Another crucial anticodon modification is s2U, which is universally found at U34 of tRNA species that also have U35 (Bjork et al. 2007). Earlier experiments in E. coli demonstrated that thiolation requires IscS to transfer the sulfur from cysteine to form a persulfide, which was transferred to MnmA, to catalyze 2-thiouridylation of substrate tRNAs (Kambampati and Lauhon 2003). Subsequent E. coli ribonucleome analysis by Suzuki and coworkers (Ikeuchi et al. 2006) resulted in the identification of five other genes involved in this pathway, designated tusA to tusE, and reconstitution experiments with purified proteins showed that inclusion of all components stimulates the biochemical reaction 100-fold. Thiolation is accomplished by transfer of sulfur from IscS to TusA, followed by sulfur transfer to TusD of the TusB–TusC–TusD complex, and likely to TusE, which forms a complex with MnmA and tRNA (Ikeuchi et al. 2006). The yeast pathway for s2U modification appears to require several similar components and a similar set of reactions using Urm1 and Uba4, which also participate in urmylation of proteins (Nakai et al. 2004, 2007, 2008; Huang et al. 2008; Schlieker et al. 2008; Leidel et al. 2009; Noma et al. 2009). A crucial role for the s2U moiety is inferred, since mutants lacking genes at the end of the pathway grow poorly, which is consistent with the near universal conservation of this modification in tRNAs with U34 and U35 (Bjork et al. 2007).
The function of s2U is decoding, rather than prevention of misreading, because overproduction of the unmodified genes encoding tRNAs with s2U can overcome the lethality caused by the total lack of mcm5s2U in an elp3 tuc1 double mutant (Bjork et al. 2007). Remarkably, although tRNALys(UUU), tRNAGln(UUG), and tRNAGlu(UUC) all have mcm5s2U, only tRNALys(UUU) overexpression is required to suppress the lethal phenotype of the elp3 tuc1 strain (Bjork et al. 2007). The s2U modification also appears to aid in the decoding of wobble G codons (Johansson et al. 2008). This is also true in mitochondria, since Tarassov and coworkers (Kamenski et al. 2007) have shown that reduced formation of the s2U moiety of cmnm5s2U34 of tRNALys(UUU) impairs translation of the two mitochondrial proteins with AAG codons in a strain unable to import tRNALys(CUU), providing strong evidence that the long-known import of tRNALys(CUU) (Martin et al. 1979) is required at a high temperature for translation of these codons.
Formation and function of wybutosine revealed
At long last, many of the particulars of the mysterious wybutosine (Wye base, yW) modification have been elucidated. Wybutosine (or its hydroxy derivative, peroxywybutosine) is a complex guanosine base modification that is found exclusively at base 37 of tRNAPhe in most, but not all, eukaryotes, and other similar wyosine family derivatives are widely found in archaeal tRNAPhe species (Waas et al. 2005; de Crecy-Lagard et al. 2010). Wybutosine is of interest because of its unusual structure (which contains an additional imidazole ring that is fused to the guanosine ring, to which is attached the α-amino-α-carboxy-propyl group of methionine, as well as several appended groups), and because of reports that reduced levels of Wye base are associated with tRNAPhe in different tumor cells (Grunberger et al. 1975; Kuchino et al. 1982). It has been known for some time that the first step in Wye base formation is catalyzed by the m1G methyltransferase Trm5 (Droogmans and Grosjean 1987; Bjork et al. 2001). Recent work has shown that subsequent biosynthesis of wybutosine involves formation of the methyl imidazole ring by Tyw1 (Waas et al. 2005; Noma et al. 2006), followed by addition of the α-amino-α-carboxy-propyl group of methionine by Tyw2 (Kalhor et al. 2005; Noma et al. 2006), and methylation of guanosine N3 by Tyw3 (Noma et al. 2006). Intriguingly, the last step of Wye base formation involves both methylation of the α-carboxy end group and methoxycarboxylation of the α-amino end group, with incorporation of carbon dioxide, all seemingly catalyzed by Tyw4 (Noma et al. 2006; Suzuki et al. 2009).
An analysis of translation has since shown that one function of the Wye base is to prevent −1 frameshifting (Waas et al. 2007). This study was done using a derivative of the slippery sequence used by the double-stranded virus SCV-LA (GGGUUUA) in which the A site codon was changed from UUA to UUU or UUC. Frameshifting was most pronounced when the last codon was UUU rather than UUC, and was progressively reduced as the Wye base was increasingly formed, since frameshifting was twofold higher than wild type in a tyw1 mutant, which has tRNAPhe with m1G37, and 1.5-fold higher than wild type in a tyw2 mutant, in which the Wye base is in an intermediate state in which only the imidazole ring is attached. This finding provides a highly satisfactory explanation of the value, and perhaps the evolution, of increased modification in the Wye base, and is consistent with previous analysis of frameshifting in vitro (Carlson et al. 2001). These results nicely complement earlier work by Bjork and colleagues (Urbonavicius et al. 2001, 2003) demonstrating that several modifications in the anticodon loop of tRNAs in E. coli, Salmonella typhimurium, or S. cerevisiae affect +1 frameshifting, but not −1 frameshifting.
The conserved G−1 of tRNAHis and novel activities of Thg1, the tRNAHis guanylyltransferase
Recent experiments have revealed a number of intriguing results about the additional G−1 residue of tRNAHis. Decades of previous analysis had shown that this additional G−1 was found uniquely in tRNAHis, that no other tRNA species (with one exception) had any nucleotide at this position, and that the additional G−1 residue is conserved by two very different mechanisms: In bacteria and some archaea, the G−1 is encoded (opposite C73) and retained during processing (Orellana et al. 1986), whereas, in eukaryotes and other archaea, the G−1 residue is added post-transcriptionally (across from A73 and C73, respectively) by tRNAHis guanylyltransferase (Cooley et al. 1982; Jahn and Pande 1991) encoded by the essential THG1 gene in yeast (Gu et al. 2003). Depletion of Thg1 leads to the accumulation of uncharged tRNAHis lacking its G−1 residue, which is consistent with biochemical data that the G−1 residue and its 5′ phosphate are important for HisRS activity (Rudinger et al. 1994; Nameki et al. 1995; Rosen et al. 2006), as well as to activation of the GCN4 pathway due to the accumulation of uncharged tRNAHis (Gu et al. 2005). Unexpectedly, however, loss of Thg1 is also associated with the delayed accumulation of m5C at C48 and C50, and with nuclear localization of a significant fraction of the tRNAHis (Gu et al. 2005).
Recent results demonstrate four additional surprising results about the G−1 residue of tRNAHis and about Thg1.
First, Thg1 from both yeast and humans is somehow associated with cell cycle progression. In yeast, thg1 mutants have a cell cycle defect at G2/M, and Thg1 interacts with Orc2 of the origin recognition complex (Rice et al. 2005), while human Thg1 is cell cycle-regulated, and its knockdown is associated with a defect in cell proliferation and with the onset of polynucleate cells (Guo et al. 2004).
Second, tRNAHis from a clade of α-proteobacteria including Sinorhizobium meliloti has been found to lack a G−1 residue, and to have a HisRS species with variations consistent with altered tRNAHis recognition, thereby proving that the G−1 residue of tRNAHis cannot be universally required (Wang et al. 2007). It is now known that, despite its near universal conservation in eukaryotes, the G−1 residue of tRNAHis is also not absolutely required for growth in eukaryotes, since yeast strains lacking Thg1 can survive, albeit poorly, without the G−1 residue, provided that both tRNAHis and its synthetase are overproduced (Preston and Phizicky 2010).
Third, it has been shown that Thg1 has several notable mechanistic similarities to, but little obvious homology with, aminoacyl tRNA synthetases, since, like many tRNA synthetases, recognition of the anticodon is necessary and sufficient for reaction at the acceptor end, and since the chemical steps of adenylylation of the 5′ phosphate of tRNA and subsequent guanylyltransfer during G−1 addition are mechanistically similar to formation of the aminoacyl adenylate and the subsequent tRNA acyltransfer during charging by tRNA synthetases (Jackman and Phizicky 2006a).
Fourth, it has been shown that Thg1 has a distinct 3′–5′ template-dependent polymerase activity in which it adds multiple guanine or cytidine nucleotides (or deoxynucleotides) to the 5′ ends of appropriate tRNA species, thus forming multiple phosphodiester bonds in a direction opposite to that of all known polymerases (Jackman and Phizicky 2006b). Although the function of this unusual 3′–5′ template-dependent polymerase activity is not yet known, it may have a repair function similar to that of the mechanistically similar, but unidentified, editing activity that repairs mismatches at the 5′ end of tRNA species in the mitochondria of Acanthamoeba castellani (Price and Gray 1999). Recent results from Jackman and coworkers (Abad et al. 2010) show that archaeal Thg1 homologs have a template-dependent uridine and guanidine nucleotide addition activity across from A73 and C73, respectively, but no G−1 addition activity across from A73. This result, together with the observation that several archaea have both an encoded G−1 residue in their tRNAHis genes and a Thg1 homolog, suggests that template-dependent 3′–5′ nucleotide addition activity is the original function of the Thg1 family of proteins, and that the known role of Thg1 in the G−1 addition across from A73 is a newer evolved function (Abad et al. 2010). Nonetheless, the essential role of Thg1 in yeast is its tRNAHis G−1 addition activity, although 3′–5′ template-dependent polymerization is observed in vivo with substrate tRNAHis species bearing C73 (Preston and Phizicky 2010).
Unexpected deamination promiscuity in editing of tRNA
Two other findings suggest expanded roles and functions of RNA-editing proteins in tRNA function. First, recent evidence suggests that the Trypanosoma brucei adenosine deaminase complex ADAT2/ADAT3 can catalyze both A-to-I editing of the wobble adenosine of tRNAThr(AGU), as well as C-to-U editing of ssDNA (Rubio et al. 2007). Since mutation of the active site of ADAT2 eliminates both A-to-I editing of tRNA and C-to-U editing of DNA, while not preventing complex formation, it seems highly likely that the single active site contains both activities. Furthermore, at least in vivo, this same protein appears to be responsible for both the C-to-U editing that occurs at residue 32 of tRNAThr and the A-to-I editing that occurs at residue 34 (Rubio et al. 2006). These findings point to a common evolutionary origin of both classes of these editing activities, and suggest the possibility that both editing activities in tRNA processing derive from one activity (Rubio et al. 2007). Second, another recent result demonstrates that tRNAs from Methanopyrus kandleri have promiscuous editing of their tRNAs at C8, to form U8 at that position, which is crucial for maintenance of the highly conserved U8:A14 reverse Hoogsteen tertiary base pair (Randau et al. 2009).
tRNA turnover as a quality control mechanism
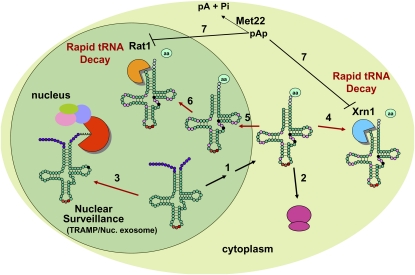
Emerging results have challenged the widely held notion that tRNA biosynthesis inevitably leads to product tRNAs that are virtually infinitely stable. tRNA half-lives are very long indeed, with estimates of 50 h in chicken muscle (Nwagwu and Nana 1980), 3 d in avian liver (Kanerva and Maenpaa 1981), and 44 h in Euglena gracilis (Karnahl and Wasternack 1992), roughly comparable with the half-life of rRNA (Nwagwu and Nana 1980; Karnahl and Wasternack 1992). However, experimental results in the last few years have led to the discovery and characterization of two pathways by which tRNAs turn over in the cell as part of quality control mechanisms that monitor the integrity of tRNA during and after biogenesis (Fig. 4).
Figure 4.
Two different tRNA degradation pathways in yeast. pre-tRNA transcribed in the nucleus is processed (black arrows) in the nucleus and the cytoplasm (steps 1) to remove the 5′ leader and 3′ trailer (purple circles), to add CCA to the 3′ end (blue circles), to remove the intron if present (not shown), and to add modifications [pink circles, as for tRNAVal(AAC)], ultimately emerging in the cytoplasm for translation (step 2). If m1A58 is not added to pre-tRNAiMet (absence of m1A indicated by black circle), this pre-tRNA is degraded by the nucelar surveillance pathway (step 3, red arrow) in which the pre-tRNA is first polyadenylated by the TRAMP complex, and then degraded from the 3′ end by the nuclear exosome. If m7G46 and m5C49 are not added to tRNAVal(AAC) (black circles), the hypomodified mature tRNA is at least partially functional, but is degraded by the RTD pathway (red arrows), by Xrn1 in the cytoplasm (step 4), or by Rat1 in the nucleus (step 6), possibly after nuclear import (step 5). (Step 7) The elevated presence of pAp in met22 mutants inhibits the RTD pathway by inhibiting both Xrn1 and Rat1.
Nuclear surveillance of pre-tRNA and degradation from the 3′ end
Anderson and coworkers (Kadaba et al. 2004, 2006) documented the existence of a pathway that monitors the quality of pre-tRNA during tRNA biogenesis. Previous work had shown that Trm6/Trm61 (Gcd10/Gcd14) was the methyltransferase that catalyzed formation of m1A58 in tRNA (Anderson et al. 2000), that the temperature sensitivity of trm6ts mutants could be suppressed by increased amounts of initiator tRNA on high-copy plasmids, and that trm6ts mutants had increased turnover of newly synthesized tRNAiMet, but not elongator tRNAMet (Anderson et al. 1998). The identification and analysis of trm6ts suppressors led to the definition of the nuclear surveillance turnover pathway in which nuclear pre-tRNAiMet lacking m1A58 is subject to polyadenylation by Trf4 of the TRAMP complex, and then degradation by Rrp6 and the nuclear exosome (Kadaba et al. 2004, 2006). Other experiments have shown that overexpression of the poly(A) polymerase Trf5 can substitute for the lack of Trf4 in the nuclear surveillance pathway (Kadaba et al. 2006), and that a mutant with a defect in the ATP-dependent RNA helicase activity of Mtr4 prevents degradation of pre-tRNAiMet, but does not perturb its formation of the TRAMP complex or polyadenylation of the pre-tRNA (Wang et al. 2008).
Two lines of evidence show that the 3′–5′ exonuclease Rex1 plays a significant role in the nuclear surveillance pathway. First, rex1 trm6 mutants have a synthetic growth phenotype that is associated with polyadenylation of those pre-tRNAiMet species and pre-tRNAVal(CAC) species that have longer 3′ trailers, and of dimeric tRNAArg–tRNAAsp transcripts (Ozanick et al. 2009). Second, Wolin and coworkers (Copela et al. 2008) have shown that unspliced pre-tRNA species are subject to the nuclear surveillance pathway, but not in rex1 mutants, suggesting that Rex1 function may generate these pre-tRNA degradation substrates, and further experiments suggest that Rex1 is in competition with La (Lhp1) protein for these 3′ ends.
The full extent of the involvement of La protein in the nuclear surveillance and/or other tRNA turnover pathways is not yet known, but it likely has some role, since overexpression of La protein can also suppress trm6ts mutants (Anderson et al. 1998). La protein acts by binding the UUU-OH sequence at the end or pre-tRNA transcripts (see Maraia and Bayfield 2006; Teplova et al. 2006 and references therein) through the use of two sites (Bayfield and Maraia 2009), has an RNA chaperone activity (Chakshusmathi et al. 2003), and appears to protect RNA from Rrp6 of the nuclear surveillance pathway in Schizosaccharomyces pombe (Huang et al. 2006). However, La also protects RNA independently of Rrp6, suggesting that it participates in another turnover pathway (Huang et al. 2006).
Detailed and elegant biochemical experiments support the claim that the TRAMP complex collaborates with the nuclear exosome to polyadenylate substrate tRNAs and degrade them from the 3′ end (LaCava et al. 2005; Vanacova et al. 2005), and show that this activity acts on a pre-tRNA transcript (LaCava et al. 2005); on an unmodified tRNAiMet transcript, but not its fully modified derivative (Vanacova et al. 2005); on a tRNAAla variant with a structural defect, but not the unmodified wild-type tRNAAla (Vanacova et al. 2005); and on tRNAiMet lacking only m1A58, but not other tRNA substrates (Schneider et al. 2007).
Rapid tRNA decay (RTD) of mature tRNA from the 5′ end
Another set of experiments has shown that mature tRNAs are also subject to turnover. Thus, trm8 trm4 mutants, which lack m7G and m5C in their tRNAs, are temperature-sensitive due to the specific degradation of tRNAVal(AAC) by an RTD pathway that is distinct from the nuclear surveillance pathway that acts on pre-tRNAiMet lacking m1A58 (Alexandrov et al. 2006). This RTD pathway appears to be general, since several different tRNA species lacking different combinations of modifications are subject to degradation by this pathway (Chernyakov et al. 2008). Moreover, degradation occurs at the level of mature tRNA, rather than pre-tRNA, since cells treated with the transcription inhibitor thiolutin undergo degradation at the same rate and degrade all of the tRNA of substrate species (Chernyakov et al. 2008). Genetic analysis suggests that tRNA degradation by the RTD pathway is catalyzed by the 5′–3′ exonucleases Rat1 and Xrn1 and requires the methionine biosynthetic enzyme Met22 (Chernyakov et al. 2008), likely because its substrate, pAp, accumulates in met22 mutants (Murguia et al. 1996) and inhibits Rat1 and Xrn1 (Dichtl et al. 1997). Intriguingly, the substrate for the RTD pathway may be aminoacylated tRNA, since tRNA degradation is accompanied by selective loss of the aminoacylated fraction of the tRNAVal(AAC) population (Alexandrov et al. 2006), and since mutations in MET22 or RAT1 and XRN1 that prevent degradation also restore the fraction of charged tRNA (Chernyakov et al. 2008). It is not yet known if and to what extent any part of the translation machinery participates in this pathway; why specific hypomodified tRNA substrates are selected for degradation by this pathway, whereas other tRNAs lacking the same modifications are spared; and the extent to which this pathway acts on other types of defective tRNAs.
tRNA cleavage pathways activated by stress and other growth conditions, and signaling by tRNA fragments
A series of exciting studies in the last few years has detailed the existence of a previously unknown pathway that occurs in a variety of organisms under specific growth conditions, and generates tRNA cleavage in the region of the anticodon loop (for review, see Thompson and Parker 2009b). Thus, Collins and coworkers (Lee and Collins 2005) have shown that starvation of Tetrahymena thermophila induces cleavage of a large number of different tRNAs that derive from the macronucleus, but not the mitochondria. tRNA cleavage occurs at variable positions in and around the anticodon loop and occasionally in the variable arm, appears to target tRNAs that are modified but lack their CCA end, is quantitatively minor, and appears to require translation (Lee and Collins 2005). Experiments in yeast demonstrate that similar cleavage of multiple tRNA species occurs in cells undergoing oxidative stress, methionine starvation, extended growth in stationary phase, and growth at high temperature, but not in cells undergoing ultraviolet (UV) stress, nitrogen starvation, or glucose starvation (Thompson et al. 2008). Furthermore, similar cleavage is observed in Arabidopsis thaliana and human cells undergoing oxidative stress (Thompson et al. 2008; Fu et al. 2009; Yamasaki et al. 2009), as well as Streptomyces coelicolor (Haiser et al. 2008) and several other organisms, generally under starvation conditions or other conditions affecting developmental change (see Thompson and Parker 2009b and references therein). Subsequent experiments show that this pathway is mediated by the RNase T2 family member Rny1, which relocalizes from the vacuole to the cytoplasm during oxidative stress and mediates cell death (Thompson and Parker 2009a). However, the activity associated with Rny1 that mediates cell death is demonstrably separate from its cleavage activity (Thompson and Parker 2009a). Intriguingly, the human Rny1 ortholog RNASET2 has a set of activities similar to those of yeast Rny1, since expression of RNASET2 in yeast restores the production of tRNA cleavage fragments in an rny1 mutant strain and restores the reduced viability of cells undergoing oxidative stress (Thompson and Parker 2009a), while expression of RNASET2 in mammalian cells acts as a tumor suppressor independent of its cleavage activity (Acquati et al. 2005; Smirnoff et al. 2006).
tRNA cleavage products are also generated by Dicer-dependent cleavage. Deep sequencing has revealed the existence of a prominent Dicer-generated tRNAIle fragment in mouse embryonic stem (ES) cells that derives from the 3′ end of the gene and part of the trailer sequence, and may arise because of alternative folding of the tRNA transcript (Babiarz et al. 2008). In addition, deep sequencing has identified a number of Dicer-generated 5′ tRNA fragments in HeLa cells that tend to arise from cleavage after residue 19 in the D-loop, and derive mostly from four major tRNA species (Cole et al. 2009). Since these RNA fragments appear to be modified at the 3′ end and bind Argonaute complexes weakly, they are not likely to act as microRNAs (miRNAs); thus, their function remains unknown.
Possible signaling effects of tRNA fragments
Two recent reports have emerged suggesting that tRNA cleavage products inhibit translation. First, Anderson and coworkers (Yamasaki et al. 2009) have shown that each of several different stress treatments of mammalian cells activates the ribonuclease angiogenin to cleave tRNA to produce tRNA-derived stress-induced RNAs (tiRNAs) that inhibit translation. Angiogenin is a potent angiogenic factor isolated from the medium of human cell lines or from plasma (Shapiro et al. 1987), with ribonuclease activity that is essential for its angiogenic activity (Shapiro and Vallee 1987). Remarkably, the 5′ tiRNAs produced by angiogenin have a distinct inhibitory effect on cell growth, since transfection of the 5′ tiRNA fragments, but not the 3′ tiRNA fragments, induces translation arrest independent of phosphorylation of eIF2α (Yamasaki et al. 2009) through formation of stress granules (Emara et al. 2010). Second, pumpkin phloem sap has been shown to contain tRNA fragments of several tRNAs, which are cleaved in either the anticodon loop or the D-loop, and which appear to inhibit translation based on reduced translation in the presence of phloem sap RNA, or of RNase A-generated fragments of yeast tRNA (Zhang et al. 2009). It remains to be determined if translation is similarly inhibited in other systems in which tRNA fragments are produced.
In another development, Dutta and colleagues (YS Lee et al. 2009) have found by deep sequencing methods that multiple different tRNA-derived fragments (tRFs) are made in prostate cancer cell lines, at least one of which has potent effects on promoting cell proliferation. These tRFs are 13–26 nt long, comprise fully 40% of the non-miRNA sequences in this size range, and derive from one of three precise regions of the tRNA: the 5′ end of mature tRNA, the 3′ end of the mature tRNA ending in the CCA sequence, or the 3′ trailer sequence beginning immediately after the 3′ end of the tRNA. One of these tRF species, tRF-1001, derives from the 3′ trailer sequence of a specific tRNASer(UGA) species and has profound effects on cell growth. tRF-1001 is highly expressed in a number of different proliferating cancer cell lines, and its expression is reduced after serum starvation or when cell density is high. Furthermore, siRNA-mediated knockdown of tRF-1001 expression is associated with decreased cell proliferation, and cell proliferation is restored by transfection of a synthetic 2′-O-methylated tRF-1001 RNA oligonucleotide that does not interfere with the siRNA treatment. Further experiments show that tRF-1001 is formed by the action of ELAC2, which encodes RNase Z and was originally identified as a candidate prostate cancer susceptibility gene (Tavtigian et al. 2001), and that tRF-1001 (and its pre-tRNA) is found almost exclusively in the cytoplasm, leading to the suggestion that pre-tRNAs can be processed either into tRNA in the nucleus, or in the cytoplasm to form tRF-1001 and related species (YS Lee et al. 2009). Since tRF-1001 does not affect expression of a reporter that should be sensitive to siRNA or miRNA, the mechanism by which it promotes cell proliferation remains to be determined. Similarly, it remains to be determined if and to what extent other tRFs regulate cellular functions, and how these tRFs are generated.
Distinct tRNA cleavage pathways as part of host defense mechanisms
These widespread tRNA cleavage pathways are distinct from a variety of tRNA cleaving toxins that act on specific sets of tRNAs and at specific positions within the tRNA anticodon loop to disable substrate tRNAs. The most well known of these is the PrrC endonuclease, which attacks tRNALys species during infection by phage T4 (Amitsur et al. 1987), but several others have been described, including the colicins and onconase (for review, see Phizicky 2008). A notable recent addition to this family is the γ-toxin of K. lactis that targets specific tRNAs in the yeast S. cerevisiae that have the mcm5s2U modification at position 34 and cleave the tRNA by formation of a 2′–3′ cyclic phosphate (Lu et al. 2005). Since Shuman and coworkers (Nandakumar et al. 2008) have shown that this cleavage pathway can be repaired by introduction of the T4 ligase and kinase, or by expression of a plant RNA ligase domain together with the yeast splicing cyclic phosphodiesterase and kinase activities, it seems plausible that any of the cleavage pathways described above are also subject to RNA repair. Intriguingly, CCA addition by nucleotidyl transferase may also act as part of a quality control mechanism, since tRNAs that are nicked are not efficient substrates for the enzyme (Dupasquier et al. 2008). It is not clear what happens to such substrates in vivo, but they could, in principle, also be repaired.
Nonconventional uses of tRNA and other signaling pathways using tRNA
Although the conventional function of tRNAs is to deliver amino acids to the translation machinery as specified by mRNA codons, it has been well established in both prokaryotic and eukaryotic cells that tRNAs serve numerous other functions. In prokaryotes, aminoacylated tRNAs serve as donors to deliver amino acids used for diverse biochemical pathways, including peptidoglycan biosynthesis, additions to lipids, production of certain antibiotics, and targeting of proteins for degradation via the N-end rule pathway (for review, see Banerjee et al. 2010; Francklyn and Minajigi 2010). Moreover, as prokaryotic organisms lack a complete set of aminoacyl-tRNA synthetases to generate all required aminoacylated tRNAs, mechanisms using noncognate aminoacyl-tRNAs function to generate the full set of cognate aminoacyl-tRNAs. For example, Glu-tRNAGln and Asp-tRNAAsn are modified to Gln-tRNAGln and Asn-tRNAAsn by amidotransferases (for review, see Feng et al. 2004). Finally, in Gram-positive bacteria, tRNAs also function as sensors to regulate gene expression in response to nutrient availability, as demonstrated clearly by the T-box riboswitch mechanism; this tRNA-dependent process employs uncharged tRNAs, which interact with mRNA leader sequences to generate anti-termination elements and thereby allow the full transcription of gene products involved in amino acid biosynthesis (for review, see Green et al. 2010).
tRNAs also serve nonconventional functions in eukaryotes. Most of these functions differ from nonconventional functions for tRNAs in prokaryotes, but at least two roles are similar among prokaryotes and eukaryotes. The first is the use of aminoacylated tRNAs as amino acid donors for N-terminal conjugation of amino acids to proteins, targeting the recipient proteins for degradation (for review, see Varshavsky 1997; Mogk et al. 2007). The second concerns the role of uncharged tRNAs in signal transduction pathways responding to nutrient deprivation. In yeast, the general amino acid response pathway responds to amino acid deprivation using uncharged tRNAs that interact with Gcn2, the protein kinase that phosphorylates translation initiation factor eIF2. Phosphorylation of eIF2 by Gcn2 results in decreased levels of general translation, but increased translation of the transcription regulator Gcn4, which in turn results in transcription of numerous genes involved in amino acid and nucleotide biosynthesis (for review, see Dever and Hinnebusch 2005). In mice, a similar Gcn2-dependent process occurs in which binding of uncharged tRNAs to Gcn2 results in an altered eating response (Hao et al. 2005; Maurin et al. 2005; for review, see Dever and Hinnebusch 2005). Surprisingly, the Gcn4 pathway is also turned on by DNA-damaging treatments in yeast that act through Mec1 and Rad53 signaling to relocalize Los1 to the cytoplasm, resulting in the accumulation of unspliced tRNA in the nucleus (Ghavidel et al. 2007). It is unknown which tRNA species activates this pathway, but Gcn4 is also activated in los1-Δ cells independently of Gcn2 (Qiu et al. 2000), so this DNA damage response pathway likely occurs by the same pathway.
There are also nonconventional roles for tRNAs that are not shared between eukaryotes and prokaryotes. For example, tRNAs have been implicated recently in regulation of apoptosis in mammalian cells (Mei et al. 2010). These studies showed that tRNAs bind cytochrome c, thereby preventing the interaction of cytochrome c with the caspase activator Apaf-1 and preventing its activation. The results provide new ways to think about how tRNA cellular levels influence cell growth and oncogenesis. Another eukaryotic-specific nonconventional function for tRNAs is the employment of tRNAs in the retroviral life cycles. Retroviruses have usurped tRNAs to serve as primers for reverse transcription of their RNA genomes (for review, see Marquet et al. 1995), and for HIV-1 minus strand transfer (Piekna-Przybylska et al. 2010 and references therein). HIV has also usurped the retrograde tRNA pathway (see below) as one mechanism to deliver the reverse-transcribed complex from the cytoplasm to the nucleus (Zaitseva et al. 2006).
The long and winding cellular road for tRNA biogenesis
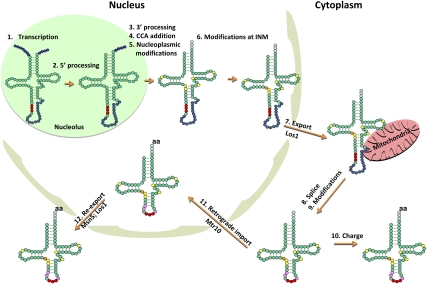
The highly unexpected set of new mechanisms of regulation of tRNA synthesis, new insights into modifications, new pathways of tRNA processing and turnover, and new tRNA cleavage pathways is matched by equally unexpected recent results describing the long and winding road by which tRNA biogenesis occurs. Indeed, as described further below, the widespread cellular distribution of gene products functioning in transcription, precursor processing, and nuclear export events for tRNAs differs substantially from the much more localized distribution of gene products functioning in analogous events for biogenesis of other RNAs. For example, for mRNA production, it appears that most of the biochemical machinery is recruited to the site of transcription; the capping, end processing, splicing, and components of the export machinery are recruited to the loci or to mRNAs cotranscriptionally (for review, see Iglesias and Stutz 2008). Similarly, as visualized by electron microscopy, pre-rRNA processing was shown to be cotranscriptional (Osheim et al. 2004), and recent kinetic studies of metabolically labeled yeast cells concluded that, with the prominent exception of cytoplasmic processing of 20S pre-rRNA to 18S mature rRNA, processing excisions and modification steps for rRNA production occur cotranscriptionally (Kos and Tollervey 2010). In stark contrast, tRNA transcription and processing occur at several distinct subcellular locations, including the nucleolus, nucleoplasm, inner nuclear membrane (INM), cytoplasm, and cytoplasmic surface of mitochondria (Fig. 5).
Figure 5.
The cell biology of tRNA biosynthesis and nuclear–cytoplasmic trafficking for intron-containing tRNAs in the yeast S. cerevisiae. tRNA transcription and 5′ end-processing occur in the nucleolus. Following 3′ end-processing, CCA addition, and various modification steps in the nucleoplasm and at the INM, intron-containing pre-tRNAs are exported to the cytoplasm via the Los1 exportin and at least one unknown pathway. After pre-tRNA splicing on the cytoplasmic surface of mitochondria, additional modifications in the cytoplasm, and aminoacylation, mature charged tRNAs can participate in protein synthesis. Cytoplasmic tRNAs are constitutively imported into nuclei, directly or indirectly, via Mtr10. Re-export of nuclear tRNAs to the cytoplasm is mediated by Los1 and Msn5 and is regulated by nutrient status; likely, Msn5-dependent re-export requires that the tRNA be appropriately structured and aminoacylated in the nucleus. (Green and red circles) Parts of the tRNA that are maintained in the mature structure; (red circles) anticodon; (purple circles) transcribed 5′ leader and 3′ trailer sequences; (dark-blue circles) intron sequence; (light-blue circles) CCA end; (yellow, orange, and pink circles) various modifications made in the nucleoplasm, at the INM, and in the cytoplasm, respectively; (aa) amino acid. Processing steps are labeled, as are the β-importin members that function in the nucleus–cytoplasm import and export steps.
tRNA transcription in the nucleolus in yeast
Although best known for its prominent role in rRNA transcription, pre-rRNA processing, and ribosome assembly, the nucleolus is also the site for tRNA transcription in yeast and pre-tRNA 5′ end processing in numerous organisms. The yeast genome contains 274 tRNA genes distributed randomly among the 16 chromosomes. Yet, as shown clearly by fluorescence in situ hybridization (FISH), tRNA genes are recruited to a single subnuclear location—the nucleolus—and are transcribed there (Thompson et al. 2003; for review, see Hopper et al. 2010; Pai and Engelke 2010). Location of the dispersed yeast tRNA genes to the nucleolus is dependent on tRNA gene transcription, as wild-type tRNA genes are located in the nucleolus ∼50% of the time, whereas transcriptionally inactivated tRNA genes are located in the nucleolus only ∼10% of the time (Thompson et al. 2003; Hopper et al. 2010). Localization of tRNA genes to the nucleolus is also dependent on intact nucleoli, as dispersion of the nucleolar structure results in distribution of tRNA genes to the nucleoplasm (Thompson et al. 2003; Wang et al. 2005).
The chromosome-condensing complex condensin is involved in tRNA gene organization. Condensin associates physically with tRNA genes, and cells with conditionally defective condensin subunits fail to cluster tRNA genes in the nucleolus (D'Ambrosio et al. 2008; Haeusler et al. 2008). Interestingly, condensin may function in clustering the tRNA genes together rather than in locating them to the nucleolus per se. This idea is supported by the observation that the nucleolar location of tRNA genes can be disrupted by treatment with nocodazole, but this treatment does not affect tRNA gene clustering (Haeusler et al. 2008). Thus, clustering of tRNA genes together and the location of the cluster to the nucleolus appear to be separable processes. Although there is little evidence for the localization of tRNA genes to the nucleolus in other organisms, tRNA gene clustering may be a conserved process, as it has been reported that dispersed tRNA genes in the fission yeast S. pombe cluster in a nuclear region close to centromeres, and that clustering at centromeres is dependent on condensin (Iwasaki et al. 2010). The roles for tRNA gene clustering and location to the nucleolus are not completely understood, but this organization clearly down-regulates transcription of closely located mRNA-encoding genes (Kendall et al. 2000; Wang et al. 2005).
tRNA 5′ end processing in the nucleolus
5′ End processing is generally the first step of the tRNA processing pathway. RNase P, the endonuclease responsible for catalyzing removal of the 5′ leader sequences of pre-tRNAs (see above), is localized to the nucleolus in yeast and vertebrate cells, although, in vertebrate cells, some of the subunits of this complex RNP are also located elsewhere (Bertrand et al. 1998; Jarrous et al. 1999). Because RNase P shares subunits with RNase MRP (Chamberlain et al. 1998), a complex RNP functioning in pre-rRNA processing in the nucleolus (Schmitt and Clayton 1993), the location of RNase P in the nucleolus could merely facilitate sharing of the subunits with RNase MRP. However, RNase P is more likely to have a more direct nucleolar function, since it has been reported to process yeast intron-encoded box C/D snoRNAs that are required for appropriate pre-rRNA processing in the nucleolus (Coughlin et al. 2008).
tRNA 3′ end processing in the nucleoplasm?
3′ End processing of pre-tRNAs generally follows 5′ end processing, and is catalyzed by both exonucleases (Rex1 in yeast) (Piper and Straby 1989; Copela et al. 2008; Ozanick et al. 2009) and the RNase Z endonuclease (Trz1 in yeast) (for review, see Vogel et al. 2005). The balance between endonucleolytic and exonucleolytic 3′ end processing depends, at least in part, on the tRNA-binding La protein in both lower and higher eukaryotes (Huang et al. 2006; Copela et al. 2008; Zhao et al. 2009). Because there is at least one yeast pre-tRNA for which 3′ end processing precedes 5′ end processing (Kufel and Tollervey 2003), and because some of the enzymes that function in 3′ pre-tRNA end processing also function in the pre-rRNA processing pathway (e.g., Rex1) (van Hoof et al. 2000), one might have predicted that pre-tRNA 3′ end processing, like 5′ end processing, would occur in the nucleolus. However, there appears to be no evidence for this, as the genome-wide GFP-tagging project conducted in yeast reports that nuclear pools of Trz1 and Lhp1 (yeast La) are primarily nucleoplasmic, not nucleolar (Huh et al. 2003).
Nuclear tRNA modification enzymes reside at various subnuclear locations
Several studies demonstrate that several different tRNA modifications occur in the nucleus in various organisms. Studies in Xenopus oocytes using a plasmid encoding yeast tRNATyr injected into oocyte nuclei showed that some modifications were found on initial tRNA transcripts, whereas others were not detected until the tRNAs were end-matured (Melton et al. 1980; Nishikura and De Robertis 1981). Since these initial transcripts and processing intermediates were restricted to the nucleus, the enzymes catalyzing modification must have resided in the nucleus. Likewise, studies employing yeast with defects in tRNA nuclear export showed that pre-tRNAs contain some, but not all, modifications (Hopper et al. 1978; Knapp et al. 1978; Etcheverry et al. 1979). Consistent with these findings, studies of individual tRNA modification proteins, as well as the genome-wide GFP-tagging database in yeast, demonstrate that a subset of the modification enzymes reside in the nucleus and lack cytoplasmic pools (Simos et al. 1996; Huh et al. 2003; for review, see Martin and Hopper 1994). For example, of the eight pseudouridine synthases, two (Pus1 and Pus7) do not appear to have cytoplasmic pools, and five of the 17 proteins involved with tRNA methylation (Trm1, Trm6, Trm8, Trm61, and Trm82) appear to be nuclear-localized.
Surprisingly, modification enzymes residing primarily in the nucleus have distinct subnuclear distributions, and can be located in the nucleolus, in the nucleoplasm, or at the INM. For example, Pus1 is rather evenly distributed throughout the nucleoplasm (Simos et al. 1996; Huh et al. 2003). In contrast, MOD5-encoded isozymes are located in the mitochondria, cytoplasm, and nucleus, and the nuclear Mod5 is concentrated in the nucleolus (Tolerico et al. 1999). Because the i6A modification catalyzed by Mod5 (Laten et al. 1978; Martin and Hopper 1982) occurs only after intron removal (Spinelli et al. 1997; for review, see Grosjean et al. 1997), which happens in the cytoplasm (Yoshihisa et al. 2003, 2007), the role of the Mod5 nucleolar pool remains unknown. Trm1 is located at yet a third intranuclear site, the INM. Although the cis-acting and some of the trans-acting factors responsible for Trm1 distribution to the nucleus and the INM have been characterized (Rose et al. 1992; Murthi and Hopper 2005; Lai et al. 2009), it remains unknown why Trm1 is located in the nucleus and, specifically, at the INM, since preventing Trm1 nuclear import does not prevent its catalysis of tRNA modification (Rose et al. 1992). In sum, reasons for the different subnuclear distributions of tRNA modification enzymes remain to be resolved.
tRNA modifications catalyzed in the cytoplasm
The Xenopus injection studies described above also showed that some modifications, primarily those located in the anticodon loop, appear only on mature tRNAs, and therefore could be catalyzed in the cytoplasm (Melton et al. 1980; Nishikura and De Robertis 1981); however, there have been few reports regarding the distribution of tRNA modification enzymes in higher eukaryotic cells. In yeast, numerous tRNA modification proteins have been reported to reside solely in the cytoplasm. For some of these, cytoplasmic location may have evolved to accommodate substrate specificity. For example, the anticodon modifications of A37 to i6A, and of C32 and G34 to Cm32 and Gm34, respectively, occur only after intron removal (Jiang et al. 1997; Pintard et al. 2002; for review, see Grosjean et al. 1997). Since, as described below, intron removal in yeast is restricted to the cytoplasm (Yoshihisa et al. 2003, 2007), enzymes that catalyze modifications that occur after splicing should be cytoplasmic, at least in yeast.
tRNA splicing on the mitochondrial cytoplasmic surface in some, but not all, organisms
Perhaps the most curious partitioning of tRNA biogenesis enzymes concerns the location of the tRNA splicing machinery in yeast. In stark contrast to the nuclear location for tRNA splicing in vertebrates (Melton et al. 1980; Lund and Dahlberg 1998; Paushkin et al. 2004), pre-tRNA splicing in yeast does not occur in the nucleus. As described above, the yeast splicing machinery is comprised of three enzymes: tRNA splicing endonuclease, tRNA ligase (Trl1), and 2′ phosphotransferase (Tpt1). Remarkably, the tRNA splicing endonuclease subunits are located on the cytoplasmic surface of mitochondria (Huh et al. 2003; Yoshihisa et al. 2003, 2007; Shaheen and Hopper 2005). The other two proteins—Trl1 and Tpt1—required for splicing are not excluded from the nucleus (Huh et al. 2003; N. Dhungel and AK Hopper, unpubl.). Nevertheless, since the pre-tRNAs that accumulate in the cytoplasm are true processing intermediates (Yoshihisa et al. 2007), it is clear that, in yeast, tRNA splicing occurs in the cytoplasm, not the nucleus.
The location of tRNA splicing in plants is not completely resolved. Expression of recombinant genes in heterologous plants provided evidence for nuclear pools of tRNA splicing endonuclease (Englert et al. 2007), consistent with pre-tRNA splicing in plants occurring in the nucleus as in vertebrates; however, other genetic studies showed that inhibition of tRNA nuclear export results in defects in intron removal (Park et al. 2005), most easily explained by a cytoplasmic location of pre-tRNA splicing in plants, as is the case for yeast. Although additional studies are required to resolve the location of tRNA splicing in the plant kingdom, it is nevertheless clear that there are different locations for pre-tRNA splicing among different organisms. Possible reasons for the different subcellular distributions of the tRNA splicing enzymes among eukaryotes include selections based on other functions of the splicing machinery (Sidrauski et al. 1996; Sidrauski and Walter 1997; Paushkin et al. 2004) and/or coordination of cellular metabolism and pre-tRNA processing in yeast, but not in vertebrate cells (for review, see Hopper and Shaheen 2008; Hopper et al. 2010).
tRNA processes occurring in both the nucleus and the cytoplasm
Cca1 addition
Although all E. coli tRNA genes encode their 3′-terminal CCA nucleotides, the E. coli genome also has a CCA nucleotidyl transferase, and its deletion causes slow growth, suggesting an important role of this enzyme in tRNA 3′ end repair in prokaryotes (Zhu and Deutscher 1987). As described above, all eukaryotic tRNA genes lack an encoded CCA and therefore require CCA addition during processing, and available evidence suggests that this step occurs in the nucleus. Injection studies in Xenopus oocytes clearly document that CCA is added to tRNAs before they are exported to the cytoplasm (Melton et al. 1980; Nishikura and De Robertis 1981; Lund and Dahlberg 1998), and it is known that the CCA addition to tRNAs is required for efficient tRNA nuclear export in vertebrate cells (Arts et al. 1998b; Lipowsky et al. 1999). The evidence also supports a role for CCA addition in the nucleus in yeast, as end-processed intron-containing pre-tRNAs located primarily in the nucleus possess CCA at their 3′ ends (Wolfe et al. 1996). Nevertheless, in addition to the nuclear pools, there are also cytoplasmic pools of CCA nucleotidyl transferase in both yeast and vertebrate cells (Solari and Deutscher 1982; Wolfe et al. 1996). These cytoplasmic pools likely serve to repair tRNA 3′ ends analogous to the function of this activity in prokaryotes, since redistribution of cytoplasmic CCA nucleotidyl transferase to the nucleus in yeast results in accumulation of mature tRNAs lacking 3′ CCA nucleotides (Wolfe et al. 1994). Thus, in eukaryotes, whereas nucleoplasmic CCA nucleotidyl transferase functions in tRNA processing, cytoplasmic CCA nucleotidyl transferase likely functions in tRNA end repair.
RTD turnover pathway
The RTD turnover pathway also appears to function in both the nucleus and the cytoplasm. Evidence to support this is the fact that hypomodified tRNAs that are subject to this pathway are not completely stabilized if either the nuclear Rat1 5′–3′ exonuclease or the orthologous cytoplasmic Xrn1 exonuclease is deleted, but near-complete stabilization is achieved by elimination of both Rat1 and Xrn1 (Chernyakov et al. 2008). In contrast to the RTD pathway, the TRAMP-mediated tRNA turnover pathway is restricted to the nucleus (for review, see Houseley et al. 2006).
Enzymes shared by more than one subcellular compartment
A common theme for tRNA processing enzymes is sharing proteins encoded by single genes between the cytoplasmic/nuclear compartments and in mitochondria. The yeast tRNA modification enzyme Trm1 provided the first example of eukaryotic genes encoding sorting isozymes—multiple proteins encoded by single genes, each with different subcellular distribution (Hopper et al. 1982). TRM1 encodes two proteins differing by possession of a 16-amino-acid N-terminal peptide: Trm1-I, the long form, is found exclusively in the mitochondria, and the short form, Trm1-II, is distributed primarily to the nuclear INM (Ellis et al. 1989; Li et al. 1989). Similar mechanisms create different isozymes for Mod5 and Cca1, resulting in the longest forms being located in mitochondria and the shorter forms being located elsewhere in cells (Wolfe et al. 1994; Tolerico et al. 1999; for review, see Martin and Hopper 1994). Although the information is still not complete, compartmental sharing appears to be characteristic of tRNA processing enzymes for yeast. For example, of the 17 known yeast genes encoding subunits of nucleoside methyltransferases, none appear to encode proteins solely targeted to mitochondria; thus, for those tRNA modifications that occur on both mitochondrial and nuclear encoded tRNAs, the mitochondrial enzymes must be encoded by the same genes that encode the cytoplasmic- and/or nuclear-located enzymes. Likewise, of the eight genes involved in tRNA pseudouridylation, only two, PUS2 and PUS9, encode activities that modify only tRNAs localized in mitochondria (Behm-Ansmant et al. 2004, 2007). The mechanisms by which the numerous dual-targeted activities are distributed to multiple subcellular compartments have not been completely determined.
Compartmental sharing is also found among some of the aminoacyl-tRNA synthetases. As first shown in yeast, a single gene encodes both cytoplasmic and mitochondrial histidyl-tRNA synthetase (Natsoulis et al. 1986). Similarly, mitochondrial and cytoplasmic alanyl-tRNA, glycyl-tRNA, and valyl-tRNA synthetases are encoded by single genes (Chatton et al. 1988; Turner et al. 2000; Tang et al. 2004). The remainder of the yeast mitochondrial and cytoplasmic tRNA synthetases appear to be encoded by separate genes. In plants, a similar situation exists; mitochondrial and chloroplast tRNA synthetases are generally encoded by the same gene, and there are multiple examples of single genes encoding both cytoplasmic and plastid activities (Duchene et al. 2005).
Amazing nuclear–cytoplasmic tRNA dynamics
A long-standing view of the nuclear–cytoplasmic dynamics for tRNAs was that tRNAs were transcribed and processed in the nucleus and then delivered to the cytoplasm for aminoacylation and function in translation. This view was challenged by the demonstration that (1) tRNA aminoacylation is not restricted to the cytoplasm (Arts et al. 1998b; Lund and Dahlberg 1998; Sarkar et al. 1999; Grosshans et al. 2000), (2) defects in tRNA aminoacylation in the nucleus cause tRNA nuclear accumulation (Lund and Dahlberg 1998; Sarkar et al. 1999; Grosshans et al. 2000; Azad et al. 2001; Steiner-Mosonyi and Mangroo 2004), and, (3) in yeast, tRNA splicing occurs not in the nucleoplasm, but rather in the cytoplasm on the surface of mitochondria (Yoshihisa et al. 2003, 2007). These observations and the fact that, in yeast, spliced tRNAs (i.e., tRNAs that had “visited” the cytoplasm) accumulate in the nucleus when aminoacylation is defective (Grosshans et al. 2000; Feng and Hopper 2002) led to the proposal and subsequent proof that tRNA subcellular traffic is not unidirectional, from the nucleus to the cytoplasm. Rather, a retrograde pathway exists by which cytoplasmic tRNAs are imported into the nucleus, and they can be re-exported to the cytoplasm in response to nutrient availability (Shaheen and Hopper 2005; Takano et al. 2005; Hurto et al. 2007; Whitney et al. 2007). This retrograde process is conserved in vertebrate cells, and HIV appears to have co-opted the process as one mechanism to deliver retrotranscribed complexes to the nucleus (Zaitseva et al. 2006; Shaheen et al. 2007). The movement of tRNA between the nucleus and the cytoplasm thus occurs in three separate steps (Fig. 5). First, newly transcribed and partially processed tRNAs are exported from the nucleus to the cytoplasm (“primary tRNA nuclear export”). Second, tRNAs are imported from the cytoplasm into the nucleus (“tRNA nuclear import”). Finally, tRNAs that were once in the cytoplasm but reside in the nucleus can re-enter the cytoplasm by “tRNA re-export” (Whitney et al. 2007).
Primary tRNA nuclear export
There is a well-defined mechanism for the export of tRNA from the nucleus to the cytoplasm that uses the Ran-GTPase pathway and the vertebrate member of the Ran-binding β-importin family, Xpo-t (Arts et al. 1998a; Kutay et al. 1998). Xpo-t orthologs in fungi (Los1 or Xpot) and plants (PSD or PAUSED) appear to function similarly (Hellmuth et al. 1998; Sarkar and Hopper 1998; Hunter et al. 2003). Xpo-t and its orthologs directly bind tRNA in the presence of Ran-GTP, and the heterotrimeric Ran-GTP•Xpo-t•tRNA complex moves through nuclear pores to the cytoplasm, where, upon Ran-GAP-mediated enhancement of hydrolysis of Ran-GTP to Ran-GDP, the complex dissociates and tRNA is released. Recently, a 3.2 Å resolution structure of S. pombe Xpot in complex with Ran-GTP and tRNA was obtained, as well as a 3.1 Å structure of the unbound Xpot (Cook et al. 2009). The structures elegantly demonstrate that Xpot wraps around tRNA, making contacts with the acceptor arm and the TΨC and D-loops, leaving the anticodon loop exposed. These studies verify earlier work using structure probing and the use of artificial substrates that demonstrated that Xpo-t binds preferentially to tRNAs with mature 5′ and 3′ termini containing a CCA end, and that alterations of tRNA sequences disrupting the cloverleaf or tertiary tRNA structure result in reduced binding to Xpo-t, but the presence of introns does not influence the binding efficiency of Xpo-t to tRNA (Arts et al. 1998b; Lipowsky et al. 1999). The interactions show that Xpo-t provides a proofreading function so that tRNAs with inappropriate structure or immature 5′ and 3′ termini fail to be delivered to the cytoplasm. The interactions also account for how unspliced tRNAs containing introns in the anticodon loop can interact with Los1 and be exported to the cytoplasm. However, the data shed little light on how aminoacylation affects tRNA nuclear export because unspliced tRNAs cannot be aminoacylated (O'Farrell et al. 1978), but, in yeast, they are nevertheless exported from the nucleus by Los1.
Two lines of evidence suggest that, at least in some organisms, there is an additional pathway to deliver tRNA to the cytoplasm that is independent of the Xpo-t orthologs, First, LOS1 in budding and fission yeast, and its plant ortholog, PSD, are each nonessential, even though tRNA nuclear export is obviously essential for these cells. Although plants missing PSD have developmental defects, growth of los1Δ and LOS1 wild-type cells is very similar (Hurt et al. 1987; Hunter et al. 2003; Li and Chen 2003). Second, some organisms, such as arthropods, do not have an Xpo-t ortholog among their β-importin family members (S Shibata et al. 2006 and references therein).
In addition to its role in nuclear export of particular phosphorylated proteins in budding yeast (Kaffman et al. 1998; DeVit and Johnston 1999; for review, see Hopper 1999) and in nuclear export of pre-miRNAs in metazoans and plants (Lund et al. 2004; Zeng and Cullen 2004; Park et al. 2005; S Shibata et al. 2006), the β-importin member Exportin-5 likely serves as a tRNA nuclear exporter in some organisms. Exportin-5 binds tRNA in the presence of Ran-GTP (Bohnsack et al. 2002; Calado et al. 2002; S Shibata et al. 2006). The structure of this complex has not yet been reported, but there is a prediction of it based on the recently reported 2.9 Å structure of pre-miRNA in complex with Exportin-5 and Ran-GTP (Okada et al. 2009). In yeast, biological experiments have documented a role for the Exportin-5 ortholog Msn5 in tRNA nuclear export. First, los1Δ msn5Δ double mutants accumulate more tRNA in the nucleus than either single mutant (Takano et al. 2005). (Since los1Δ msn5Δ double mutants are hearty, there must be at least one other unidentified exporter for tRNA nuclear export in yeast.) Second, as described further below, Msn5 functions in the tRNA re-export part of the retrograde process (Eswara et al. 2009; Murthi et al. 2010). Likewise, in Drosophila, where the Exportin-t ortholog is absent, the Exportin-5 ortholog dmExp5 serves as a tRNA nuclear exporter in addition to serving for pre-miRNA nuclear export (S Shibata et al. 2006). So, in yeast and flies, Exportin-5 homologs function in tRNA nuclear export. This is in contrast to the biological role in other organisms. For example, despite the ability of vertebrate Exportin-5 to bind tRNA, it has not been considered a major contributor to tRNA nuclear–cytoplasmic dynamics for vertebrates because Xpo-t exports the majority of the tRNA nuclear pool to the cytoplasm in vertebrate cells (Arts et al. 1998b; Lipowsky et al. 1999), and because Exportin-5 has only a small effect on the tRNA nuclear pool (Bohnsack et al. 2002; Calado et al. 2002). Similarly, the plant Exportin-5 ortholog HST has been reported not to function in tRNA nuclear export (Park et al. 2005). In sum, Exportin-5 homologs appear to function in tRNA nuclear export in some, but not all, organisms.
Retrograde tRNA nuclear import
One member of the yeast β-importin family, Mtr10 (vertebrate TNPO3 or TRN-SR2), is implicated in retrograde tRNA nuclear import. Deletion of MTR10 causes defects in tRNA nuclear accumulation (Shaheen and Hopper 2005; Hurto et al. 2007; Whitney et al. 2007). Whether Mtr10 has a direct interaction with tRNA is unknown, but its role may be indirect, based on well-characterized interactions of Mtr10 and TNPO3 with proteins, especially RNA-binding proteins (for review, see Pemberton and Paschal 2005). tRNAs accumulate in the nucleus upon nutrient deprivation (Shaheen and Hopper 2005; Hurto et al. 2007; Whitney et al. 2007). However, the nuclear import process is not nutrient-sensitive, as it has been shown to be a constitutive process (Shaheen and Hopper 2005; Takano et al. 2005; Zaitseva et al. 2006; Murthi et al. 2010).
Retrograde tRNA re-export
Since tRNA nuclear import is constitutive, but the retrograde pathway is nutrient-sensitive, the re-export step is likely to be regulated and nutrient-responsive. Interestingly, two lines of evidence suggest that Los1 functions in both the tRNA primary and the re-export processes. First, cytoplasmic tRNAs accumulate in nuclei in los1Δ heterokaryons (Shaheen and Hopper 2005; Takano et al. 2005). Second, the nuclear pool of tRNAs in los1Δ cells is markedly reduced when tRNA nuclear import is reduced in mtr10Δ los1Δ cells (Murthi et al. 2010). Los1-mediated nuclear export is unlikely to be responsible for nutrient-dependent or aminoacylation-dependent tRNA re-export because Los1 can export pre-tRNA that is not aminoacylated.
As described above, the yeast β-importin member Msn5 functions in tRNA nuclear export. As deletion of MSN5 has no effect on nuclear export of intron-containing pre-tRNAs, Msn5's role in nuclear tRNA export is restricted to the re-export pathway, at least for those tRNAs encoded by intron-containing genes (Eswara et al. 2009; Murthi et al. 2010), and is the likely tRNA exporter responding to nutrient availability. It is unknown how Msn5 interacts and exports mature tRNAs from the nucleus to the cytoplasm, but not transcripts that contain introns. It is also unknown how Msn5 could be nutrient-responsive and/or export primarily aminoacylated tRNAs. One proposal is that it would cooperate with another protein able to distinguish mature tRNAs from intron-containing pre-tRNAs, and aminoacylated from uncharged tRNAs. The translation elongation factor Tef1/2 (vertebrate eEF1-a) is a candidate for such a protein because it interacts only with mature aminoacylated tRNA, has been implicated in tRNA nuclear export (Grosshans et al. 2000; McGuire and Mangroo 2007), and is able to move into and out of the yeast nucleus (Murthi et al. 2010). Other yeast proteins including Utp9 (Eswara et al. 2009), Utp8 (McGuire et al. 2009), and Sol1/2 (Shen et al. 1996; Stanford et al. 2004) have also been implicated in the nuclear–cytoplasmic dynamics of mature tRNAs, but further studies are required to learn the roles of these proteins in tRNA re-export.
Future prospects
The pace of progress in our understanding of tRNA processing, tRNA trafficking, and tRNA function is truly staggering, since almost all of the highlighted information in this review is new since 2003 (Hopper and Phizicky 2003), and since we omitted so many results from the study of organisms other than yeast and from organelles. The future promises to be equally revealing. For example, we can expect that every tRNA modification pathway will be worked out in yeast and in several other model organisms, as well as in humans, and we will know most of the functions in some detail. In yeast, the recent finding that sua5 mutants lack the universally conserved threonylcarbamoyladenosine modification (t6A) has opened the door to study of this ancient modification (El Yacoubi et al. 2009). The increasing application of synthetic genetic and chemical genetic arrays and other similar methods for analyzing complex genetic phenotypes promises to yield great insights into tRNA biology (Gustavsson and Ronne 2008; Wilmes et al. 2008). Increased use of deep sequencing methods and microarray-based analysis will undoubtedly contribute significantly new information on tRNA expression, signaling, and translation (Netzer et al. 2009). The increased sophistication of crystallographic analysis (Murphy et al. 2004; Ogle and Ramakrishnan 2005) and in vitro translation assays (Kothe and Rodnina 2007; Zaher and Green 2009) promises to add great dimension to our understanding of tRNA translation efficiency and fidelity, and ever more sophisticated assay systems will allow an increasingly clear picture of translation in vivo (Salas-Marco and Bedwell 2005; Kramer and Farabaugh 2007) and subcellular tRNA trafficking pathways. In addition, further study of the involvement of tRNA and tRNA fragments in signaling pathways and stress response pathways will undoubtedly lead to new links between tRNA function and other global cellular response systems.
Acknowledgments
This work was supported by grants from the NIH: GM27930 to A.K.H., and GM52347 to E.M.P. We thank Elizabeth Grayhack for comments on the manuscript, and are grateful to the students and post-doctoral fellows in the Hopper and Phizicky laboratories for many stimulating conversations.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1956510.
Note added in proof
Recent results demonstrate that the tRNA m5C methyltransferase Dnmt2, which modifies residue 38 of substrate tRNAs (Goll et al. 2006), has a prominent role in the stress response (Schaefer et al. 2010). Drosophila Dnmt2 mutants are significantly less viable in response to oxidative or heat stress, and substrate tRNAs in these mutants are more sensitive to stress-induced cleavage and cleavage by angiogenin (Schaefer et al. 2010). Since this sensitivity to cleavage by angiogenin is also observed in tRNA from mouse embyonic fibroblasts derived from the corresponding mouse mutant (Schaefer et al. 2010), these results suggest that tRNA modification may play a conserved role in modulation of the stress response.
References
- Abad MG, Rao BS, Jackman JE 2010. Template-dependent 3′–5′ nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc Natl Acad Sci 107: 674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah U, Cullen PJ 2009. The tRNA modification complex elongator regulates the Cdc42-dependent mitogen-activated protein kinase pathway that controls filamentous growth in yeast. Eukaryot Cell 8: 1362–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquati F, Possati L, Ferrante L, Campomenosi P, Talevi S, Bardelli S, Margiotta C, Russo A, Bortoletto E, Rocchetti R, et al. 2005. Tumor and metastasis suppression by the human RNASET2 gene. Int J Oncol 26: 1159–1168 [PubMed] [Google Scholar]
- Aebi M, Kirchner G, Chen JY, Vijayraghavan U, Jacobson A, Martin NC, Abelson J 1990. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem 265: 16216–16220 [PubMed] [Google Scholar]
- Alexandrov A, Martzen MR, Phizicky EM 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8: 1253–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96 [DOI] [PubMed] [Google Scholar]
- Amitsur M, Levitz R, Kaufmann G 1987. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J 6: 2499–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG 1998. The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 12: 3650–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Hinnebusch AG 2000. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1- methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci 97: 5173–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, Ekstein J, Rubin BY 2001. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet 68: 753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansmant I, Motorin Y, Massenet S, Grosjean H, Branlant C 2001. Identification and characterization of the tRNA:Ψ 31-synthase (Pus6p) of Saccharomyces cerevisiae. J Biol Chem 276: 34934–34940 [DOI] [PubMed] [Google Scholar]
- Ares M Jr, Grate L, Pauling MH 1999. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA 5: 1138–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW 1998a. Identification of a nuclear export receptor for tRNA. Curr Biol 8: 305–314 [DOI] [PubMed] [Google Scholar]
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW 1998b. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 17: 7430–7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrom SU, Bystrom AS 1994. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell 79: 535–546 [DOI] [PubMed] [Google Scholar]
- Azad AK, Stanford DR, Sarkar S, Hopper AK 2001. Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol Biol Cell 12: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R, Chen S, Dare K, Gilreath M, Praetorius-Ibba M, Raina M, Reynolds NM, Rogers T, Roy H, Yadavalli SS, et al. 2010. tRNAs: Cellular barcodes for amino acids. FEBS Lett 584: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayfield MA, Maraia RJ 2009. Precursor-product discrimination by La protein during tRNA metabolism. Nat Struct Mol Biol 16: 430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Planta RJ, Grosjean H 1997. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of Ψ55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res 25: 4493–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ 2007. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 28: 860–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Urban A, Ma X, Yu YT, Motorin Y, Branlant C 2003. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite–multisubstrate RNA:Ψ-synthase also acting on tRNAs. RNA 9: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Grosjean H, Massenet S, Motorin Y, Branlant C 2004. Pseudouridylation at position 32 of mitochondrial and cytoplasmic tRNAs requires two distinct enzymes in Saccharomyces cerevisiae. J Biol Chem 279: 52998–53006 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Branlant C, Motorin Y 2007. The Saccharomyces cerevisiae Pus2 protein encoded by YGL063w ORF is a mitochondrial tRNA:Ψ27/28-synthase. RNA 13: 1641–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR 1998. Nucleolar localization of early tRNA processing. Genes Dev 12: 2463–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Xu J, Johnson RC, Schimmel P, de Crecy-Lagard V 2002. Identification of the tRNA-dihydrouridine synthase family. J Biol Chem 277: 25090–25095 [DOI] [PubMed] [Google Scholar]
- Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP 2001. A primordial tRNA modification required for the evolution of life? EMBO J 20: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR, Huang B, Persson OP, Bystrom AS 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13: 1245–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguta M 2009. Control of RNA polymerases I and III by the TOR signaling pathway. Cell Cycle 8: 4023–4024 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21: 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisnard S, Lagniel G, Garmendia-Torres C, Molin M, Boy-Marcotte E, Jacquet M, Toledano MB, Labarre J, Chedin S 2009. H2O2 activates the nuclear localization of Msn2 and Maf1 through thioredoxins in Saccharomyces cerevisiae. Eukaryot Cell 8: 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde BS, Namavar Y, Barth PG, Poll-The BT, Nurnberg G, Becker C, van Ruissen F, Weterman MA, Fluiter K, te Beek ET, et al. 2008. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat Genet 40: 1113–1118 [DOI] [PubMed] [Google Scholar]
- Calado A, Treichel N, Muller EC, Otto A, Kutay U 2002. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 21: 6216–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96 [DOI] [PubMed] [Google Scholar]
- Calvin K, Xue S, Ellis C, Mitchell MH, Li H 2008. Probing the catalytic triad of an archaeal RNA splicing endonuclease. Biochemistry 47: 13659–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canino G, Bocian E, Barbezier N, Echeverria M, Forner J, Binder S, Marchfelder A 2009. Arabidopsis encodes four tRNase Z enzymes. Plant Physiol 150: 1494–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BA, Mushinski JF, Henderson DW, Kwon SY, Crain PF, Lee BJ, Hatfield DL 2001. 1-Methylguanosine in place of Y base at position 37 in phenylalanine tRNA is responsible for its shiftiness in retroviral ribosomal frameshifting. Virology 279: 130–135 [DOI] [PubMed] [Google Scholar]
- Castano JG, Tobian JA, Zasloff M 1985. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNAMet(i) (3′ pre-tRNase). J Biol Chem 260: 9002–9008 [PubMed] [Google Scholar]
- Cavaille J, Chetouani F, Bachellerie JP 1999. The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA 5: 66–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J 22: 6562–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev 12: 1678–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton B, Walter P, Ebel JP, Lacroute F, Fasiolo F 1988. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J Biol Chem 263: 52–57 [PubMed] [Google Scholar]
- Chen C, Tuck S, Bystrom AS 2009. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 5: e1000561 doi: 10.1371/journal.pgen.1000561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM 2008. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′–3′ exonucleases Rat1 and Xrn1. Genes Dev 22: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho IM, Lai LB, Susanti D, Mukhopadhyay B, Gopalan V 2010. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc Natl Acad Sci doi: 10.1073/pnas.1005556107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesla M, Boguta M 2008. Regulation of RNA polymerase III transcription by Maf1 protein. Acta Biochim Pol 55: 215–225 [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AG, Fukuhara N, Jinek M, Conti E 2009. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461: 60–65 [DOI] [PubMed] [Google Scholar]
- Cooley L, Appel B, Soll D 1982. Post-transcriptional nucleotide addition is responsible for the formation of the 5′ terminus of histidine tRNA. Proc Natl Acad Sci 79: 6475–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela LA, Fernandez CF, Sherrer RL, Wolin SL 2008. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 14: 1214–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin DJ, Pleiss JA, Walker SC, Whitworth GB, Engelke DR 2008. Genome-wide search for yeast RNase P substrates reveals role in maturation of intron-encoded box C/D small nucleolar RNAs. Proc Natl Acad Sci 105: 12218–12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM, McCraith SM, Zillmann M, Kierzek R, Michaud N, LaReau RD, Turner DH, Phizicky EM 1993. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1″–2″ cyclic phosphate. Science 261: 206–208 [DOI] [PubMed] [Google Scholar]
- Culver GM, McCraith SM, Consaul SA, Stanford DR, Phizicky EM 1997. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J Biol Chem 272: 13203–13210 [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F 2008. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22: 2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P, Sprinzl M, Watanabe K, Albani M, Kersten H 1979. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res 6: 1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crecy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, et al. 2010. Biosynthesis of wyosine derivatives in tRNA: An ancient and highly diverse pathway in Archaea. Mol Biol Evol doi: 10.1093/molbev/msq096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Sierra-Gallay IL, Pellegrini O, Condon C 2005. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature 433: 657–661 [DOI] [PubMed] [Google Scholar]
- Desai N, Lee J, Upadhya R, Chu Y, Moir RD, Willis IM 2005. Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J Biol Chem 280: 6455–6462 [DOI] [PubMed] [Google Scholar]
- Dever TE, Hinnebusch AG 2005. GCN2 whets the appetite for amino acids. Mol Cell 18: 141–142 [DOI] [PubMed] [Google Scholar]
- DeVit MJ, Johnston M 1999. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol 9: 1231–1241 [DOI] [PubMed] [Google Scholar]
- Dichtl B, Stevens A, Tollervey D 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J 16: 7184–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK 1987. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol 7: 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Segni G, Gastaldi S, Tocchini-Valentini GP 2008. Cis- and trans-splicing of mRNAs mediated by tRNA sequences in eukaryotic cells. Proc Natl Acad Sci 105: 6864–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droogmans L, Grosjean H 1987. Enzymatic conversion of guanosine 3′ adjacent to the anticodon of yeast tRNAPhe to N1-methylguanosine and the wye nucleoside: Dependence on the anticodon sequence. EMBO J 6: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A 2004. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res 32: 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchene AM, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters NM, Zaepfel M, Marechal-Drouard L, Small ID 2005. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci 102: 16484–16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupasquier M, Kim S, Halkidis K, Gamper H, Hou YM 2008. tRNA integrity is a prerequisite for rapid CCA addition: Implication for quality control. J Mol Biol 379: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant PC, Davis DR 1999. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J Mol Biol 285: 115–131 [DOI] [PubMed] [Google Scholar]
- Ellis SR, Morales MJ, Li JM, Hopper AK, Martin NC 1986. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J Biol Chem 261: 9703–9709 [PubMed] [Google Scholar]
- Ellis SR, Hopper AK, Martin NC 1989. Amino-terminal extension generated from an upstream AUG codon increases the efficiency of mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferases. Mol Cell Biol 9: 1611–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V 2009. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 37: 2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P 2010. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285: 10959–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke DR, Gegenheimer P, Abelson J 1985. Nucleolytic processing of a tRNAArg–tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem 260: 1271–1279 [PubMed] [Google Scholar]
- Englert M, Latz A, Becker D, Gimple O, Beier H, Akama K 2007. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie 89: 1351–1365 [DOI] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ, Bystrom AS 2006. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 24: 139–148 [DOI] [PubMed] [Google Scholar]
- Eswara MB, McGuire AT, Pierce JB, Mangroo D 2009. Utp9p facilitates Msn5p-mediated nuclear reexport of retrograded tRNAs in Saccharomyces cerevisiae. Mol Biol Cell 20: 5007–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etcheverry T, Colby D, Guthrie C 1979. A precursor to a minor species of yeast tRNASer contains an intervening sequence. Cell 18: 11–26 [DOI] [PubMed] [Google Scholar]
- Feng W, Hopper AK 2002. A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomycescerevisiae. Proc Natl Acad Sci 99: 5412–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Sheppard K, Namgoong S, Ambrogelly A, Polycarpo C, Randau L, Tumbula-Hansen D, Soll D 2004. Aminoacyl-tRNA synthesis by pre-translational amino acid modification. RNA Biol 1: 16–20 [PubMed] [Google Scholar]
- Fichtner L, Frohloff F, Burkner K, Larsen M, Breunig KD, Schaffrath R 2002. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol Microbiol 43: 783–791 [DOI] [PubMed] [Google Scholar]
- Fichtner L, Jablonowski D, Schierhorn A, Kitamoto HK, Stark MJ, Schaffrath R 2003. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol 49: 1297–1307 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Shatkin AJ 1983. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell 32: 547–557 [DOI] [PubMed] [Google Scholar]
- Francklyn CS, Minajigi A 2010. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett 584: 366–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583: 437–442 [DOI] [PubMed] [Google Scholar]
- Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD 2010. Human AlkB homolog ABH8 is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol 30: 2449–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furter R, Snaith M, Gillespie DE, Hall BD 1992. Endonucleolytic cleavage of a long 3′-trailer sequence in a nuclear yeast suppressor tRNA. Biochemistry 31: 10817–10824 [DOI] [PubMed] [Google Scholar]
- Garber RL, Altman S 1979. In vitro processing of B. mori transfer RNA precursor molecules. Cell 17: 389–397 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Gerber AP, Keller W 1999. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286: 1146–1149 [DOI] [PubMed] [Google Scholar]
- Gerber A, Grosjean H, Melcher T, Keller W 1998. Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J 17: 4780–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A 2007. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell 131: 915–926 [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398 [DOI] [PubMed] [Google Scholar]
- Green NJ, Grundy FJ, Henkin TM 2010. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett 584: 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer CL, Peebles CL, Gegenheimer P, Abelson J 1983. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell 32: 537–546 [DOI] [PubMed] [Google Scholar]
- Grosjean H, Szweykowska-Kulinska Z, Motorin Y, Fasiolo F, Simos G 1997. Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: A review. Biochimie 79: 293–302 [DOI] [PubMed] [Google Scholar]
- Grosshans H, Hurt E, Simos G 2000. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev 14: 830–840 [PMC free article] [PubMed] [Google Scholar]
- Grunberger D, Weinstein IB, Mushinski JF 1975. Deficiency of the Y base in a hepatoma phenylalanine tRNA. Nature 253: 66–67 [DOI] [PubMed] [Google Scholar]
- Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM 2003. tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev 17: 2889–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM 2005. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol Cell Biol 25: 8191–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857 [DOI] [PubMed] [Google Scholar]
- Guo D, Hu K, Lei Y, Wang Y, Ma T, He D 2004. Identification and characterization of a novel cytoplasm protein ICF45 that is involved in cell cycle regulation. J Biol Chem 279: 53498–53505 [DOI] [PubMed] [Google Scholar]
- Gustavsson M, Ronne H 2008. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA 14: 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22: 2204–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser HJ, Karginov FV, Hannon GJ, Elliot MA 2008. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res 36: 732–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Sharp JW, Ross-Inta CM, McDaniel BJ, Anthony TG, Wek RC, Cavener DR, McGrath BC, Rudell JB, Koehnle TJ, et al. 2005. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science 307: 1776–1778 [DOI] [PubMed] [Google Scholar]
- Harding HP, Lackey JG, Hsu HC, Zhang Y, Deng J, Xu RM, Damha MJ, Ron D 2008. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA 14: 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A 2009. The making of tRNAs and more—RNase P and tRNase Z. Prog Mol Biol Transl Sci 85: 319–368 [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 18: 6374–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Giege R, Florentz C 1999. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 38: 13338–13346 [DOI] [PubMed] [Google Scholar]
- Ho CK, Rauhut R, Vijayraghavan U, Abelson J 1990. Accumulation of pre-tRNA splicing ‘2/3’ intermediates in a Saccharomyces cerevisiae mutant. EMBO J 9: 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzle A, Fischer S, Heyer R, Schutz S, Zacharias M, Walther P, Allers T, Marchfelder A 2008. Maturation of the 5S rRNA 5′ end is catalyzed in vitro by the endonuclease tRNase Z in the archaeon H. volcanii. RNA 14: 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W 2008. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135: 462–474 [DOI] [PubMed] [Google Scholar]
- Hopper AK 1999. Nucleocytoplasmic transport: Inside out regulation. Curr Biol 9: R803–R806 doi: 10.1016/S0960-9822(99)80494-1 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM 2003. tRNA transfers to the limelight. Genes Dev 17: 162–180 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Shaheen HH 2008. A decade of surprises for tRNA nuclear–cytoplasmic dynamics. Trends Cell Biol 18: 98–104 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Banks F, Evangelides V 1978. A yeast mutant which accumulates precursor tRNAs. Cell 14: 211–219 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Schultz LD, Shapiro RA 1980. Processing of intervening sequences: A new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell 19: 741–751 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Furukawa AH, Pham HD, Martin NC 1982. Defects in modification of cytoplasmic and mitochondrial transfer RNAs are caused by single nuclear mutations. Cell 28: 543–550 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Pai DA, Engelke DR 2010. Cellular dynamics of tRNAs and their genes. FEBS Lett 584: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Bystrom AS 2005. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Bayfield MA, Intine RV, Maraia RJ 2006. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol 13: 611–618 [DOI] [PubMed] [Google Scholar]
- Huang B, Lu J, Bystrom AS 2008. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 14: 2183–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R 2009. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev 23: 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Hunter CA, Aukerman MJ, Sun H, Fokina M, Poethig RS 2003. PAUSED encodes the Arabidopsis exportin-t ortholog. Plant Physiol 132: 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt DJ, Wang SS, Lin YH, Hopper AK 1987. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol 7: 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurto RL, Tong AH, Boone C, Hopper AK 2007. Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae. Genetics 176: 841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Stutz F 2008. Regulation of mRNP dynamics along the export pathway. FEBS Lett 582: 1987–1996 [DOI] [PubMed] [Google Scholar]
- Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T 2006. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol Cell 21: 97–108 [DOI] [PubMed] [Google Scholar]
- Ishii R, Minagawa A, Takaku H, Takagi M, Nashimoto M, Yokoyama S 2005. Crystal structure of the tRNA 3′ processing endoribonuclease tRNase Z from Thermotoga maritima. J Biol Chem 280: 14138–14144 [DOI] [PubMed] [Google Scholar]
- Ishii R, Minagawa A, Takaku H, Takagi M, Nashimoto M, Yokoyama S 2007. The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes. Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K 2010. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell 21: 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski D, Frohloff F, Fichtner L, Stark MJ, Schaffrath R 2001. Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol Microbiol 42: 1095–1105 [DOI] [PubMed] [Google Scholar]
- Jablonowski D, Fichtner L, Stark MJ, Schaffrath R 2004. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol Biol Cell 15: 1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM 2006a. tRNAHis guanylyltransferase adds G-1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA 12: 1007–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Phizicky EM 2006b. tRNAHis guanylyltransferase catalyzes a 3′–5′ polymerization reaction that is distinct from G-1 addition. Proc Natl Acad Sci 103: 8640–8645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Montange RK, Malik HS, Phizicky EM 2003. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn D, Pande S 1991. Histidine tRNA guanylyltransferase from Saccharomyces cerevisiae. II. Catalytic mechanism. J Biol Chem 266: 22832–22836 [PubMed] [Google Scholar]
- Jarrous N, Wolenski JS, Wesolowski D, Lee C, Altman S 1999. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J Cell Biol 146: 559–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HQ, Motorin Y, Jin YX, Grosjean H 1997. Pleiotropic effects of intron removal on base modification pattern of yeast tRNAPhe: An in vitro study. Nucleic Acids Res 25: 2694–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, Bystrom AS 2004. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA 10: 712–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS 2008. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 28: 3301–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL 2007. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell 26: 367–379 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Rank NM, O'Neill EM, Huang LS, O'Shea EK 1998. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396: 482–486 [DOI] [PubMed] [Google Scholar]
- Kalhor HR, Clarke S 2003. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol 23: 9283–9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor HR, Penjwini M, Clarke S 2005. A novel methyltransferase required for the formation of the hypermodified nucleoside wybutosine in eucaryotic tRNA. Biochem Biophys Res Commun 334: 433–440 [DOI] [PubMed] [Google Scholar]
- Kambampati R, Lauhon CT 2003. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42: 1109–1117 [DOI] [PubMed] [Google Scholar]
- Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin RP, Tarassov I 2007. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol Cell 26: 625–637 [DOI] [PubMed] [Google Scholar]
- Kanerva PA, Maenpaa PH 1981. Codon-specific serine transfer ribonucleic acid degradation in avian liver during vitellogenin induction. Acta Chem Scand B 35: 379–385 [DOI] [PubMed] [Google Scholar]
- Karnahl U, Wasternack C 1992. Half-life of cytoplasmic rRNA and tRNA, of plastid rRNA and of uridine nucleotides in heterotrophically and photoorganotrophically grown cells of Euglena gracilis and its apoplastic mutant W3BUL. Int J Biochem 24: 493–497 [DOI] [PubMed] [Google Scholar]
- Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S 1992. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry 31: 1040–1046 [DOI] [PubMed] [Google Scholar]
- Kendall A, Hull MW, Bertrand E, Good PD, Singer RH, Engelke DR 2000. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc Natl Acad Sci 97: 13108–13113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikovska E, Svard SG, Kirsebom LA 2007. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci 104: 2062–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom LA 2007. RNase P RNA mediated cleavage: Substrate recognition and catalysis. Biochimie 89: 1183–1194 [DOI] [PubMed] [Google Scholar]
- Knapp G, Beckmann JS, Johnson PF, Fuhrman SA, Abelson J 1978. Transcription and processing of intervening sequences in yeast tRNA genes. Cell 14: 221–236 [DOI] [PubMed] [Google Scholar]
- Kos M, Tollervey D 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelecky B, Pohl E, Vogel A, Schilling O, Meyer-Klaucke W 2006. The crystal structure of the zinc phosphodiesterase from Escherichia coli provides insight into function and cooperativity of tRNase Z-family proteins. J Bacteriol 188: 1607–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelawala L, Grayhack EJ, Phizicky EM 2008. Identification of yeast tRNA Um44 2′-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNASer species. RNA 14: 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe U, Rodnina MV 2007. Codon reading by tRNAAla with modified uridine in the wobble position. Mol Cell 25: 167–174 [DOI] [PubMed] [Google Scholar]
- Kramer EB, Farabaugh PJ 2007. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Greenblatt JF 2001. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol Cell Biol 21: 8203–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y, Borek E, Grunberger D, Mushinski JF, Nishimura S 1982. Changes of post-transcriptional modification of wye base in tumor-specific tRNAPhe. Nucleic Acids Res 10: 6421–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J, Tollervey D 2003. 3′-Processing of yeast tRNA(Trp) precedes 5′-processing. RNA 9: 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D 1998. Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lai TP, Stauffer KA, Murthi A, Shaheen HH, Peng G, Martin NC, Hopper AK 2009. Mechanism and a peptide motif for targeting peripheral proteins to the yeast inner nuclear membrane. Traffic 10: 1243–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski FA, Fire AZ, RajBhandary UL, Sharp PA 1983. Characterization of tRNA precursor splicing in mammalian extracts. J Biol Chem 258: 11974–11980 [PubMed] [Google Scholar]
- Laten H, Gorman J, Bock RM 1978. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res 5: 4329–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecointe F, Simos G, Sauer A, Hurt EC, Motorin Y, Grosjean H 1998. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of Ψ 38 and Ψ 39 in tRNA anticodon loop. J Biol Chem 273: 1316–1323 [DOI] [PubMed] [Google Scholar]
- Lee SR, Collins K 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280: 42744–42749 [DOI] [PubMed] [Google Scholar]
- Lee J, Moir RD, Willis IM 2009. Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J Biol Chem 284: 12604–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M 2009. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458: 228–232 [DOI] [PubMed] [Google Scholar]
- Li Y, Altman S 2004. In search of RNase P RNA from microbial genomes. RNA 10: 1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen X 2003. PAUSED, a putative exportin-t, acts pleiotropically in Arabidopsis development but is dispensable for viability. Plant Physiol 132: 1913–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP 1994. The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J Biol Chem 269: 6064–6071 [PubMed] [Google Scholar]
- Li Z, Deutscher MP 1996. Maturation pathways for E. coli tRNA precursors: A random multienzyme process in vivo. Cell 86: 503–512 [DOI] [PubMed] [Google Scholar]
- Li Z, Deutscher MP 2002. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA 8: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JM, Hopper AK, Martin NC 1989. N2,N2-Dimethylguanosine-specific tRNA methyltransferase contains both nuclear and mitochondrial targeting signals in Saccharomyces cerevisiae. J Cell Biol 109: 1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I, Mathy N, Pellegrini O, Condon C 2006. Structure of the ubiquitous 3′ processing enzyme RNase Z bound to transfer RNA. Nat Struct Mol Biol 13: 376–377 [DOI] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Izaurralde E, Kutay U, Schafer S, Gross HJ, Beier H, Gorlich D 1999. Coordination of tRNA nuclear export with processing of tRNA. RNA 5: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS 2005. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 11: 1648–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082–2085 [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Maraia RJ, Bayfield MA 2006. The La protein–RNA complex surfaces. Mol Cell 21: 149–152 [DOI] [PubMed] [Google Scholar]
- Marck C, Grosjean H 2003. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: Evolutionary implications. RNA 9: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet R, Isel C, Ehresmann C, Ehresmann B 1995. tRNAs as primer of reverse transcriptases. Biochimie 77: 113–124 [DOI] [PubMed] [Google Scholar]
- Marquez SM, Harris JK, Kelley ST, Brown JW, Dawson SC, Roberts EC, Pace NR 2005. Structural implications of novel diversity in eucaryal RNase P RNA. RNA 11: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez SM, Chen JL, Evans D, Pace NR 2006. Structure and function of eukaryotic Ribonuclease P RNA. Mol Cell 24: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ 2008. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 133: 78–89 [DOI] [PubMed] [Google Scholar]
- Martin NC, Hopper AK 1982. Isopentenylation of both cytoplasmic and mitochondrial tRNA is affected by a single nuclear mutation. J Biol Chem 257: 10562–10565 [PubMed] [Google Scholar]
- Martin NC, Hopper AK 1994. How single genes provide tRNA processing enzymes to mitochondria, nuclei and the cytosol. Biochimie 76: 1161–1167 [DOI] [PubMed] [Google Scholar]
- Martin RP, Schneller JM, Stahl AJ, Dirheimer G 1979. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry 18: 4600–4605 [DOI] [PubMed] [Google Scholar]
- Maurin AC, Jousse C, Averous J, Parry L, Bruhat A, Cherasse Y, Zeng H, Zhang Y, Harding HP, Ron D, et al. 2005. The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab 1: 273–277 [DOI] [PubMed] [Google Scholar]
- McGuire AT, Mangroo D 2007. Cex1p is a novel cytoplasmic component of the Saccharomyces cerevisiae nuclear tRNA export machinery. EMBO J 26: 288–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Keates RA, Cook S, Mangroo D 2009. Structural modeling identified the tRNA-binding domain of Utp8p, an essential nucleolar component of the nuclear tRNA export machinery of Saccharomyces cerevisiae. Biochem Cell Biol 87: 431–443 [DOI] [PubMed] [Google Scholar]
- Mehlgarten C, Schaffrath R 2003. Mutant casein kinase I (Hrr25p/Kti14p) abrogates the G1 cell cycle arrest induced by Kluyveromyces lactis zymocin in budding yeast. Mol Genet Genomics 269: 188–196 [DOI] [PubMed] [Google Scholar]
- Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X 2010. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell 37: 668–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton DA, De Robertis EM, Cortese R 1980. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature 284: 143–148 [DOI] [PubMed] [Google Scholar]
- Minagawa A, Takaku H, Takagi M, Nashimoto M 2004. A novel endonucleolytic mechanism to generate the CCA 3′ termini of tRNA molecules in Thermotoga maritima. J Biol Chem 279: 15688–15697 [DOI] [PubMed] [Google Scholar]
- Minagawa A, Ishii R, Takaku H, Yokoyama S, Nashimoto M 2008. The flexible arm of tRNase Z is not essential for pre-tRNA binding but affects cleavage site selection. J Mol Biol 381: 289–299 [DOI] [PubMed] [Google Scholar]
- Mogk A, Schmidt R, Bukau B 2007. The N-end rule pathway for regulated proteolysis: Prokaryotic and eukaryotic strategies. Trends Cell Biol 17: 165–172 [DOI] [PubMed] [Google Scholar]
- Mohan A, Whyte S, Wang X, Nashimoto M, Levinger L 1999. The 3′ end CCA of mature tRNA is an antideterminant for eukaryotic 3′-tRNase. RNA 5: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Lee J, Haeusler RA, Desai N, Engelke DR, Willis IM 2006. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc Natl Acad Sci 103: 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Grosjean H 1999. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA 5: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Keith G, Simon C, Foiret D, Simos G, Hurt E, Grosjean H 1998. The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA 4: 856–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S 1988. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336: 179–181 [DOI] [PubMed] [Google Scholar]
- Murawski M, Szczesniak B, Zoladek T, Hopper AK, Martin NC, Boguta M 1994. maf1 mutation alters the subcellular localization of the Mod5 protein in yeast. Acta Biochim Pol 41: 441–448 [PubMed] [Google Scholar]
- Murguia JR, Belles JM, Serrano R 1996. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J Biol Chem 271: 29029–29033 [DOI] [PubMed] [Google Scholar]
- Murphy FVt, Ramakrishnan V, Malkiewicz A, Agris PF 2004. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat Struct Mol Biol 11: 1186–1191 [DOI] [PubMed] [Google Scholar]
- Murthi A, Hopper AK 2005. Genome-wide screen for inner nuclear membrane protein targeting in Saccharomyces cerevisiae: Roles for N-acetylation and an integral membrane protein. Genetics 170: 1553–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthi A, Shaheen HH, Huang HY, Preston MA, Lai TP, Phizicky EM, Hopper AK 2010. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol Biol Cell 21: 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Umeda N, Suzuki T, Nakai M, Hayashi H, Watanabe K, Kagamiyama H 2004. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J Biol Chem 279: 12363–12368 [DOI] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Lill R, Suzuki T, Hayashi H 2007. Thio modification of yeast cytosolic tRNA is an iron–sulfur protein-dependent pathway. Mol Cell Biol 27: 2841–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Hayashi H 2008. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem 283: 27469–27476 [DOI] [PubMed] [Google Scholar]
- Nameki N, Asahara H, Shimizu M, Okada N, Himeno H 1995. Identity elements of Saccharomyces cerevisiae tRNA(His). Nucleic Acids Res 23: 389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S 2008. RNA repair: An antidote to cytotoxic eukaryal RNA damage. Mol Cell 31: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M, Tamura M, Kaspar RL 1999a. Minimum requirements for substrates of mammalian tRNA 3′ processing endoribonuclease. Biochemistry 38: 12089–12096 [DOI] [PubMed] [Google Scholar]
- Nashimoto M, Wesemann DR, Geary S, Tamura M, Kaspar RL 1999b. Long 5′ leaders inhibit removal of a 3′ trailer from a precursor tRNA by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res 27: 2770–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis G, Hilger F, Fink GR 1986. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell 46: 235–243 [DOI] [PubMed] [Google Scholar]
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. 2009. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462: 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL 2001. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 7: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K, De Robertis EM 1981. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J Mol Biol 145: 405–420 [DOI] [PubMed] [Google Scholar]
- Noma A, Kirino Y, Ikeuchi Y, Suzuki T 2006. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J 25: 2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A, Sakaguchi Y, Suzuki T 2009. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res 37: 1335–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund ME, Johansson JO, von Pawel-Rammingen U, Bystrom AS 2000. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA 6: 844–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwagwu M, Nana M 1980. Ribonucleic acid synthesis in embryonic chick muscle, rates of synthesis and half-lives of transfer and ribosomal RNA species. J Embryol Exp Morphol 56: 253–267 [PubMed] [Google Scholar]
- O'Farrell PZ, Cordell B, Valenzuela P, Rutter WJ, Goodman HM 1978. Structure and processing of yeast precursor tRNAs containing intervening sequences. Nature 274: 438–445 [DOI] [PubMed] [Google Scholar]
- Oficjalska-Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, Sentenac A, Lefebvre O 2006. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell 22: 623–632 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T 2009. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326: 1275–1279 [DOI] [PubMed] [Google Scholar]
- Orellana O, Cooley L, Soll D 1986. The additional guanylate at the 5′ terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol Cell Biol 6: 525–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16: 943–954 [DOI] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell 3: 109–118 [DOI] [PubMed] [Google Scholar]
- Ow MC, Kushner SR 2002. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev 16: 1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick SG, Wang X, Costanzo M, Brost RL, Boone C, Anderson JT 2009. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res 37: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai DA, Engelke DR 2010. Spatial organization of genes as a component of regulated expression. Chromosoma 119: 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW 1999. RNase P RNAs from some Archaea are catalytically active. Proc Natl Acad Sci 96: 7803–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou A, Gross HJ 1996. Pre-tRNA 3′-processing in Saccharomyces cerevisiae. Purification and characterization of exo- and endoribonucleases. Eur J Biochem 242: 747–759 [DOI] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS 2005. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR 2004. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117: 311–321 [DOI] [PubMed] [Google Scholar]
- Peebles CL, Gegenheimer P, Abelson J 1983. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell 32: 525–536 [DOI] [PubMed] [Google Scholar]
- Pellegrini O, Nezzar J, Marchfelder A, Putzer H, Condon C 2003. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis. EMBO J 22: 4534–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6: 187–198 [DOI] [PubMed] [Google Scholar]
- Perwez T, Kushner SR 2006. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol Microbiol 60: 723–737 [DOI] [PubMed] [Google Scholar]
- Phizicky EM 2008. Waste not, want not: A case for tRNA repair. Nat Struct Mol Biol 15: 783–784 [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Alfonzo JD 2010. Do all modifications benefit all tRNAs? FEBS Lett 584: 265–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Schwartz RC, Abelson J 1986. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem 261: 2978–2986 [PubMed] [Google Scholar]
- Phizicky EM, Consaul SA, Nehrke KW, Abelson J 1992. Yeast tRNA ligase mutants are nonviable and accumulate tRNA splicing intermediates. J Biol Chem 267: 4577–4582 [PubMed] [Google Scholar]
- Piekna-Przybylska D, DiChiacchio L, Mathews DH, Bambara RA 2010. A sequence similar to tRNA 3 Lys gene is embedded in HIV-1 U3-R and promotes minus-strand transfer. Nat Struct Mol Biol 17: 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B 2002. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J 21: 1811–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper PW, Straby KB 1989. Processing of transcripts of a dimeric tRNA gene in yeast uses the nuclease responsible for maturation of the 3′ termini upon 5 S and 37 S precursor rRNAs. FEBS Lett 250: 311–316 [DOI] [PubMed] [Google Scholar]
- Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, Hopper AK, Sentenac A, Boguta M 2001. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol 21: 5031–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, Phizicky EM 2010. The requirement for the highly conserved G-1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase. RNA 16: 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH, Gray MW 1999. A novel nucleotide incorporation activity implicated in the editing of mitochondrial transfer RNAs in Acanthamoeba castellanii. RNA 5: 302–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranam RS, Attardi G 2001. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol Cell Biol 21: 548–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B 2005. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol Cell Biol 25: 4359–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz J, Florentz C, Benseler F, Giege R 1994. A single methyl group prevents the mischarging of a tRNA. Nat Struct Biol 1: 580–582 [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Anderson J, Bjork GR, Sarkar S, Hopper AK, Hinnebusch AG 2000. Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the α subunit of eukaryotic translation initiation factor 2. Mol Cell Biol 20: 2505–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Chen CZ, Collins RN 2005. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell 17: 841–853 [DOI] [PubMed] [Google Scholar]
- Randau L, Calvin K, Hall M, Yuan J, Podar M, Li H, Soll D 2005a. The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge–helix–bulge motifs of joined tRNA halves. Proc Natl Acad Sci 102: 17934–17939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau L, Munch R, Hohn MJ, Jahn D, Soll D 2005b. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature 433: 537–541 [DOI] [PubMed] [Google Scholar]
- Randau L, Schroder I, Soll D 2008. Life without RNase P. Nature 453: 120–123 [DOI] [PubMed] [Google Scholar]
- Randau L, Stanley BJ, Kohlway A, Mechta S, Xiong Y, Soll D 2009. A cytidine deaminase edits C to U in transfer RNAs in Archaea. Science 324: 657–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redko Y, Li de Lasierra-Gallay I, Condon C 2007. When all's zed and done: The structure and function of RNase Z in prokaryotes. Nat Rev Microbiol 5: 278–286 [DOI] [PubMed] [Google Scholar]
- Reina JH, Azzouz TN, Hernandez N 2006. Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS ONE 1: e134 doi: 10.1371/journal.pone.0000134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice TS, Ding M, Pederson DS, Heintz NH 2005. The highly conserved tRNAHis guanylyltransferase Thg1p interacts with the origin recognition complex and is required for the G2/M phase transition in the yeast Saccharomyces cerevisiae. Eukaryot Cell 4: 832–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins J, Veras I, Cabarcas S, Willis I, Schramm L 2007. Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int J Biol Sci 3: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AM, Joyce PB, Hopper AK, Martin NC 1992. Separate information required for nuclear and subnuclear localization: Additional complexity in localizing an enzyme shared by mitochondria and nuclei. Mol Cell Biol 12: 5652–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen AE, Brooks BS, Guth E, Francklyn CS, Musier-Forsyth K 2006. Evolutionary conservation of a functionally important backbone phosphate group critical for aminoacylation of histidine tRNAs. RNA 12: 1315–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmanith W, Karwan RM 1998. Characterization of human mitochondrial RNase P: Novel aspects in tRNA processing. Biochem Biophys Res Commun 247: 234–241 [DOI] [PubMed] [Google Scholar]
- Rubio MA, Ragone FL, Gaston KW, Ibba M, Alfonzo JD 2006. C to U editing stimulates A to I editing in the anticodon loop of a cytoplasmic threonyl tRNA in Trypanosoma brucei. J Biol Chem 281: 115–120 [DOI] [PubMed] [Google Scholar]
- Rubio MA, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GA, Papavasiliou FN, Alfonzo JD 2007. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc Natl Acad Sci 104: 7821–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger J, Florentz C, Giege R 1994. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues −1 and 73 but is dependent on the RNA context. Nucleic Acids Res 22: 5031–5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM 2005. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol 348: 801–815 [DOI] [PubMed] [Google Scholar]
- Salia SR, Singh SK, Gurha P, Gupta R 2003. Two reactions of Haloferax volcanii RNA splicing enzymes: Joining of exons and circularization of introns. RNA 9: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta MP, Tongprasit W, Sethi H, Chin CS, Stolc V 2006. Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc Natl Acad Sci 103: 4192–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK 1998. tRNA nuclear export in Saccharomyces cerevisiae: In situ hybridization analysis. Mol Biol Cell 9: 3041–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Azad AK, Hopper AK 1999. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci 96: 14366–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24: 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer S, Rosch S, Marchfelder A 2002. Assigning a function to a conserved group of proteins: The tRNA 3′- processing enzymes. EMBO J 21: 2769–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer S, Rosch S, Marchfelder A 2003. Recombinant RNase Z does not recognize CCA as part of the tRNA and its cleavage efficieny is influenced by acceptor stem length. Biol Chem 384: 333–342 [DOI] [PubMed] [Google Scholar]
- Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL 2008. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci 105: 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 13: 7935–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D 2007. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell 27: 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta R, Vainauskas S, Yarian C, Sochacka E, Malkiewicz A, Guenther RH, Koshlap KM, Agris PF 2000. Modified constructs of the tRNA TΨC domain to probe substrate conformational requirements of m(1)A(58) and m(5)U(54) tRNA methyltransferases. Nucleic Acids Res 28: 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci 102: 11290–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK 2007. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci 104: 8845–8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Vallee BL 1987. Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc Natl Acad Sci 84: 2238–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Strydom DJ, Olson KA, Vallee BL 1987. Isolation of angiogenin from normal human plasma. Biochemistry 26: 5141–5146 [DOI] [PubMed] [Google Scholar]
- Shen WC, Stanford DR, Hopper AK 1996. Los1p, involved in yeast pre-tRNA splicing, positively regulates members of the SOL gene family. Genetics 143: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata HS, Minagawa A, Takaku H, Takagi M, Nashimoto M 2006. Unstructured RNA is a substrate for tRNase Z. Biochemistry 45: 5486–5492 [DOI] [PubMed] [Google Scholar]
- Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J, Yoneda Y 2006. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res 34: 4711–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Nakamura M, Anai S, De Velasco M, Tanaka M, Tsujikawa K, Ouji Y, Konishi N 2009. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res 69: 3157–3164 [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039 [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P 1996. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87: 405–413 [DOI] [PubMed] [Google Scholar]
- Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt EC 1996. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J 15: 2270–2284 [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, Liebert CB, Chadwick B, Idelson M, Reznik L, et al. 2001. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet 68: 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff P, Roiz L, Angelkovitch B, Schwartz B, Shoseyov O 2006. A recombinant human RNASET2 glycoprotein with antitumorigenic and antiangiogenic characteristics: Expression, purification, and characterization. Cancer 107: 2760–2769 [DOI] [PubMed] [Google Scholar]
- Solari A, Deutscher MP 1982. Subcellular localization of the tRNA processing enzyme, tRNA nucleotidyltransferase, in Xenopus laevis oocytes and in somatic cells. Nucleic Acids Res 10: 4397–4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M, Kawamura F, Sekine Y 2007. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science 318: 450–453 [DOI] [PubMed] [Google Scholar]
- Spinelli SL, Consaul SA, Phizicky EM 1997. A conditional lethal yeast phosphotransferase (tpt1) mutant accumulates tRNAs with a 2′-phosphate and an undermodified base at the splice junction. RNA 3: 1388–1400 [PMC free article] [PubMed] [Google Scholar]
- Spinelli SL, Malik HS, Consaul SA, Phizicky EM 1998. A functional homolog of a yeast tRNA splicing enzyme is conserved in higher eukaryotes and in Escherichia coli. Proc Natl Acad Sci 95: 14136–14141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS 2005. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 33: D139–D140 doi: 10.1093/nar/gki012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford DR, Whitney ML, Hurto RL, Eisaman DM, Shen WC, Hopper AK 2004. Division of labor among the yeast Sol proteins implicated in tRNA nuclear export and carbohydrate metabolism. Genetics 168: 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Mosonyi M, Mangroo D 2004. The nuclear tRNA aminoacylation-dependent pathway may be the principal route used to export tRNA from the nucleus in Saccharomyces cerevisiae. Biochem J 378: 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studte P, Zink S, Jablonowski D, Bar C, von der Haar T, Tuite MF, Schaffrath R 2008. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol Microbiol 69: 1266–1277 [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A, Immanuel D, Arndt KT 1991. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol 11: 2133–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Noma A, Suzuki T, Ishitani R, Nureki O 2009. Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4. Nucleic Acids Res 37: 2910–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku H, Minagawa A, Takagi M, Nashimoto M 2003. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res 31: 2272–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T 2005. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142 [DOI] [PubMed] [Google Scholar]
- Tang HL, Yeh LS, Chen NK, Ripmaster T, Schimmel P, Wang CC 2004. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J Biol Chem 279: 49656–49663 [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, Dayananth P, et al. 2001. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet 27: 172–180 [DOI] [PubMed] [Google Scholar]
- Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ 2006. Structural basis for recognition and sequestration of UUU(OH) 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell 21: 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LD, Daniels CJ 1990. Recognition of exon–intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J Biol Chem 265: 18104–18111 [PubMed] [Google Scholar]
- Thompson DM, Parker R 2009a. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R 2009b. Stressing out over tRNA cleavage. Cell 138: 215–219 [DOI] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR 2003. Nucleolar clustering of dispersed tRNA genes. Science 302: 1399–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolerico LH, Benko AL, Aris JP, Stanford DR, Martin NC, Hopper AK 1999. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics 151: 57–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J, Di Como CJ, Herrero E, De La Torre-Ruiz MA 2002. Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J Biol Chem 277: 43495–43504 [DOI] [PubMed] [Google Scholar]
- Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A 2005. Crystal structure of the RNA component of bacterial ribonuclease P. Nature 437: 584–587 [DOI] [PubMed] [Google Scholar]
- Towpik J, Graczyk D, Gajda A, Lefebvre O, Boguta M 2008. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J Biol Chem 283: 17168–17174 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, Abelson JN 1997. The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89: 849–858 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Paushkin SV, Patel M, Li H, Peltz SW 2006. Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature 441: 375–377 [DOI] [PubMed] [Google Scholar]
- Tsai HY, Pulukkunat DK, Woznick WK, Gopalan V 2006. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc Natl Acad Sci 103: 16147–16152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RJ, Lovato M, Schimmel P 2000. One of two genes encoding glycyl-tRNA synthetase in Saccharomyces cerevisiae provides mitochondrial and cytoplasmic functions. J Biol Chem 275: 27681–27688 [DOI] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM 2002. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell 10: 1489–1494 [DOI] [PubMed] [Google Scholar]
- Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J 20: 4863–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Stahl G, Durand JM, Ben Salem SN, Qian Q, Farabaugh PJ, Bjork GR 2003. Transfer RNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting. RNA 9: 760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189 doi: 10.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 19: 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zyl W, Huang W, Sneddon AA, Stark M, Camier S, Werner M, Marck C, Sentenac A, Broach JR 1992. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Cell Biol 12: 4946–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A 1997. The N-end rule pathway of protein degradation. Genes Cells 2: 13–28 [DOI] [PubMed] [Google Scholar]
- Vogel A, Schilling O, Spath B, Marchfelder A 2005. The tRNase Z family of proteins: Physiological functions, substrate specificity and structural properties. Biol Chem 386: 1253–1264 [DOI] [PubMed] [Google Scholar]
- Waas WF, de Crecy-Lagard V, Schimmel P 2005. Discovery of a gene family critical to wyosine base formation in a subset of phenylalanine-specific transfer RNAs. J Biol Chem 280: 37616–37622 [DOI] [PubMed] [Google Scholar]
- Waas WF, Druzina Z, Hanan M, Schimmel P 2007. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J Biol Chem 282: 26026–26034 [DOI] [PubMed] [Google Scholar]
- Waldron C, Lacroute F 1975. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol 122: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SC, Engelke DR 2006. Ribonuclease P: The evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol 41: 77–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MJ, Davis NW, Gegenheimer P 1988. Novel mechanisms for maturation of chloroplast transfer RNA precursors. EMBO J 7: 1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Haeusler RA, Good PD, Thompson M, Nagar S, Engelke DR 2005. Silencing near tRNA genes requires nucleolar localization. J Biol Chem 280: 8637–8639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sobral BW, Williams KP 2007. Loss of a universal tRNA feature. J Bacteriol 189: 1954–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jia H, Jankowsky E, Anderson JT 2008. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA 14: 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zheng XF 2009. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle 8: 4085–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Tsang CK, Zheng XF 2009. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J 28: 2220–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Martinez J 2007. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 447: 222–226 [DOI] [PubMed] [Google Scholar]
- Wen T, Oussenko IA, Pellegrini O, Bechhofer DH, Condon C 2005. Ribonuclease PH plays a major role in the exonucleolytic maturation of CCA-containing tRNA precursors in Bacillus subtilis. Nucleic Acids Res 33: 3636–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ML, Hurto RL, Shaheen HH, Hopper AK 2007. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell 18: 2678–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson ML, Crary SM, Jackman JE, Grayhack EJ, Phizicky EM 2007. The 2′-O-methyltransferase responsible for modification of yeast tRNA at position 4. RNA 13: 404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis IM, Moir RD 2007. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci 32: 51–53 [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. 2008. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell 32: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL 2008. 3′ End processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS, Petrakis TG, Ethelberg S, Tokunaga M, Erdjument-Bromage H, Tempst P, Svejstrup JQ 2001. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J Biol Chem 276: 32743–32749 [DOI] [PubMed] [Google Scholar]
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci 99: 3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell 4: 123–128 [DOI] [PubMed] [Google Scholar]
- Wolfe CL, Lou YC, Hopper AK, Martin NC 1994. Interplay of heterogeneous transcriptional start sites and translational selection of AUGs dictate the production of mitochondrial and cytosolic/nuclear tRNA nucleotidyltransferase from the same gene in yeast. J Biol Chem 269: 13361–13366 [PubMed] [Google Scholar]
- Wolfe CL, Hopper AK, Martin NC 1996. Mechanisms leading to and the consequences of altering the normal distribution of ATP(CTP):tRNA nucleotidyltransferase in yeast. J Biol Chem 271: 4679–4686 [DOI] [PubMed] [Google Scholar]
- Xing F, Martzen MR, Phizicky EM 2002. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA 8: 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing F, Hiley SL, Hughes TR, Phizicky EM 2004. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J Biol Chem 279: 17850–17860 [DOI] [PubMed] [Google Scholar]
- Xue S, Calvin K, Li H 2006. RNA recognition and cleavage by a splicing endonuclease. Science 312: 906–910 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Altman S 2007. A noncoding RNA in Saccharomyces cerevisiae is an RNase P substrate. RNA 13: 682–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89: 393–402 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891 [DOI] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T 2003. Possibility of cytoplasmic pre-tRNA splicing: The yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T 2007. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells 12: 285–297 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009. Quality control by the ribosome following peptide bond formation. Nature 457: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva L, Myers R, Fassati A 2006. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol 4: e332 doi: 10.1371/journal.pbio.0040332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR 2004. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res 32: 4776–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sun L, Kragler F 2009. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol 150: 378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Su W, Yuan S, Huang Y 2009. Functional conservation of tRNase ZL among Saccharomyces cerevisiae, Schizosaccharomyces pombe and humans. Biochem J 422: 483–492 [DOI] [PubMed] [Google Scholar]
- Zhu L, Deutscher MP 1987. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J 6: 2473–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M, Gorovsky MA, Phizicky EM 1991. Conserved mechanism of tRNA splicing in eukaryotes. Mol Cell Biol 11: 5410–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M, Gorovsky MA, Phizicky EM 1992. HeLa cells contain a 2′-phosphate-specific phosphotransferase similar to a yeast enzyme implicated in tRNA splicing. J Biol Chem 267: 10289–10294 [PubMed] [Google Scholar]
- Zofallova L, Guo Y, Gupta R 2000. Junction phosphate is derived from the precursor in the tRNA spliced by the archaeon Haloferax volcanii cell extract. RNA 6: 1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]