Abstract
Piwi proteins are modified by symmetric dimethylation of arginine (sDMA), and the methylarginine-dependent interaction with Tudor domain proteins is critical for their functions in germline development. Cocrystal structures of an extended Tudor domain (eTud) of Drosophila Tudor with methylated peptides of Aubergine, a Piwi family protein, reveal that sDMA is recognized by an asparagine-gated aromatic cage. Furthermore, the unexpected Tudor-SN/p100 fold of eTud is important for sensing the position of sDMA. The structural information provides mechanistic insights into sDMA-dependent Piwi–Tudor interaction, and the recognition of sDMA by Tudor domains in general.
Keywords: Tudor, Aubergine, Piwi, arginine methylation, structure
Piwi proteins are a conserved subfamily of Argonaute proteins that, together with small RNAs with which they associate (piRNA [Piwi-interacting RNA]), safeguard germline development by silencing transposons (Aravin and Hannon 2008; Lin and Yin 2008). Certain arginine residues at the N termini of Piwi proteins are symmetrically dimethylated by PRMT5, and the post-translational modification is required for interaction with Tudor domain proteins (Chen et al. 2009; Kirino et al. 2009; Nishida et al. 2009; Reuter et al. 2009; Vagin et al. 2009; Vasileva et al. 2009; Wang et al. 2009; Siomi et al. 2010). In Drosophila, the Piwi family protein Aub is symmetrically dimethylated at Arg11, Arg13, and Arg15, and loss of methylation at these sites disrupts the interaction with the maternal effect protein Tud and reduces association with piRNA (Kirino et al. 2009, 2010; Nishida et al. 2009). However, a mechanistic understanding of methylarginine-dependent Tud–Aub interaction is lacking. At present, no structure of any protein in complex with a symmetric dimethylation of arginine (sDMA)-modified protein/peptide has been reported. The best characterized sDMA–Tudor interaction to date is the binding of sDMA-modified spliceosomal Sm proteins by the Tudor domain of Survival Motor Neuron (SMN) (Brahms et al. 2001; Friesen et al. 2001; Sprangers et al. 2003), although an understanding of their binding mode in atomic resolution details is still lacking. Interestingly, several Tudor domains have been shown to bind histone tails with methylated lysine residues, and an understanding of the structural basis for methyllysine recognition by Tudor domains has been developed (Botuyan et al. 2006; Huang et al. 2006; Kim et al. 2006). Thus, a pressing matter for understanding the biological functions of Tudor domain proteins is to elucidate the structural determinants responsible for Tudor domain's binding preference for an sDMA or methyllysine, which necessitates the determination of a Tudor domain structure in complex with an sDMA ligand.
Results and Discussion
Tudor domains 7–11 are sufficient for germ cell formation and Aubergine binding
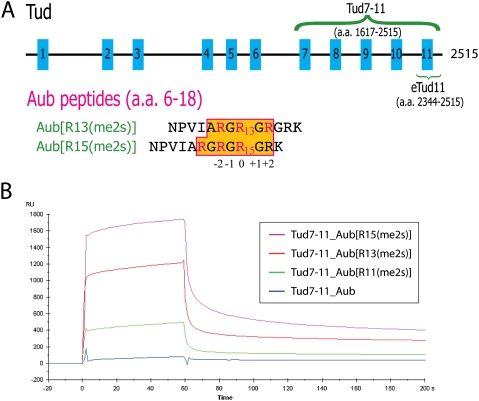
Tud is a large protein (2515 amino acids) comprised of 11 copies of a defined sequence motif, which can be found in many eukaryotic proteins and has been named the Tudor domain (Fig. 1A; Golumbeski et al. 1991; Ponting 1997; Talbot et al. 1998; Thomson and Lasko 2005). Here, we first show that the five C-terminal tandem Tudor domains (Tud7–11) are necessary and sufficient for Aub binding and germ cell formation. It was shown previously that domains 2–6 of Tud are nonessential for these functions (Arkov et al. 2006; Kirino et al. 2010). Figure 1B shows that the bacterially expressed Tud7–11 protein was able to bind biotin-labeled Aub peptides containing a single sDMA residue at the 11th, 13th, or 15th positions, respectively, as determined using a Surface Plasmon Resonance (SPR) assay. It appears that Tud7–11 binds to Aub peptides containing a single sDMA substitution at the 11th, 13th, and 15th positions with increasing binding affinities (15th > 13th > 11th), although precise binding constants cannot be deduced in the absence of knowledge about their binding stoichiometries. Equally important, a transgene encoding Tud7–11 is functional in polar granule and germ cell formation (Fig. 2A; Supplemental Figs. S1, S2), and Tud7–11 colocalizes with Aub to the posterior pole of oocytes and early embryos (Fig. 2I; Supplemental Fig. S1). Thus, we conclude that Tud7–11 contains essential elements required for germ cell formation and sDMA-dependent interaction with Aub. Mutations of a pair of conserved aromatic residues in the putative sDMA-binding pockets of each of the five Tudor domains indicate that the integrity of Tud9, Tud10, and Tud11 are essential for germ cell formation (Fig. 2C-F; Supplemental Figs. S1, S2). Furthermore, Aub localization requires functional Tud, implicating a possible involvement of each of the five Tudor domains in binding Aub (Fig. 2J–L; Supplemental Figs. S1 and S2). Direct binding of sDMA-containing Aub peptides to a Tud fragment encompassing Tud11 (eTud11 [extended Tudor domain encompassing Tud11]) (see below) was confirmed using an SPR assay, and binding characteristics qualitatively resembled those of Tud7–11 (Supplemental Fig. S3).
Figure 1.
Tudor domains interacting with Aubergine. (A, top) A schematic representation of 11 Tudor domains in Drosophila Tud. Each blue box represents a Tudor domain, and the length of each domain and the spacing between them are drawn approximately to scale. (Bottom) Aub peptides used for cocrystallization. One peptide, R13(me2s), has a symmetrically dimethylated Arg13, and the other, R15(me2s), has a methylated Arg15. Three known arginine methylation sites are colored pink in the amino acid sequence. The ordered amino acids in the two structures are shown in the boxed area with an orange background. The alignment of the two sequences reflects an identical Tud-binding mode involving amino acids located in the region between the −2 and +2 positions. (B) Binding curves of Tud7–11 to Aub measured using SPR. Curves from top to bottom represent measurements of the same concentration of Tud7–11 (5 μM) flown through biosensor chips coupled with Btn-Aub[R15(me2s)] (magenta), Btn-Aub[R13(me2s)] (red), Btn-Aub[R11(me2s)] (green), and Btn-Aub (blue) peptides, respectively. The vertical and horizontal axes show the Biacore response unit and the time scale (in seconds).
Figure 2.
Mutants in single Tudor domains affect germ cell formation and Aub localization. (A–F) Stage 4 embryos stained with anti-Vasa (green) to detect germ cell formation. tudor mutant with a wild-type (A) and single Tud domain mutant (B–F) tud7–11 transgene. (A) Wild-type transgene. In B–F, the second and fourth aromatic residues in a single Tudor domain were changed to alanines in the tud7-11 transgene (for amino acids mutated, see Fig. 4, below; Materials and Methods). Although all transgenes were inserted into the same genomic region and point mutations were generated in equivalent amino acids, protein expression level and phenotypic outcome vary (see also Supplemental Figs. S1, S2). (G–L) Aub localization in the wild-type (G), tudor mutant (H), tudor mutant with a wild-type (I), Tudor domain 8 (J), Tudor domain 9 (K), and Tudor domain 10 (L) single-mutant tud7–11 transgene. Stage 10 oocytes stained with Aub (green) and FasIII (red) antibodies. Note that Tud7–11 is sufficient for localization of Aub to the posterior pole of the oocyte, but only the full-length tudor transgene restores Aub localization to the nuage of nurse cells. Quantitation of localized Tud7–11 and Aub proteins are shown in Supplemental Figure S1. All egg chambers and embryos are oriented anterior to the left.
Overall structure of eTud11
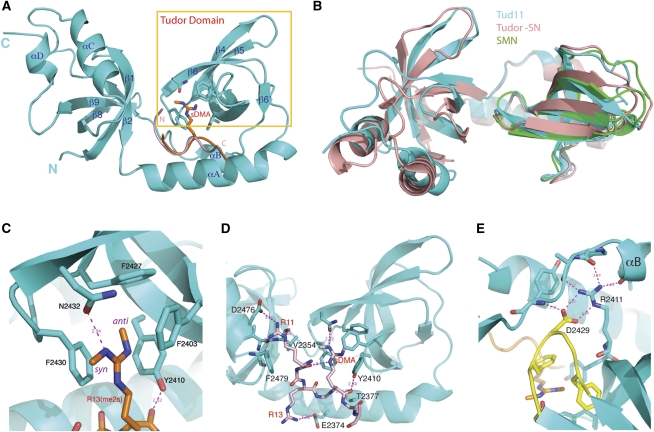
eTud11 (amino acids 2344–2515) was crystallized alone and in complex with synthetic Aub peptides (amino acids 6–18) carrying a single sDMA at residue 13 or 15 (Fig. 1A). Surprisingly, both the 1.8 Å native and the 2.8 Å complex structures show that the Tudor domain is embedded in a single α/β structure formed by N-terminal and C-terminal flanking sequences (Fig. 3A). Two N-terminal anti-parallel β strands (β1 and β2) form an oligonucleotide-binding (OB)-fold module with three β strands (β7–β9) and an α helix (αC) next to the C-terminal end of the Tudor domain. The Tudor domain is connected to the OB fold via a long helix at the N-terminal end (αA), and a short helix (αB) within a linker loop at the C-terminal end. Although previously unsuspected, the structure of eTud11 closely resembles that of the eTud domain of Tudor-SN/p100 (Fig. 3B), which is a multifunctional protein involved in transcriptional regulation and mRNA and small RNA biogenesis (Shaw et al. 2007; Friberg et al. 2009). The structures of eTud11 and Tudor-SN can be superimposed with a root-mean-square (r.m.s.) deviation of 1.84 Å, using 116 Cα atoms from both the Tudor and OB-fold domains as references. Interestingly, the eTud of Tudor-SN has also been shown to preferentially recognize sDMA peptides (Friberg et al. 2009).
Figure 3.
Structure of eTud11–Aub complexes. (A) A ribbon representation of the overall structure of eTud11 (cyan). Residues involved in binding sDMA of Aub are shown in a stick model, and the canonical Tudor domain is enclosed in a yellow box. The sDMA residue is shown in a stick model and colored brown, and the peptide backbones are shown as a coil: brown for that of R13(me2s), and light pink for that of R15(me2s). (B) Superposition of the structures of eTud11 (cyan), Tudor-SN (salmon), and the Tudor domain of SMN (green). (C) An enlarged view of the sDMA-binding pocket. Residues forming the gated aromatic cage for sDMA binding are shown in a stick model. The anti-methyl and syn methyl groups attached to the Nω and Nω′ of sDMA are indicated, and magenta dashed lines represent hydrogen bonds. (D) R15(me2s) interacts with both the Tudor and OB-fold domains. The peptide is shown in a stick model (light pink, labeled in red), and so are the eTud11 residues (labeled in black) involved in the interaction. Magenta dashed lines indicate hydrogen bonds. (E) A detailed view of the interaction network involving the conserved arginine and aspartic acid residues in the eTud domain. The β5–β6 loop is colored in yellow.
Recognition of sDMA
The Tudor domain is composed mainly of four β strands (β3–β6), and the overall fold of Tud11 is very similar to that of SMN (Fig. 3B). Four aromatic residues—Phe2403 on the β3–β4 loop, Tyr2410 on β4, and Phe2427 and Phe2430 on the β5–β6 loop—form the sDMA-binding cage (Fig. 3C). Tyr2410 forms the back wall, and Phe2427 ceils the cage. Interestingly, a highly conserved asparagine, Asn2432, is located at the entry of the aromatic cage, forming the outer edge of the ceiling. The complex structures with the Aub R13(me2s) and R15(me2s) peptides (Aub peptides with symmetrically dimethylated Arg13 and Arg15, respectively) contain six and seven ordered amino acids (amino acids 10–15 and 11–17), respectively, and in both structures the sDMA residues are placed unambiguously (Supplemental Fig. S4). The two structures show an almost identical sDMA-binding mode; hence, the R13(me2s) structure will be used for description hereafter. The side chain of R13(me2s) enters the aromatic cage from the bottom (Fig. 3C). The methyl groups attached to the ω and ω′ amino groups of R13(me2s) have different conformations. One methyl group is on the opposite side of the guanidino carbon–Nω bond (anti-conformation), while the other is on the same side (syn conformation) (Fig. 3C). The plane of the R13(me2s) guanidino group is approximately parallel to the phenyl ring of Phe2430, and lies half way between Phe2403 and Phe2430. The Nω methyl group in the anti-conformation faces to the inside of the cage, and is 3.4–4.0 Å away from all four aromatic residues making up the cage. The positively charged ω amino group is engaged in cation–π interactions with all four aromatic residues, at distances of ∼3.6 Å, 3.7 Å, 3.8 Å, and 5.2 Å from the center of the benzene rings of Phe2403, Tyr2410, Phe2430, and Phe2427, respectively. The Nω′ methyl group in syn conformation points away from the aromatic cage, while it still enjoys hydrophobic interactions with Phe2430 and Phe2403 at a distance of 3.6–4.6 Å away. The Nω′ amino group makes a hydrogen bond with the hydroxyl group of Asn2432, in addition to being in a position for cation–π interactions with Phe2430 and Phe2403. The sole hydrogen bond involving the side chain of R13(me2s) appears to be crucial for sDMA recognition. The wild-type eTud11 protein binds to the R13(me2s) Aub peptide with a dissociation constant (KD) of 48 μM, as determined by isothermal titration calorimetry (ITC), while substituting Asn2432 with an alanine (N2432A) resulted in a KD of 1.6 mM—a >30-fold reduction of binding affinity compared with the wild-type protein (Supplemental Fig. S5; Supplemental Table S2). Thus, Asn2432 is an integral part of a gated aromatic cage required for sDMA binding.
Sensing the sDMA position by the OB-fold domain
Tud7–11 and eTud11 bind to the same Aub peptide methylated at different arginines with apparently different affinities (Fig. 1B; Supplemental Fig. S3). Compared with the 48 μM KD of R13(me2s), Aub peptides with R11(me2s) and R15(me2s) bind to eTud11 with KDs of 71 μM and 6 μM, respectively (Supplemental Fig. S5; Supplemental Table S2). The above results are in reasonable agreement with the values of 37 μM and 14 μM for R13(me2s) and R15(me2s), respectively, derived from SPR measurements (Supplemental Fig. S6). An examination of the complex structure with R13(me2s) and R15(me2s) peptides reveals a common binding mode involving amino acids occupying positions within two residues (−2 to +2 positions) from the sDMA site (Figs. 1A, 3D): First, the hydroxyl group of Tyr2410 of Tud makes a hydrogen bond with the carbonyl of sDMA in both structures. Second, the peptide backbone is bound in a shallow channel formed along αA. The peptide backbone packs against αA mainly via van der Waals interactions, with a hydrogen bond between the hydroxyl group of Thr2377 and the carbonyl of a glycine at the −1 position. Third, the arginine residue at the −2 positions, which is Arg11 in the R13(me2s) complex and Arg13 in the R15(me2s) complex, points away from the protein and makes a hydrogen bond with Glu2374 (Fig. 3D). It is expected that most sDMA sites of Piwi proteins will bind Tudor domains in a similar manner, as they occur in repetitive RG/RA motifs as that in Aub (Chen et al. 2009; Vagin et al. 2009).
Regions further away from the sDMA residue are responsible for different binding affinities of Aub peptides methylated at R13 or R15. The principal difference occurs at the position four residues N-terminal (−4 position) to the sDMA site. In the R13(me2s) structure, the −4 residue is an isoleucine, which is disordered in the structure. The corresponding residue in the R15(me2s) structure is Arg11, which is in an extended conformation and binds in a shallow cleft between β1 and αC in the OB-fold domain (Fig. 3D). The Nε atom of Arg11 makes a hydrogen bond with the carbonyl of Val2354 of β1, and the amino groups contact the hydroxyl group of Asp2476 and the carbonyl of Val2353 also by hydrogen bonding. The aliphatic part of Arg11 makes van de Waals contacts with Phe2479 on αC and Val2354. Thus, the structure identifies Arg11 as one major determinant responsible for the differential binding affinities of the R13(me2s) and R15(me2s) peptides, and the OB-fold domain of eTud11 distinguishes the methylation sites of Aub.
Conservation of Tudor-SN fold among Piwi-binding Tudor domains
It is unexpected that the structure of the eTud11 domain resembles that of Tudor-SN. We wondered if the Tudor-SN fold is prevalent among Tudor domain proteins, especially those that bind to Piwi proteins. The only other Piwi-binding Tudor domain structure known is that of Tdrkh (Chen et al. 2009). Interestingly, it also contains two helices corresponding to αA and αC, in addition to the canonical Tudor domain. However, the protein used for crystallization was truncated immediately before αA and after αC; thus, whether an OB fold domain is present next to the Tdrkh Tudor domain is not known structurally. Our analyses also implicated the involvement of Tud7–11 in Aub binding (Fig. 2; Supplemental Fig. S1), and secondary structure analyses using the PredictProtein server (Rost et al. 2004) showed that each of the five Tudor domains are flanked by the same pattern of secondary structure element distribution (data not shown), suggesting that they all have an overall fold similar to that of eTud11. Furthermore, the above domains all have absolutely conserved arginine and aspartic acid residues corresponding to Arg2411 and Asp2429 of eTud11, respectively (Fig. 4). Asp2429 is located in the β5–β6 loop, where three of the five sDMA-binding residues reside. In both the eTud11 and Tdrkh structures, the two oppositely charged residues contact each other via charge and hydrogen bond interactions (Fig. 3E). The charged pair also makes one hydrogen bond with a residue at the C-terminal end of αB, and three with the main chain groups of amino acids in the loop between αB and the OB-fold domain, thus stabilizing the conformation of this region. Interestingly, it was observed previously that mutations of the corresponding arginine in Tud10 impaired the function of Tud (Arkov et al. 2006), suggesting that the interaction network involving this residue is important for the integrity of the eTud fold. In contrast, the canonical Tudor domain of SMN lacks both residues (Fig. 4), and they are also absent in methyl-lysine-binding Tudor domains (Huang et al. 2006). Additionally, two highly conserved residues, Tyr2363 and Gln2365, interact with the β5–β6 loop via hydrogen bond and van der Waals interactions, which stabilizes the sDMA-binding loop. These findings strongly suggest that the YR-DF/YGN combination is a signature motif for Piwi-binding eTud domains.
Figure 4.
Structure-guided sequence alignment. Conserved residues are indicated in red, and the residues of the canonical Tudor domain are highlighted in yellow. A red asterisk indicates the position of Tud11 residues forming the binding pocket for sDMA, a green triangle indicates the position of residues conserved in eTud domains, and the blue arrows indicate aromatic residues mutated in the Tudor transgenes analyzed in Supplemental Figure S1. At the top of the sequences, a schematic representation of the secondary structure elements of eTud11 is shown, and every 10 residues are indicated with a plus sign (+). Numbers to the left and right of the sequence indicate the numbering of the end residues in the context of the full-length proteins.
We have evidence that Tud10 indeed has a Tudor-SN fold (H Liu and RM Xu, unpubl.), and we expect the rest of the Tud7–11 modules to follow suit. From sequence alignment, it appears that only Tud9 and Tud10 lack an intact four-aromatic-residue cage; both Tud9 and Tud10 have a leucine at the corresponding position of Phe2427. Additionally, Tud10 has a serine at the position of Phe2403 (Fig. 4). It is not yet tested if these alterations would disable sDMA binding. Nevertheless, it is clear that multiple sDMA-binding cages are present in Tud. Our initial study indicates that the eTud modules of Tud7–11 form a compact structure, and multiple putative sDMA-binding pockets are exposed. Accordingly, one Tud molecule appears to be capable of binding several Aubergine molecules simultaneously. The overall binding stoichiometry and precise binding preferences of individual eTud domains await further experimental elucidation.
Materials and methods
Proteins and peptides
Tud7–11 (amino acids 1617–2515) and eTud11 (amino acids 2344–2515) fragments were chosen based on secondary structure analyses using the PredictProtein server (Rost et al. 2004). Recombinant proteins were expressed in the Rosetta strain of Escherichia coli using the pET28-Smt3 vector. Detailed procedures of protein preparation can be found in the Supplemental Material.
Aub peptides used for cocrystallization and binding assays were purchased from SciLight Biotechnology. They all have the same amino acid sequence, NPVIARGR(13)GR(15)GRK (amino acids 6–18 of Aub), but differ in arginine methylation and addition of an N-terminal biotin group. Aub, Aub[R11(me2s)], Aub[R13(me2s)] and Aub[R15(me2s)] stand for Aub peptides with unmethylated arginines, symmetrically dimethylated Arg11, Arg13, and Arg15, respectively. N-terminal Biotin-labeled peptides were used for SPR binding assays. Btn-Aub, Btn-Aub[R11(me2s)], Btn-Aub[R13(me2s)], and Btn-Aub[R15(me2s)] correspond to the aforementioned peptides, but with an extra N-terminal biotin group.
SPR experiments were carried out in a manner similar to that in Huang et al. (2006), while a more detailed description can be found in the Supplemental Material. A detailed description of ITC experiments and results can also be found in the Supplemental Material.
Crystallization and structure determination
Native crystals of eTud11 were grown by the hanging drop vapor diffusion method at 16°C in a solution containing 1.2 M sodium citrate. The eTud11 complexes with Aub[R13(me2s)] and Aub[R15(me2s)] were prepared by mixing the protein and the peptide at a 1:10 molar ratio, and the cocrystals grew at 16°C in a solution containing 30% PEG8000, 0.2 M sodium acetate, and 0.1 M sodium cacodylate (pH 6.0).
The 1.8 Å native data set was collected at the X29 beamline of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory (BNL). The SeMet MAD data sets were collected at the X12C beamline of the NSLS. Diffraction data for the eTud11–Aub peptide complexes were collected at the 17U beamline of Shanghai Synchrotron Radiation Facility (SSRF). All data were processed using HKL2000 software (Otwinowski and Minor 1997). The native crystal belongs to the C2 space group, and has two protein molecules per asymmetric unit. The structure was solved by the SeMet MAD method using the PHENIX program suite (Adams et al. 2010). The cocrystals with the Aub peptides belong to the P43212 space group, and they all have one protein–peptide complex per asymmetric unit. The complex structures were solved by molecular replacement using the Phaser program (McCoy et al. 2007), using the native structure as the search model. Coot (Emsley and Cowtan 2004) was used for model building, refinement was carried out using CNS (Brunger et al. 1998), and figures were prepared using Pymol (http://www.pymol.org). Detailed statistics of the crystallographic analyses are shown in Supplemental Table 1. The Protein Data Bank (PDB) codes for the native, R13(me2s), and R15(me2s) complex structures are 3NTK, 3NTH, and 3NTI, respectively.
P-element transformation and site-directed mutagenesis
tudor cDNA from the 4849 nucleotide (nt) to 7545 nt was cloned into the pBluescript II vector. A translation start codon was generated preceding the aspartic acid (amino acids 1617). Coding region was amplified by PCR and cloned into the intermediate vector, pCasper-nos, to adopt a nanos promoter, 5′ untranslated region (UTR), and one HA tag (Wang and Lehmann 1991). Fragments then were subcloned into the destination vector, pattB-K10 3′UTR, and targeted to the P2 locus for insertion through the phiC31 integrase system (Genetic Services, Inc.) (Groth et al. 2004). Tud(D7–11) wild-type and mutant transgenes were crossed into tudor mutant background [tud1/Df(2R)PurP133] to test for phenotype (also see the Supplemental Material).
Whole-mount antibody staining of ovaries
Ovaries were fixed and stained following published procedures (Navarro et al. 2004). Primary antibodies were as follows: 1:1000 rabbit anti-Aub (gift from G. Hannon, Cold Spring Harbor Laboratory), 1:200 mouse anti-HA (Covance), 1:25 mouse anti-FasIII (Developmental Studies Hybridoma Bank). Secondary antibodies were as follows: Cy3 conjugated (Jackson Immunoresearch) and Alexa 488 (Molecular Probes) at a dilution of 1:500 for 2 h at room temperature; DAPI in a dilution of 1:10,000 in PBST for 10 min, and mounted in VectaShield mounting medium (Vector Laboratories). Images were analyzed using a Zeiss 510 LSM confocal microscope.
Acknowledgments
We thank Yuanyuan Chen for assistance with ITC and SPR experiments, NSLS and SSRF beamline scientists for technical support during data collection, and Joy Fleming for comments on the manuscript. This work was supported by grants from the Ministry of Science and Technology (2009CB825501 and 2006CB910903 to R.M.X. and W.G.), the Natural Science Foundation of China (90919029 and 3098801 to R.M.X.), Chinese Academy of Sciences (CAS), and the NIH (GM063716). R.M.X. holds a CAS-Novo Nordisk Great Wall Professorship, and R.L. is an investigator of the Howard Hughes Medical Institute. R.M.X. is an adjunct professor at the Skirball Institute of Biomolecular Medicine, New York University School of Medicine.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1956010.
Supplemental material is available at http://www.genesdev.org.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ 2008. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol 73: 283–290 [DOI] [PubMed] [Google Scholar]
- Arkov AL, Wang JY, Ramos A, Lehmann R 2006. The role of Tudor domains in germline development and polar granule architecture. Development 133: 4053–4062 [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7: 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, et al. 2009. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci 106: 20336–20341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Friberg A, Corsini L, Mourao A, Sattler M 2009. Structure and ligand binding of the extended Tudor domain of D. melanogaster Tudor-SN. J Mol Biol 387: 921–934 [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G 2001. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell 7: 1111–1117 [DOI] [PubMed] [Google Scholar]
- Golumbeski GS, Bardsley A, Tax F, Boswell RE 1991. tudor, a posterior-group gene of Drosophila melanogaster, encodes a novel protein and an mRNA localized during mid-oogenesis. Genes Dev 5: 2060–2070 [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM 2006. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312: 748–751 [DOI] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT 2006. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z 2010. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 16: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Yin H 2008. A novel epigenetic mechanism in Drosophila somatic cells mediated by Piwi and piRNAs. Cold Spring Harb Symp Quant Biol 73: 273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R 2004. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol 6: 427–435 [DOI] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al. 2009. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 28: 3820–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Ponting CP 1997. Tudor domains in proteins that interact with RNA. Trends Biochem Sci 22: 51–52 [DOI] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J 2004. The PredictProtein server. Nucleic Acids Res 32: W321–W326 doi: 10.1093/nar/gkh377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw N, Zhao M, Cheng C, Xu H, Saarikettu J, Li Y, Da Y, Yao Z, Silvennoinen O, Yang J, et al. 2007. The multifunctional human p100 protein ‘hooks’ methylated ligands. Nat Struct Mol Biol 14: 779–784 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Mannen T, Siomi H 2010. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev 24: 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M 2003. High-resolution X-ray and NMR structures of the SMN Tudor domain: Conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol 327: 507–520 [DOI] [PubMed] [Google Scholar]
- Talbot K, Miguel-Aliaga I, Mohaghegh P, Ponting CP, Davies KE 1998. Characterization of a gene encoding survival motor neuron (SMN)-related protein, a constituent of the spliceosome complex. Hum Mol Genet 7: 2149–2156 [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P 2005. Tudor and its domains: Germ cell formation from a Tudor perspective. Cell Res 15: 281–291 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA 2009. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R 2009. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol 19: 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lehmann R 1991. Nanos is the localized posterior determinant in Drosophila. Cell 66: 637–647 [DOI] [PubMed] [Google Scholar]
- Wang J, Saxe JP, Tanaka T, Chuma S, Lin H 2009. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol 19: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]