Abstract
The enzyme homocitrate synthase (HCS) catalyzes the first step in lysine biosynthesis, and early biochemical data placed it in the cytoplasm or mitochondria, where most amino acid synthesis occurs. It was therefore surprising when refined fractionation techniques and specific immunoreagents clearly demonstrated its localization to the nucleus. These observations raised the question of whether HCS had a function within the nucleus independent of lysine synthesis. We demonstrate that HCS encoded by LYS20 in yeast is linked to the key process of DNA damage repair through the essential MYST family histone acetyltransferase Esa1 and the H2A.Z histone variant. This discovery indicates that HCS has a role in addition to amino acid synthesis, and that it functions in nuclear activities involving chromatin regulation that are distinct from its previously established role in lysine biosynthesis. The chromatin-linked roles are dependent on nuclear localization of Lys20, but are independent of HCS catalytic activity. Thus, Lys20 appears to have evolved as a bifunctional protein that connects cellular metabolism with chromatin functions.
Keywords: Lysine, histone, DNA damage, yeast, Htz1, bifunctional protein
Histone acetyltransferases (HATs) modulate chromatin functions by acetylating lysines on histones, transcription factors, and other substrates. Among HATs, the MYST family is highly conserved, and notably includes multiple essential enzymes in organisms ranging from yeast to humans (for review, see Lafon et al. 2007). One well-studied MYST enzyme in yeast, Esa1 (Smith et al. 1998; Clarke et al. 1999), acetylates a specific subset of lysine residues on the four core histones, along with the H2A.Z histone variant encoded by HTZ1 (Babiarz et al. 2006; Keogh et al. 2006; Millar et al. 2006). Esa1 is the catalytic subunit of the yeast NuA4 and piccolo complexes (Allard et al. 1999; Boudreault et al. 2003; Lafon et al. 2007), and has functional interactions with many other genes that encode chromatin-modifying enzymes (Kobor et al. 2004; Krogan et al. 2004; Lin et al. 2008). In addition, Esa1 has been implicated in diverse chromatin-mediated processes, including DNA damage sensing and repair, transcriptional silencing, and cell cycle control (for review, see Doyon and Côté 2004; Lafon et al. 2007), although not all of these functions may require its catalytic activity (Decker et al. 2008). Mutations in ESA1 cause sensitivity to DNA double-stranded breaks induced by the topoisomerase I inhibitor camptothecin (Bird et al. 2002). Mutation of the histone H4 lysine residues targeted by Esa1 likewise results in camptothecin sensitivity (CPTs). These observations have been interpreted to mean that lysine acetylation by Esa1 is required for resistance to camptothecin. Much remains to be learned about Esa1's role in DNA repair and other nuclear processes, and its acetylation of H2A.Z is one recent area of focus.
Both Esa1 (Babiarz et al. 2006; Keogh et al. 2006; Millar et al. 2006) and the key transcriptional HAT Gcn5 (Babiarz et al. 2006) target H2A.Z as a substrate for acetylation. They also share other mutant phenotypes, such as sensitivity to DNA damage (Choy and Kron 2002). H2A.Z has been implicated in boundary formation in silent chromatin, and is also found dispersed throughout the genome (Meneghini et al. 2003; Shia et al. 2006; Raisner and Madhani 2008). Like esa1 conditional mutants, null mutants of HTZ1 are sensitive to DNA-damaging agents (Kobor et al. 2004; Krogan et al. 2004), implying a role for H2A.Z in resistance to genotoxins. A mechanism for this resistance is not yet established (Kalocsay et al. 2009).
A dosage suppressor screen initially identified LYS20 as a weak suppressor of the esa1 mutant temperature sensitivity. Lys20 and the closely related Lys21 isozyme have been studied extensively for their roles in lysine biosynthesis (for review, see Xu et al. 2006). Individually, null mutants of LYS20 and LYS21 are prototrophic for lysine; only the double-null mutant requires lysine for growth. The enzymes catalyze the first and rate-limiting step in lysine biosynthesis by combining an acetyl group from acetyl CoA with α-ketoglutarate, an intermediate in the Krebs cycle, to make homocitrate. Both enzymes are feedback-inhibited by lysine and catalyze the same reaction, although with different kinetics and sensitivity to cell metabolism (for review, see Xu et al. 2006; Quezada et al. 2008).
Biochemical fractionation monitored by homocitrate synthase (HCS)-specific antibodies and immunofluorescence microscopy place both Lys20 and Lys21 predominantly within the nucleus in a chromatin-bound, not freely diffusible form (Chen et al. 1997). This is an unusual localization, as the other enzymes in the lysine biosynthetic pathway are located in either the cytoplasm or the mitochondria, as HCS itself had been reported in earlier studies (for review, see Jones and Fink 1982).
The studies presented here define a new role for Lys20 in chromatin function that provides a rationale for the nuclear location of HCS. Overexpression of LYS20 suppressed the DNA damage sensitivity of esa1 strains. The suppression was completely dependent on HTZ1. Furthermore, deletion of LYS20 and LYS21 suppressed the DNA damage sensitivity of htz1Δ, but only if Esa1 was functional. These effects appear mediated through the DNA damage checkpoint, which may be sensitized by the LYS20 LYS21 double deletions, as reflected by levels of Rad53 phosphorylation. In vitro assays revealed that Lys20 has weak HAT activity directed toward H4. Furthermore, Lys20 is associated in vivo with the HAT Gcn5. Importantly, Lys20's contributions to DNA repair are dependent on its nuclear localization, yet independent of its catalytic activity. Thus, Lys20 has dual metabolic and nuclear roles that further connect Esa1 and H2A.Z through histone acetylation and DNA damage.
Results
LYS20 overexpression suppresses esa1 DNA damage phenotypes
LYS20 was identified as a dosage suppressor of the temperature sensitivity of a catalytically compromised esa1-414 allele of the essential Esa1 HAT (C Chang, A Clarke, and L Pillus, in prep.). When evaluated for effects on the DNA damage sensitivity of the same strain, increased gene dosage of LYS20 proved to be a strong suppressor, as indicated by restored growth on camptothecin (Fig. 1A). Suppression of esa1 mutant phenotypes was also observed upon overexpression of LYS21 (data not shown), yet was not as robust as that observed with LYS20. We therefore focused analysis on LYS20 as the stronger suppressor and the one more likely to provide unambiguous data.
Figure 1.
LYS20 suppresses the CPTs of esa1 mutants. This suppression is specific for esa1, and is dependent on the histone variant H2A.Z. (A) LYS20 suppresses the CPTs of multiple alleles of esa1. Wild-type cells, esa1-414 mutants, and esa1-K256Q, Y325N mutants were transformed with empty vector or the LYS20 2μm plasmid. LYS20 is transcribed by its endogenous promoter, but is expressed at elevated levels due to increased copy number of the plasmid. The camptothecin plate contains 20 μg/mL camptothecin in DMSO. (B) Overexpression of LYS20 does not suppress the CPTs of a gcn5Δ mutant. Wild-type (WT) cells were transformed with either empty vector or overexpression plasmid containing LYS20. The gcn5Δ cells were transformed with either empty vector, LYS20 overexpression plasmid, or GCN5 overexpression plasmid. Plates were synthetic complete (SC) medium lacking uracil and were incubated at 30°C. The camptothecin plate contains 20 μg/mL camptothecin in DMSO. Assays were performed as in A. (C) LYS20 suppression of the CPTs of esa1 mutants is dependent on HTZ1. The esa1 htz1Δ double mutant was transformed with either empty vector or LYS20 overexpression plasmid. All plates are synthetic complete medium lacking uracil. The camptothecin plate contains 30 μg/mL of camptothecin in DMSO.
Of note, simply overexpressing LYS20 had no effect on either growth or CPTs of wild-type cells. Furthermore, the effect on ESA1 mutants was not allele-specific, as similar suppression was observed with the independent esa1-K256Q,Y325N allele, which is also CPTs (Fig. 1A). The observed effect is not due to differences in LYS20 expression, as Lys20 protein levels do not change in the esa1 mutant (Supplemental Fig. S1).
Because multiple chromatin modifications contribute to DNA damage repair, and loss of the HAT encoded by GCN5 also results in CPTs (Choy and Kron 2002), specificity of LYS20-mediated suppression was evaluated. In gcn5Δ mutants, resistance to camptothecin was restored by transformation with GCN5, yet not upon LYS20 overexpression (Fig. 1B). Thus, LYS20-mediated suppression of loss of HAT activity in response to DNA damage appeared specific for the essential Esa1 HAT, and is not a general means of restoring defective repair functions.
A key role for the H2A.Z histone variant in esa1 mutants
Because Esa1 acetylates the H2A variant H2A.Z (Babiarz et al. 2006; Keogh et al. 2006; Millar et al. 2006), and both esa1 and htz1Δ mutants are sensitive to camptothecin, the esa1 htz1Δ double mutant was evaluated. This strain was even more CPTs than either individual mutant (Fig. 1C), suggesting that the two genes contribute independently to repair of DNA damage. Supporting this interpretation, CPTs of the esa1 htz1Δ mutants could not be suppressed by LYS20 overexpression (Fig. 1C). Thus, LYS20-mediated suppression of esa1 DNA repair defects depends on HTZ1.
In contrast to the protective effect conferred by LYS20 overexpression in esa1 mutants, it was possible that deletion of either HCS-encoding gene in otherwise wild-type cells might result in sensitivity to camptothecin. However, no sensitivity was observed upon deletion of LYS20 or LYS21; instead, deletion of either gene provided resistance to damage induced by camptothecin (Fig. 2A). The lys20Δ lys21Δ mutant was comparably resistant. The esa1 lys20Δ lys21Δ triple mutant was sensitive, demonstrating that esa1's defect was epistatic to the resistance afforded by loss of HCS. Next, the htz1Δ lys20Δ lys21Δ mutant was evaluated. In this case, deletion of LYS20 and LYS21 suppressed the CPTs of htz1Δ (Fig. 2B), in contrast to the esa1 result. These observations suggest that HCS might be involved in two pathways in response to DNA damage that are distinguished by functional interaction with the variant histone H2A.Z and the Esa1 HAT.
Figure 2.
Deletion of LYS20 and LYS21 suppresses htz1Δ DNA damage sensitivity. (A) Resistance to camptothecin is dependent on ESA1. Strains were assayed as in Figure 1. The YPD-based camptothecin (CPT) plate contains 40 μg/mL camptothecin in DMSO. (B) Deletion of LYS20 and LYS21 suppresses htz1Δ DNA damage sensitivity. All plates were incubated at 33°C. All plates are YPD-based. The camptothecin (CPT) plates contain 20 μg/mL camptothecin in DMSO, and were incubated 5 d longer than the growth control. (C) Suppression is dependent on ESA1. Independent strain isolates were assayed as in Figure 1. Plates were incubated at 30°C. All plates are YPD-based. The camptothecin (CPT) plate contains 20 μg/mL camptothecin in DMSO. Vertical bars indicate platings of two independent triple mutants. (D) Suppression is evident with HU-induced damage. Strains were assayed as before on YPD or synthetic complete as growth control, or on 0.1 M HU. The HU plate was incubated an additional 3 d in the top panel, and an additional 5 d in the bottom panel. Plates were incubated at 30°C. Vertical bars indicate platings of two independent triple mutants.
As shown in Figure 1C, the suppression of esa1 CPTs by overexpression of LYS20 is dependent on HTZ1. This raises the question of whether the suppression of htz1Δ CPTs is reciprocally dependent on ESA1 function. To address this question, the quadruple esa1 htz1Δ lys20Δ lys21Δ mutant was generated and tested for DNA damage sensitivity (Fig. 2C). Because of the extreme hypersensitivity of htz1Δ esa1 cells, lower concentrations of drug were used than in earlier assays. As seen in the bottom row of Figure 2C, the quadruple mutant remained as sensitive as the esa1 htz1Δ double mutant, thus demonstrating that Esa1 activity is required for the rescue of htz1Δ mutants by deletion of LYS20 and LYS21 upon DNA damage caused by inhibition of topoisomerase I.
DNA damage can be induced in many ways. In contrast to camptothecin, hydroxyurea (HU) inactivates ribonucleotide reductase, ultimately leading to accumulation of double-stranded breaks near DNA replication forks (for review, see Saban and Bujak 2009). To evaluate the specificity of damage sensitivity and resistance, analysis was performed with HU as the drug challenge (Fig. 2D). As for camptothecin, the lys20Δ lys21Δ double mutant was more resistant than wild-type cells. Similar to our observations with camptothecin, and, as reported previously, htz1Δ mutants are sensitive to HU (Kobor et al. 2004). This sensitivity is suppressed by elimination of LYS20 and LYS21, and, in parallel to Figure 2C, the extreme sensitivity of the esa1 htz1Δ mutant cannot be suppressed by the elimination of LYS20 and LYS21.
It remained a possibility that the suppressive effect of deleting LYS20 and LYS21, thereby preventing all HCS activity, was due to lysine auxotrophy. To evaluate this possibility, a lys2Δ htz1Δ strain that requires lysine for growth due to loss of an independent step in the biosynthetic pathway was examined. In this case, suppression of DNA damage sensitivity was not observed (Supplemental Fig. S2). Thus, the restored growth in the presence of at least two DNA-damaging agents imparted by deletion of LYS20 or LYS21 is independent of the known synthetic function of these genes.
A molecular link between LYS20 and the DNA damage checkpoint response
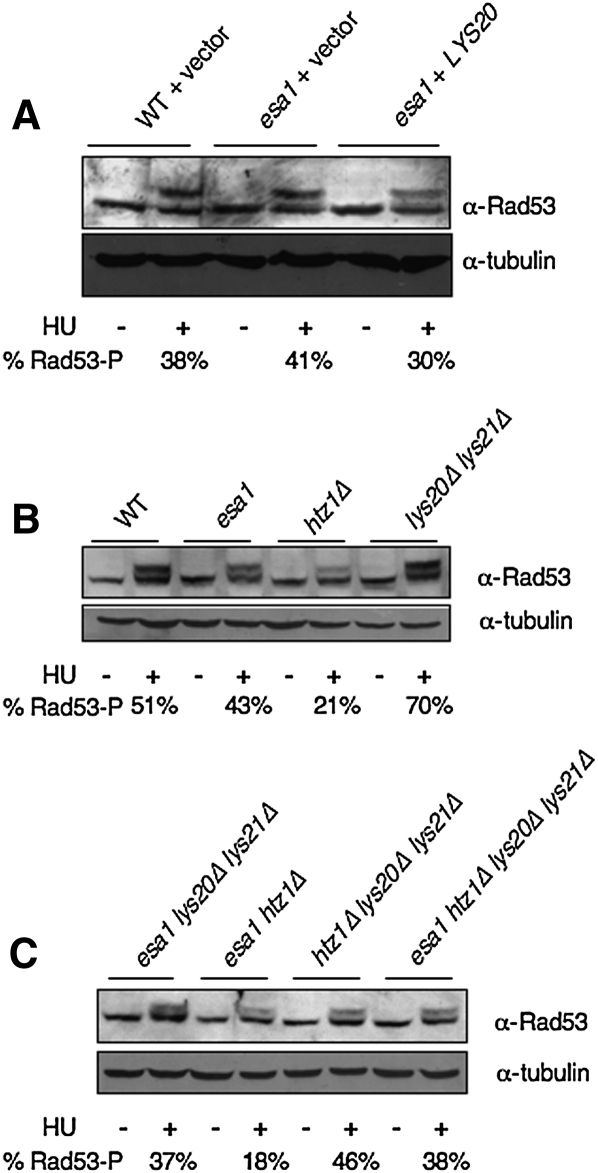
DNA damage repair can fail at multiple steps (sensing of damage or checkpoint activation), or by defects in the repair process itself. There are molecular hallmarks for many of these steps. For example, phosphorylation of Rad53 is a key indicator of DNA damage checkpoint activation. This is readily monitored by immunoblotting for Rad53, which is hyperphosphorylated upon checkpoint activation, and thereby migrates with decreased electrophoretic mobility (Sanchez et al. 1996).
A Rad53 phosphorylation shift is visible in wild-type cells upon incubation with HU (Fig. 3A). Normal Rad53 phosphorylation is also seen in esa1 mutants, and phosphorylation does not change significantly upon overexpression of LYS20. This indicates that esa1 mutants activate the Rad53 checkpoint normally, so their CPTs and rescue by LYS20 must result from a different mechanism. By comparison, lys20Δ lys21Δ double mutants have increased Rad53 phosphorylation upon induction of damage by HU (Fig. 3B). If this indicates improved activation or hyperactivation of the checkpoint, it could explain the protective effects seen in lys20Δ lys21Δ mutants (Fig. 2A).
Figure 3.
Rad53 phosphorylation status during DNA damage and suppression. (A) The Rad53 phosphorylation shift upon HU-induced DNA damage is not significantly affected by overexpression of LYS20. Extracts were prepared for cells with the indicated genotypes. Immunoblots with anti-Rad53 antibody are shown. In samples treated with HU, the proportion of phosphorylated Rad53 was quantified, and is indicated as a percentage of total Rad53. (B) The htz1Δ mutants had reduced Rad53 phosphorylation upon DNA damage. (C) The htz1Δ mutants had reduced Rad53 phosphorylation upon DNA damage. This defect was suppressed by deletion of LYS20 and LYS21.
In contrast, HTZ1 mutants were reported recently to be defective in Rad53 phosphorylation in response to DNA damage (Kalocsay et al. 2009). In Figure 3B, this defect is observed, whereas, in the htz1Δ lys20Δ lys21Δ triple mutant, DNA damage-induced phosphorylation is restored, thereby correlating a molecular marker of DNA repair with suppression of DNA damage (Fig. 3C).
Since esa1 mutants have no defect in Rad53 phosphorylation, it was noteworthy that esa1 htz1Δ mutants had reduced Rad53 phosphorylation when damage was induced (Fig. 3C). This defect was partially suppressed by deletion of LYS20 and LYS21. In the quadruple mutant, deletion of LYS20 and LYS21 can still restore Rad53 phosphorylation to htz1Δ cells. Therefore, mutation of esa1 must confer sensitivity to DNA damage by a Rad53 phosphorylation-independent mechanism, and suppression by overexpression of LYS20 does not proceed via a Rad53-dependent mechanism. Since suppression of htz1Δ damage is correlated with Rad53 phosphorylation, yet suppression of esa1 damage is not, it appears that HCS functions in DNA damage by more than one mechanism.
Defining catalytic and chromatin functions of Lys20
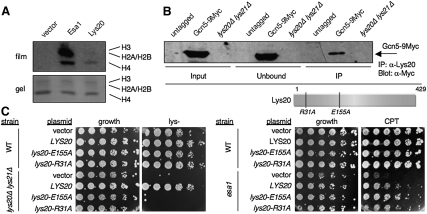
Given Lys20's nuclear localization on chromatin, its newly discovered role in DNA damage reported here, and its acetyl CoA binding for catalysis as part of the HCS reaction, we considered the possibility that the enzyme might function as a noncanonical HAT. To test this hypothesis, in vitro HAT assays were performed with recombinant Lys20 protein, using Esa1 as the positive control. In these assays, enzymes were incubated with histone substrates and [3H] acetyl CoA. Assays using recombinant core histones as substrate yielded activity for Esa1 but not for Lys20 (data not shown). Assays performed with commercially prepared calf thymus histones were also negative. However, when histones purified from yeast were used as substrate, HAT activity was observed for both Esa1 and Lys20. Lys20 activity was directed toward histone H4, and was stimulated by the addition of lysine (Fig. 4A).
Figure 4.
Toward a nuclear function for Lys20. (A) Lys20 has HAT activity in vitro. Recombinant Lys20, Esa1, or empty vector extract was added to purified yeast histones and incubated with [3H]-acetyl-CoA and 5 mM lysine. The reaction was analyzed by 18% SDS-PAGE and by autoradiography. “Vector” indicates samples prepared from cells transformed with the empty vector. Lysine provided modest stimulation of Lys20 activity. (B) Lys20 interacts with Gcn5-9Myc. Immunoprecipitations of whole-cell extracts from the indicated strain were performed with anti-HCS antibody. The precipitated material was blotted to detect Gcn5-9Myc. The lys20Δ lys21Δ strain is not Myc-tagged. (C) HCS catalytic activity is not required for nuclear functions of Lys20. Mutant alleles that are defective in lysine biosynthesis remain competent for suppression of DNA damage. The camptothecin (CPT) plate was incubated 3 d longer than the control plate.
These characteristics of Lys20 suggest it may act as a bifunctional protein. The requirement for yeast histones as substrate implies that Lys20 may require some pre-existing modification(s) on the histones to be active as a HAT. Esa1 HAT activity in these assays is robust, presumably due to the fact that Esa1 is a global HAT, acetylating multiple lysine residues spread over core and variant histones. Lys20, in contrast, may have more restricted activity, and may target many fewer residues or only one residue. Activity in the assay appears directed primarily toward histone H4, but this may be in the context of H2A.Z-containing nucleosomes, since suppression studies demonstrated that Lys20's activity in vivo was dependent on this histone variant (Fig. 1C). The absolute requirement for histones purified from yeast as substrate has thus far prevented definition of substrate site specificity. However, we tested several Esa1 H4 target residues under suppressing conditions by immunoblotting with isoform-specific antibodies, but no definitive target for Lys20 has yet been uncovered (data not shown).
Because Lys20 had been reported previously to be chromatin-bound (Chen et al. 1997), we considered that it might be associated with nuclear components that participate in processes under study here. To test this possibility, Lys20 was assayed for coimmunoprecipitation with known chromatin-associated proteins. Under the conditions tested with epitope-tagged proteins or protein-specific antisera, there was no evidence for association with histones H3, H4, Htz1, or Esa1 (data not shown). However, Lys20 and the HAT Gcn5 did coprecipitate (Fig. 4B). The association is specific, and is not due to the epitope tag on Gcn5. Lys20 was precipitated efficiently by the anti-Lys20 antibody in both tagged and untagged strains, and no signal was observed in the untagged lys20Δ lys21Δ negative control.
These data reveal that not only is Lys20 in the nucleus, a fact that was known previously, but it is also associated with the HAT Gcn5. Because we found that LYS20 and gcn5Δ did not interact functionally, as LYS20 did with esa1 (Fig. 1B), this physical interaction might be unexpected. However, it has been argued (Collins et al. 2007) that molecules interacting physically often do not do so genetically, thereby demonstrating that functional and physical interaction networks can be distinct. Given the Lys20–Gcn5 physical interaction, we tested whether recombinant Lys20 protein might potentiate the activity of Gcn5 in vitro. Under the conditions tested, Gcn5 HAT activity was not stimulated by Lys20 addition (Supplemental Fig. S3).
In defining Lys20's nuclear role, it was important to determine if its role in lysine biosynthesis is separable from its functions in repair. To this end, point mutants were constructed to abolish the HCS activity of Lys20 that were based on the crystal structure of the Schizosaccharomyces pombe HCS (Bulfer et al. 2009). Two residues were selected (R31 and E155), both of which lie in the active site and have been shown to be essential in vitro and in vivo for catalysis. The residues were mutated to alanine and assayed for HCS activity by testing for cell growth in the absence of lysine. When the mutation-bearing plasmids were introduced into wild-type cells, there was no interference with growth, indicating that the mutants are recessive and do not interfere with the function of the wild-type protein. In the lys20Δ lys21Δ strain, there was no growth, demonstrating that the mutants were unable to sustain HCS activity (Fig. 4C). The mutants were then tested to see whether they could still suppress the CPTs of esa1. The mutants could suppress (Fig. 4C). Therefore, the catalytically inactive alleles of LYS20 demonstrate that it is possible to distinguish two functions for HCS, and that its function in biosynthesis is not required for its role in the repair of DNA damage.
Nuclear localization of Lys20 is required for its DNA repair functions
Since Lys20's role in lysine synthesis can be distinguished from its role in DNA damage repair, is it possible to link one or both of these roles to the protein's localization to the nucleus? To answer this question, two approaches were taken.
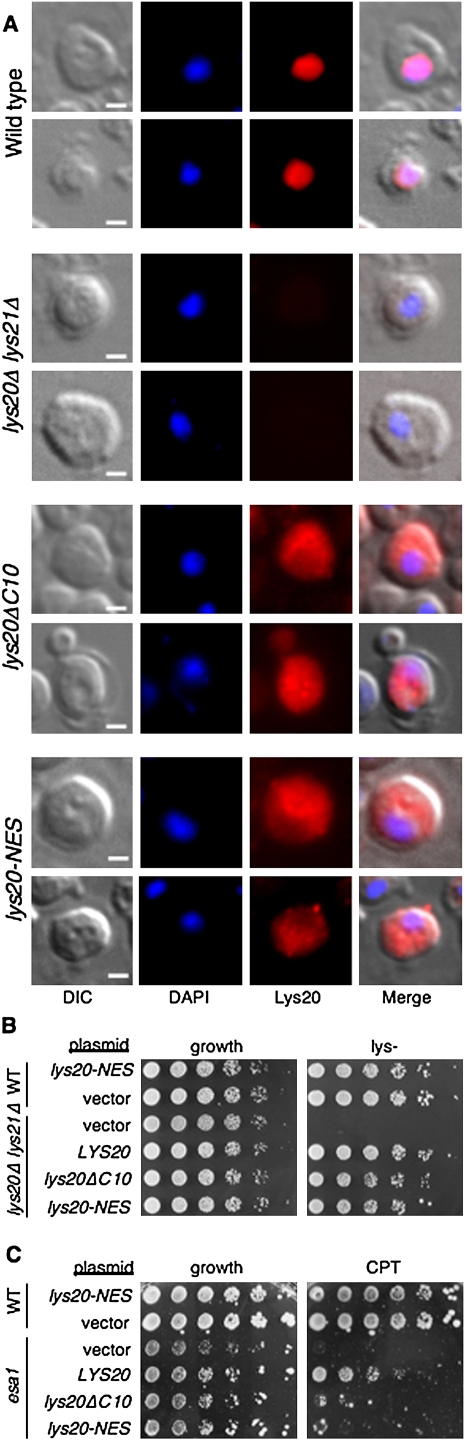
First, in an effort to understand how Lys20 is localized in vivo, we considered that, at a predicted molecular mass of 47 kDa, Lys20 is above the limit defined for free diffusion through the nuclear pore and accumulation in the nucleus (for review, see Terry et al. 2007). Uptake of proteins may be facilitated by other nuclear import molecules and by integral nuclear localization sequences (NLSs) that serve to drive the import of proteins to the nucleus after their synthesis in the cytoplasm. Several distinct classes of NLSs have been characterized, some of which are short tags that are rich in charged lysine and arginine residues (Hicks and Raikhel 1995). The PSORT algorithm (Horton and Nakai 1997) was used to search for an NLS in Lys20. A potential NLS of the Pat7 class was found beginning with the proline 10 residues from the C terminus of the protein. To test the functional significance of this potential NLS, the lys20ΔC10 construct was created that truncated the protein to remove the PAAKRTK putative localization signal and the final three amino acids of the protein.
In a second independent approach, a nuclear export sequence (NES) was added to Lys20 (lys20-NES). By analogy with NLSs, NESs function to facilitate shuttling of proteins from the nucleus to the cytoplasm (Moroianu 1999). The NES selected was a variant of the robust signal defined in PKI that can drive export of heterologous proteins (Gadal et al. 2001).
Both constructs—lys20ΔC10 and lys20-NES—were expressed in wild-type, lys20Δ lys21Δ, and esa1 cells. They were tested for localization by immunofluorescence microscopy, and for function in lysine biosynthesis and repair of DNA damage.
Using the antibody that first defined its nuclear localization (Chen et al. 1997), we found that wild-type Lys20 was nuclear, as reported, with no signal in the lys20Δ lys21Δ cells. In contrast, neither lys20-NES nor lys20-ΔC10 was restricted to the nucleus (Fig. 5A). Expression tests for both constructs assessed by immunoblotting demonstrated amounts of protein comparable with wild-type Lys20 (data not shown).
Figure 5.
Nuclear functions of Lys20 depend on nuclear localization. (A) Mutants were constructed to remove a putative NLS at the C terminus of Lys20 (lys20ΔC10), and to add a potent NES to the wild-type Lys20 protein (lys20-NES). Immunofluorescence microscopy demonstrated that the wild-type protein was enriched in the nucleus, whereas the two mutant proteins are enriched in the cytoplasm. Cells are stained with DAPI for DNA, and Lys20 is seen in red. Representative cells are shown for each strain. Bar, 1 μm. (B) The lys20-NES and lys20ΔC10 cytoplasmic proteins are competent for HCS activity, and do not interfere with normal Lys20 function when plated on lys− medium. (C) In contrast, when tested for suppression of DNA damage, lys20ΔC10 and lys20-NES do not strongly suppress the esa1 CPTs to LYS20 levels.
When assayed for HCS activity, both lys20ΔC10 and lys20-NES were fully competent to support growth in the absence of lysine (Fig. 5B). When assayed on camptothecin, esa1's sensitivity was not suppressed to wild-type levels (Fig. 5C). Thus, Lys20 metabolic activity does not depend on its nuclear localization, but efficient DNA damage suppression does require intact localization.
Discussion
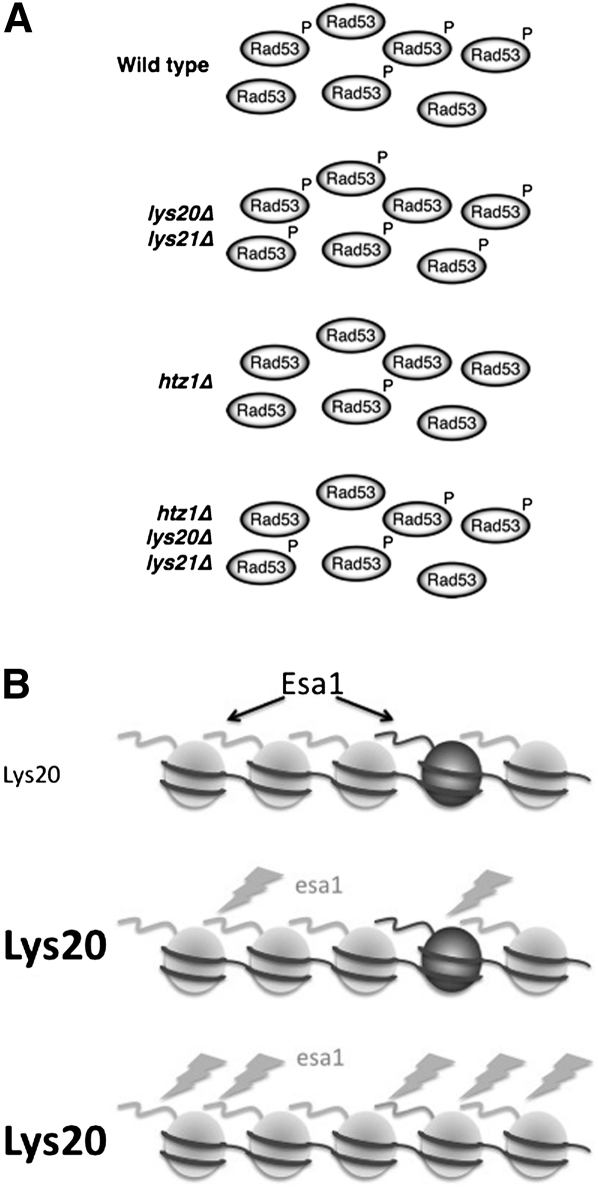
We demonstrate that LYS20, which encodes HCS, interacts functionally with the essential HAT Esa1 in vivo. The HTZ1 gene encoding the H2A variant H2A.Z mediates this interaction, and also interacts with LYS20 through a functionally distinct mechanism. All three genes are linked to the repair of DNA damage. These findings define a previously unappreciated role for Lys20 that explains the long-standing mystery of its nuclear localization. In suppressing htz1Δ CPTs, the LYS20 LYS21 deletion restores DNA damage-induced hyperphosphorylation of the Rad53 checkpoint protein (Fig. 6A). However, it appears that the step affected by esa1 DNA damage sensitivity is downstream from Rad53, because Rad53 phosphorylation levels are not perturbed in esa1 mutants and esa1 lys20Δ lys21Δ triple mutants are still sensitive to DNA damage.
Figure 6.
Roles for the bifunctional protein Lys20 in chromatin and DNA damage repair. (A) Suppression of htz1Δ CPTs by deletion of LYS20 and LYS21 may be mediated through Rad53 phosphorylation upon DNA damage. Wild-type (WT) cells respond by phosphorylating Rad53, indicated by a pool of phosphorylated protein. The lys20Δ lys21Δ cells have increased Rad53 phosphorylation relative to wild-type. In contrast, htz1Δ mutants are defective in Rad53 phosphorylation, whereas suppression of the htz1Δ mutants by lys20Δ lys21Δ is correlated with restoration of Rad53 phosphorylation. (B) Esa1 ordinarily acetylates nucleosomal H4 and H2A (light gray) and the H2A.Z variant Htz1 (dark gray). When Esa1 activity is diminished, increased Lys20 can facilitate DNA damage repair, a process dependent on Htz1.
It appears that, through H2A.Z, LYS20 is limiting for yet promotes the function of some activity that can facilitate DNA repair when Esa1's activity is diminished (Fig. 6B). Additionally, Lys20 may activate a function that Esa1 helps that contributes to the DNA damage sensitivity of htz1Δ mutants. Lys20's roles may be grounded in its potential dual catalytic roles as HCS and a noncanonical HAT. In this case, unlike the global HAT Esa1, Lys20 may catalyze acetylation only under specialized circumstances. When overexpressed in the mutants, Lys20 may act to rescue DNA damage defects by restoring acetylation to promote repair. It is also possible that acetylation may be of nonhistone substrates. Indeed, recent proteomic studies identify many acetylated proteins (Lin et al. 2009). Future work will be required to validate these potential substrates and the enzymes responsible for their modification. A separate possibility is raised by a recent report that the damage sensitivity of htz1Δ mutants is the result of the remodeling activity of the SWR complex (Halley et al. 2010). Determining how Lys20 intersects with SWR components and the damage they induce in the absence of Htz1 will be of interest.
A lysine-induced switch?
Lysine itself is a regulator of Lys20 and Lys21 through feedback inhibition (Feller et al. 1999). It has been demonstrated recently (Bulfer et al. 2010) that the mechanism used for feedback inhibition by the functionally conserved S. pombe HCS is competitive inhibition, where lysine competes with α-ketoglutarate for access to the active site, effectively shutting down the HCS reaction at high levels of lysine. It is possible that, when lysine inhibits HCS activity, it may stimulate a switch to enhance HAT activity, as observed in vitro (Fig. 4A). We note that two point mutants that abolish Lys20's sensitivity to lysine, rendering it resistant to feedback inhibition (Feller et al. 1999), still largely suppress the CPTs of esa1 (Supplemental Fig. S4). In this case, although the switch for the HCS reaction is lost, it appears that the distinct nuclear functions for Lys20 remain active.
Noncanonical HATS and metabolic enzymes in the nucleus
Studies of the global repertoire of histone acetylation by mass spectrometry indicate that many lysine residues on histones are acetylated. For many of these, the enzymes responsible for acetylation have not yet been identified. Whereas most research and mutational analysis has focused on N-terminal residues, residues in the globular domain and C-terminal tails are also acetylated (for review, see Mersfelder and Parthun 2006).
For example, H3K56, a residue that lies at the protein:DNA interface of the nucleosome surface, is acetylated and functions in cell cycle regulation and DNA damage sensitivity (Masumoto et al. 2005; Celic et al. 2006). Rtt109, a noncanonical HAT, is responsible for this modification (Driscoll et al. 2007; Han et al. 2007). For other less-well-defined modified residues, it remains an open question of whether they are additional targets of known global HATs, such as Esa1, or if they are targets of noncanonical HATs, Lys20 potentially among them.
Residues in the globular and C-terminal domains of histones are likely to be as important for cell viability as those in the N-terminal tails. Recently, 18 non-N-terminal residues were shown to be essential for cell viability (Matsubara et al. 2007; Nakanishi et al. 2008). Although none is a lysine residue, six encode residues potentially subject to post-translational modification, and may contribute to additional cross-talk and regulation by noncanonical histone-modifying enzymes.
The mammalian enzyme NCOAT may serve as a paradigm for Lys20. NCOAT was first identified as an O-GlcNAc transferase that was subsequently shown to have HAT activity (Toleman et al. 2004). Discovery of its HAT activity made it one of a small but growing number of enzymes that have a role in chromatin in addition to distinct metabolic functions.
The arginine biosynthetic enzyme Arg5 is another. It is an acetylglutamate kinase that catalyzes the second step in ornithine biosynthesis, and is a nuclear and mitochondrial transcription factor as well (Hall et al. 2004). Yet another example is the kinase Gal1 that catalyzes a key step in catabolism of galactose. Gal1 moonlights as a general activator of GAL gene transcription (for review, see Gancedo and Flores 2008).
Cellular metabolism, in the form of glycolytic activity and acetyl CoA levels, also directly influences global levels of histone acetylation, providing additional links between metabolic enzymes and modification of chromatin (Takahashi et al. 2006). Of note is that the enzymes responsible for these effects in both yeast and humans have distinct nuclear pools (Friis et al. 2009; Wellen et al. 2009). Other connections between transcription and metabolism relate specifically to Esa1. Already known to acetylate some nonhistone substrates, Esa1 was also reported to acetylate Pck1, a metabolic enzyme. This directly connects Esa1 to regulation of cellular metabolism (Lin et al. 2009). Whether Lys20 shares the ability to modify nonhistone substrates remains to be determined.
Independent studies have begun to link metabolism to DNA damage. Mutants in the gluconeogenesis enzyme Fbp1, for example, are resistant to MMS-induced DNA damage, potentially through a mechanism tied to metabolic flux (Kitanovic and Wolfl 2006). Another parallel for Lys20, in terms of DNA repair, may be the Krebs cycle enzyme fumarase, which was discovered recently to have DNA repair functions dependent on its catalytic activity (Yogev et al. 2010). Checkpoint activation is impaired in fumarase mutants, and mutants are sensitive to DNA damage, in contrast to lys20Δ. Nevertheless, this study provides further evidence linking DNA damage to a metabolic enzyme.
Lys7, now called Ccs1, is the copper chaperone for the major superoxide dismutase (SOD) enzyme in yeast that functions in detoxifying reactive oxygen species (Gamonet and Lauquin 1998). Mutants of CCS1 are very sensitive to radical-induced DNA damage, as the function of SOD is compromised. For as-yet-undiscovered reasons, ccs1Δ mutants are lysine auxotrophs. Indeed, sod1Δ mutants are also auxotrophic for both lysine and methionine. One proposed mechanism for the sod1Δ lysine auxotrophy is that one of the enzymes in the biosynthetic pathway may simply be very sensitive to the presence of reactive oxygen species (Slekar et al. 1996). However, our data argue for a tighter link between DNA damage and lysine biosynthesis: lys20Δ lys21Δ mutants are not only resistant to DNA damage, but the agents used to induce the damage are not radical generators. Instead, the resistance of lys20Δ lys21Δ mutants seems to be specific to DNA double-stand break-inducing agents such as HU and camptothecin, as resistance does not extend to UV-induced damage (data not shown).
Implications for the evolution of bifunctional proteins and chromatin regulation
Linking two cellular processes to one enzyme, as shown here for Lys20, raises evolutionary implications. The ability to bind and use a molecule common to many processes, such as acetyl CoA, confers on an enzyme the possibility to contribute to multiple pathways and simultaneously links those pathways. Like the discovery of the transcriptional activation functions of Arg5 and Gal1, and NCOAT's HAT activity, bifunctionality expands the number of reactions that can be catalyzed by any genome. The appreciation of bifunctional or “moonlighting” enzymes is growing in humans (for review, see Jeffery 2003). Bifunctionality provides a mechanism for increasing the number of possible enzymatic reactions conserved between species, even if individual enzymes are not. Indeed, in evolution, it may be more effective to modify an existing protein to take on additional functions, rather than to evolve an entirely new molecule for each new challenge.
Bifunctional proteins may often be regulated by localization. In organisms with many cell types, one protein may have distinct functions when expressed in different cells or tissues. For example, in addition to its role in chromatin, histone H4 has a role in sepsis in humans (Xu et al. 2009), and acts as an antimicrobial agent when secreted from human sebocytes (Lee et al. 2009). Similarly, Lys20 can fully participate in chromatin functions only when its nuclear localization is intact.
Analysis of the essential Esa1 HAT led to the discovery that the Lys20 HCS, previously known only for its role in amino acid synthesis, has an additional role in chromatin and in modulating the effects of DNA damage. Lys20 also associates physically with a nucleosome assembly factor, Nap1 (Krogan et al. 2006). Other nucleosome assembly factors are known to function in both chromatin assembly and HAT activation (Selth and Svejstrup 2007). Thus, it seems likely that the range of nuclear functions for HCS and other metabolic enzymes will continue to expand with further study.
Materials and methods
Yeast methods, media, and strains
The strains used were generated for this study unless otherwise noted (Supplemental Table S1). Plates containing camptothecin were buffered with 100 mM potassium phosphate (pH 7.5), and 5 mg/mL camptothecin stock dissolved in DMSO was added after autoclaving (Nitiss and Wang 1988). Growth control plates were buffered, and contained an equivalent amount of DMSO. HU plates were YPD-based and 100 mM in HU (Zhou and Elledge 1992). The concentration of camptothecin or HU was selected to optimize for the different dynamic ranges of growth for each mutant or mutant combination. Details for each experiment are in the figure legends. For dilution assays, cells were grown in YPD or the indicated selective medium at 30°C to stationary phase, normalized to A6001, and plated in fivefold serial dilutions.
Plasmids
The plasmids are listed in Supplemental Table S1. The bacterial expression constructs in the pRSET vector system (Invitrogen) are pLP820 (pRSETc), pLP831 (ESA1), and pLP1934 (LYS20). Lys20E155A was created by direct mutagenesis of pLP1412 with oLP1305 and oLP1306. Lys20R31A was created by direct mutagenesis of pLP1412 with oLP1303 and oLP1304. Lys20ΔC10 (pLP2391) was created by PCR with oLP944 and oLP1363, digested with EcoRI and BamHI, and ligated into pRS202. pLP2392 (Lys20ΔC10 without a stop codon) was created by PCR with oLP 944 and oLP1364. The product was digested with EcoRI and BamHI, and ligated into pRS202. Lys20ΔC10-NES (pLP2402) was created by annealing oLP1390 and oLP1391, which comprised a variant (ELALKLAGLDINLI) of the strong NES from PKI (Gadal et al. 2001) and a stop codon; digesting this fragment with BamHI and XbaI; and ligating into pLP2392, which contained Lys20ΔC10 from which the stop codon was removed. Lys20-NES (pLP2404) was created by PCR with oLP944 and oLP1399. This fragment, Lys20 without a stop codon, was digested with EcoRI and BamHI and subcloned into pLP2402 to create pLP2404. pLP2295 (Lys20S385F, Lys20fbr1) was created by direct mutagenesis of pLP1412 with oLP1055 and oLP1056. Lys20R276K (pLP2296, Lys20fbr2) was created by direct mutagenesis of pLP1412 with oLP1057 and oLP1058. Constructs were confirmed by sequencing.
Protein immunoblots
Cells were grown to mid-log phase and exposed to 0.1 M HU for 90 min. Cells were harvested and TCA extracts were made. Whole-cell extracts equivalent to A600 = 0.5 were separated on 8% SDS-PAGE gel, transferred to PVDF membrane, and immunoblotted with anti-Rad53 antibody (Santa Cruz Biotechnology) and anti-goat secondary antibody (Xymed), followed by detection with ECL Plus (GE Healthcare Amersham).
HAT assays
HAT assays were performed for 30 min at room temperature using standard methods (Clarke et al. 1999), with yeast histones as substrate (see below). The reaction was evaluated by 18% SDS-PAGE. Gels were transferred to nitrocellulose for 1 h at 100 V. Nitrocellulose was exposed to Kodak MS film at −80°C with a Kodak LE transcreen to detect incorporation of radioactivity. Gels after transfer were stained according to standard Coomassie Brilliant Blue protocols. For recombinant protein, bacterial extracts were prepared from Escherichia coli B834 DE3 cells containing the pRARE plasmid (Novagen) and ESA1, LYS20, or vector plasmids. Cells were induced with 1 mM IPTG for 3 h at 30°C.
Preparation of yeast histones
Yeast cells were grown to log phase and spheroplasted. Histones were extracted with 0.25 N HCl, precipitated with 20% TCA, and resuspended in 10% glycerol and 50 mM Tris (pH 8) (Lo et al. 2004; Vaquero et al. 2006).
Immunoprecipitations
For immunoprecipitations, cells, including the Gcn5-9Myc strain, were grown to log phase and lysed by bead beating. Immunoprecipitations were carried out overnight at 4°C with anti-Lys20 antibodies (generous gifts of J. Aris) used at dilutions of 1:50. Seventy microliters of Protein A Sepharose CL-4B beads (GE HealthCare) was added and rocked for 3–4 h at room temperature. Western blotting was carried out as above and blotting with anti-Myc 9E-10 antibody (1:5000; gift of R. Hampton) to detect Gcn5-Myc. The secondary antibody (anti-mouse IgG HRP; Promega) was used at 1:5000. Chemiluminescent detection was done with Pierce ECL Western blotting substrate. The percentage of phosphorylated Rad53 was determined with ImageQuant software.
Immunofluorescence microscopy
Cells were grown to mid-log phase and fixed in 3.7% formaldehyde, then spheroplasted. After being applied to slides, cells were fixed at −20°C in methanol, then in acetone. Slides were blocked for 1 h in 1% BSA and incubated overnight at 4°C in anti-Lys20 antibody (1:50; a generous gift of J. Aris). The secondary antibody, Texas Red-conjugated AffiniPure goat anti-mouse IgG (1:50; Jackson ImmunoResearch Laboratories), was applied for 2 h at 37°C, then cells were DAPI-stained for 1 h at 37°C. Mounting medium was applied (Vectashield, Vector Laboratories), and images were collected on an Axiovert 200M microscope (Carl Zeiss MicroImaging, Inc.) with a 100× 1.3 NA objective. Images were captured using a monochrome digital camera (Axiocam, Carl Zeiss MicroImaging, Inc.), and data were analyzed with Axiovision software (Carl Zeiss MicroImaging, Inc.).
Acknowledgments
We appreciate the contributions of A. Clarke, E. Spedale, and V. Le to the early stages of the project, and R. Otsuka for technical assistance. J. Aris, J. Babiarz, C. Chang, E. Dubois, R. Dutnall, P. Laybourn, J. Huh, S. Bulfer, R. Trievel, J. Rine, E. Dubois, R. Kamakaka, R. Gardner, R. Hampton, and F. Robert provided reagents and advice. M. Niwa and A. Bicknell provided assistance with microscopy. M. Koch, R. Garza, S. Jacobsen, B. Emerson, B. Mendelsohn, F. Solomon, and C. Chang read the manuscript and provided feedback. This work was initiated with support from NIH 5T32GM007240, GM54649, and GM90177.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1935910.
Supplemental material is available at http://www.genesdev.org.
References
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Côté J 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Halley JE, Rine J 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev 20: 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415 [DOI] [PubMed] [Google Scholar]
- Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev 17: 1415–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfer SL, Scott EM, Couture JF, Pillus L, Trievel RC 2009. Crystal structure and functional analysis of homocitrate synthase, an essential enzyme in lysine biosynthesis. J Biol Chem 284: 35769–35780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfer SL, Scott EM, Pillus L, Trievel RC 2010. Structural basis for L-lysine feedback inhibition of homocitrate synthase. J Biol Chem 285: 10446–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A 2006. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol 16: 1280–1289 [DOI] [PubMed] [Google Scholar]
- Chen S, Brockenbrough JS, Dove JE, Aris JP 1997. Homocitrate synthase is located in the nucleus in the yeast Saccharomyces cerevisiae. J Biol Chem 272: 10839–10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Kron SJ 2002. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol Cell Biol 22: 8215–8225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol 19: 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Decker PV, Yu DY, Iizuka M, Qiu Q, Smith MM 2008. Catalytic-site mutations in the MYST family histone acetyltransferase Esa1. Genetics 178: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Côté J 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev 14: 147–154 [DOI] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Ramos F, Pierard A, Dubois E 1999. In Saccharomyces cerevsiae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur J Biochem 261: 163–170 [DOI] [PubMed] [Google Scholar]
- Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC 2009. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res 37: 3969–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamonet F, Lauquin GJ 1998. The Saccharomyces cerevisiae LYS7 gene is involved in oxidative stress protection. Eur J Biochem 251: 716–723 [DOI] [PubMed] [Google Scholar]
- Gancedo C, Flores CL 2008. Moonlighting proteins in yeasts. Microbiol Mol Biol Rev 72: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M 2004. Regulation of gene expression by a metabolic enzyme. Science 306: 482–484 [DOI] [PubMed] [Google Scholar]
- Halley JE, Kaplan T, Wang AY, Kobor MS, Rine J 2010Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol 8: e1000401 doi: 10.1371/journal.pbio.1000401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655 [DOI] [PubMed] [Google Scholar]
- Hicks GR, Raikhel NV 1995. Protein import into the nucleus: An integrated view. Annu Rev Cell Dev Biol 11: 155–188 [DOI] [PubMed] [Google Scholar]
- Horton P, Nakai K 1997. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc Int Conf Intell Syst Mol Biol 5: 147–152 [PubMed] [Google Scholar]
- Jeffery CJ 2003. Moonlighting proteins: Old proteins learning new tricks. Trends Genet 19: 415–417 [DOI] [PubMed] [Google Scholar]
- Jones E, Fink G 1982. Regulation of amino acid and nucleotide biosynthesis in yeast. In The molecular biology of the yeast Saccharomyces cerevisiae: Metabolism and gene expression (ed. Strathhern J et al. ), pp. 181–299 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 33: 335–343 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev 20: 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanovic A, Wolfl S 2006. Fructose-1,6-bisphosphatase mediates cellular responses to DNA damage and aging in Saccharomyces cerevisiae. Mutat Res 594: 135–147 [DOI] [PubMed] [Google Scholar]
- Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol 2: e131 doi: 10.1371/journal.pbio.0020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, et al. 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci 101: 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Lafon A, Chang CS, Scott EM, Jacobson SJ, Pillus L 2007. MYST opportunities for growth control: Yeast genes illuminate human cancer gene functions. Oncogene 26: 5373–5384 [DOI] [PubMed] [Google Scholar]
- Lee DY, Huang CM, Nakatsuji T, Thiboutot D, Kang SA, Monestier M, Gallo RL 2009. Histone H4 is a major component of the antimicrobial action of human sebocytes. J Invest Dermatol 129: 2489–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD 2008. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev 22: 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J, Tao SC, Qian J, Zhao Y, Boeke JD, et al. 2009. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136: 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Henry KW, Schwartz MF, Berger SL 2004. Histone modification patterns during gene activation. Methods Enzymol 377: 130–153 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298 [DOI] [PubMed] [Google Scholar]
- Matsubara K, Sano N, Umehara T, Horikoshi M 2007. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 12: 13–33 [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Mersfelder EL, Parthun MR 2006. The tale beyond the tail: Histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res 34: 2653–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Xu F, Zhang K, Grunstein M 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev 20: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J 1999. Nuclear import and export pathways. J Cell Biochem Suppl 75: 76–83 [DOI] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol 15: 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss J, Wang JC 1988. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc Natl Acad Sci 85: 7501–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada H, Aranda C, DeLuna A, Hernandez H, Calcagno ML, Marin-Hernandez A, Gonzalez A 2008. Specialization of the paralogue LYS21 determines lysine biosynthesis under respiratory metabolism in Saccharomyces cerevisiae. Microbiology 154: 1656–1667 [DOI] [PubMed] [Google Scholar]
- Raisner RM, Madhani HD 2008. Genomewide screen for negative regulators of sirtuin activity in Saccharomyces cerevisiae reveals 40 loci and links to metabolism. Genetics 179: 1933–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saban N, Bujak M 2009. Hydroxyurea and hydroxamic acid derivatives as antitumor drugs. Cancer Chemother Pharmacol 64: 213–221 [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271: 357–360 [DOI] [PubMed] [Google Scholar]
- Selth L, Svejstrup JQ 2007. Vps75, a new yeast member of the NAP histone chaperone family. J Biol Chem 282: 12358–12362 [DOI] [PubMed] [Google Scholar]
- Shia WJ, Li B, Workman JL 2006. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev 20: 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slekar KH, Kosman DJ, Culotta VC 1996. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J Biol Chem 271: 28831–28836 [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci 95: 3561–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD 2006. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell 23: 207–217 [DOI] [PubMed] [Google Scholar]
- Terry LJ, Shows EB, Wente SR 2007. Crossing the nuclear envelope: Hierarchical regulation of nucleocytoplasmic transport. Science 318: 1412–1416 [DOI] [PubMed] [Google Scholar]
- Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE 2004. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem 279: 53665–53673 [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D 2006. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 20: 1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Andi B, Qian J, West AH, Cook PF 2006. The α-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem Biophys 46: 43–64 [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang X, Pelayo R, Monestier M, Ammollo CT, Semeraro F, Taylor FB, Esmon NL, Lupu F, Esmon CT 2009. Extracellular histones are major mediators of death in sepsis. Nat Med 15: 1318–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O, Yogev O, Singer E, Shaulian E, Goldberg M, Fox TD, Pines O 2010. Fumarase: A mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol 8: e1000328 doi: 10.1371/journal.pbio.1000328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Elledge SJ 1992. Isolation of crt mutants constitutive for transcription of the DNA damage inducible gene RNR3 in Saccharomyces cerevisiae. Genetics 131: 851–866 [DOI] [PMC free article] [PubMed] [Google Scholar]