Abstract
The canonical microRNA (miRNA) biogenesis pathway requires two RNaseIII enzymes: Drosha and Dicer. To understand their functions in mammals in vivo, we engineered mice with germline or tissue-specific inactivation of the genes encoding these two proteins. Changes in proteomic and transcriptional profiles that were shared in Dicer- and Drosha-deficient mice confirmed the requirement for both enzymes in canonical miRNA biogenesis. However, deficiency in Drosha or Dicer did not always result in identical phenotypes, suggesting additional functions. We found that, in early-stage thymocytes, Drosha recognizes and directly cleaves many protein-coding messenger RNAs (mRNAs) with secondary stem–loop structures. In addition, we identified a subset of miRNAs generated by a Dicer-dependent but Drosha-independent mechanism. These were distinct from previously described mirtrons. Thus, in mammalian cells, Dicer is required for the biogenesis of multiple classes of miRNAs. Together, these findings extend the range of function of RNaseIII enzymes beyond canonical miRNA biogenesis, and help explain the nonoverlapping phenotypes caused by Drosha and Dicer deficiency.
Keywords: RNaseIII enzymes, microRNA targets, microRNA processing, endonucleolytic cleavage
MicroRNAs (miRNAs) are small RNAs that inhibit protein-coding messenger RNAs (mRNAs) through translational repression and degradation. In animals, target recognition occurs through incomplete base-pairing. Base-pairing with the 5′ end of the miRNA appears essential for target recognition, while a greater degree of flexibility is permitted at the 3′ end (Lewis et al. 2003, 2005). miRNAs function as part of the effector RNA-induced silencing complex (RISC) with members of the Argonaute family of proteins (Peters and Meister 2007). How miRNAs repress gene expression remains unclear, but likely involves multiple mechanisms, including inhibition of ribosome progression and mRNA deadenylation (Filipowicz et al. 2008).
The biogenesis of miRNAs involves two processing steps that have been largely defined in cell-based and biochemical studies. Primary miRNA (pri-miRNA) transcripts are first cleaved by the nuclear “microprocessor” complex, which contains the RNaseIII enzyme Drosha and its dsRNA-binding partner, Dgcr8/Pasha, resulting in release of the short stem–loop pre-miRNA (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). These pre-miRNAs are then exported to the cytoplasm, where they are further processed by another RNaseIII enzyme, Dicer, and its dsRNA-binding partner, Tarbp2/Loquacious, to remove the loop structure. This liberates the mature miRNA duplexes for loading into the RISC (Chendrimada et al. 2005).
Genetic ablation of Dicer has clearly demonstrated its requirement in vivo and, by inference, the requirement for miRNAs. In mice, Dicer is required for stem cell proliferation (Murchison et al. 2005) and the differentiation and/or function of many tissues, including germ cells (Murchison et al. 2007; Tang et al. 2007) and neurons (Cuellar et al. 2008). In contrast to the large number of studies on Dicer, less is known about the requirements for Drosha. By comparing mice in which LoxP-flanked alleles of Rnasen (encoding Drosha) or Dicer1 were specifically inactivated in Foxp3+ regulatory T cells (Tregs), we confirmed that the two enzymes do indeed function in the same pathway (Chong et al. 2008). Cells engineered to be deficient for either enzyme lacked mature miRNAs. Furthermore, deletion of either Rnasen or Dicer1 in Tregs resulted in identical phenotypes. These mice died by 3 wk of age from systemic inflammation. Thus, the identical phenotypes caused by Drosha and Dicer deficiency suggest that the function of these enzymes in Tregs may be restricted to the same pathway; that is, the production of miRNAs.
In cell types other than Tregs, the function of Dicer appears not to be limited to miRNA biogenesis. Dicer is also required for the generation of siRNAs derived from endogenous dsRNA transcripts. The production of small RNAs from SINE and simple repeat elements in embryonic stem cells, and from pseudogene and retrotransposon-derived dsRNAs in oocytes, have all been shown to be Dicer-dependent (Calabrese et al. 2007; Tam et al. 2008; Watanabe et al. 2008).
In Drosophila melanogaster and Caenorhabditis elegans, the biogenesis of a subset of miRNAs has been found to be dependent on the splicing machinery rather than the microprocessor (Okamura et al. 2007; Ruby et al. 2007). Like canonical miRNAs, these “mirtrons” are encoded within short stem–loop structures. However, these stem–loops are instead located within short introns of protein-coding genes, which are released upon mRNA splicing.
While Dicer is known to have a role in the biogenesis of multiple classes of small RNAs, it has not been known whether the function of Drosha is limited to generation of miRNAs, or whether mirtron-independent miRNAs are always dependent on Drosha and Dicer. It was found recently that miR-451 is generated through a process that is Dicer-independent, but requires the catalytic activity of the Argonaute protein Ago2 (Cheloufi et al. 2010; Cifuentes et al. 2010). In this study, we investigated the impact of Drosha and Dicer deficiency in several cell lineages, including T cells at different stages of development and cultured primary fibroblasts. We found that, while Drosha and Dicer are essential for canonical miRNA biogenesis, they also have specialized activities in independent pathways.
Results
Nonredundant effects of Drosha and Dicer deficiency early in T-cell development
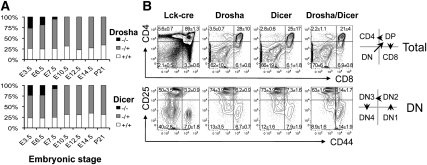
To determine the functions of Drosha and Dicer in vivo, we investigated the consequence of targeted gene deletion in the germline or in specific tissues by means of Cre/LoxP technology. Germline Dicer deficiency causes lethality early in embryogenesis (Bernstein et al. 2003). We found that Drosha deficiency also resulted in embryonic lethality by embryonic day 7.5 (E7.5) in gestation (Fig. 1A). The finding of identical phenotypes caused by Dicer and Drosha deficiency is consistent with the two enzymes having functions in the same pathway (miRNA biogenesis), and indicates a requirement for miRNAs in the post-implantation embryo.
Figure 1.
Overlapping and nonoverlapping phenotypes caused by Drosha and Dicer deficiency. (A) Embryonic lethality in germline Drosha- and Dicer-deficient mice. Shown is the frequency of the indicated genotypes at each stage in embryogenesis. Between 12 and 38 embryos were genotyped at each developmental stage. (B) Conditional deletion of the genes encoding Drosha and Dicer with Lck-cre early in thymocyte development causes a block at the DN3 stage. Shown are flow cytometric plots of CD4 versus CD8 expression on CD90+ total thymocytes (top row), and CD44 versus CD25 on CD4−CD8−TCR−CD90+ “double-negative” thymocyes (bottom row). The quadrant values represent the mean ± SD of three sets of mice analyzed at 6 wk of age.
Drosha and Dicer are expressed throughout T-cell development, with expression highest at the early double-negative (DN) stages, so-called because these progenitors lack expression of the cell surface molecules CD4 and CD8 (Supplemental Fig. S1). To investigate the function of these enzymes in T-cell development in the adult, LoxP-flanked alleles of Rnasen and Dicer1 were inactivated at the DN2 stage by breeding with Lck-cre mice. Quantitative RT–PCR (qRT-PCR) of cells at the subsequent DN3 stage confirmed a loss of >95% of Drosha or Dicer mRNA (data not shown). Furthermore, there was loss of 95%–98% of mature miRNAs, such as miR-125b and miR-181a, while there was a >20-fold accumulation of their pri-miRNA transcripts (data not shown). Both RnasenF/F Lck-cre and Dicer1F/F Lck-cre mice displayed marked reductions in thymocyte numbers (Supplemental Fig. S2). This was due to a block in T-cell development at the DN3 stage (Fig. 1B). The appearance of later-stage thymocytes was a result of cells that had escaped cre-mediated deletion (data not shown).
Interestingly, RnasenF/F Dicer1F/F Lck-cre double-deficient mice displayed a more severe block, with cells also accumulating at the DN2 stage (Fig. 1B). This could be due to more rapid miRNA depletion in the absence of both enzymes. Alternatively, this could reflect nonredundant effects of Drosha and Dicer deficiency, which would suggest that Drosha and/or Dicer may have functions in addition to miRNA biogenesis in these cells.
We showed previously that deletion of either Rnasen or Dicer1 late in T-cell development with CD4-cre resulted in a preferential loss of the Treg population (Chong et al. 2008). Unlike deletion early in T-cell development, no additive effect was observed when the two genes were deleted simultaneously with CD4-cre (Supplemental Fig. S3). Double- and single-deficient mice displayed similar reductions in the Treg population, suggesting that Drosha and Dicer function may be restricted to canonical miRNA biogenesis in these cells.
Transcriptional and proteomic profiling reveals nonredundant effects of Drosha and Dicer deficiency on gene expression in distinct cell types
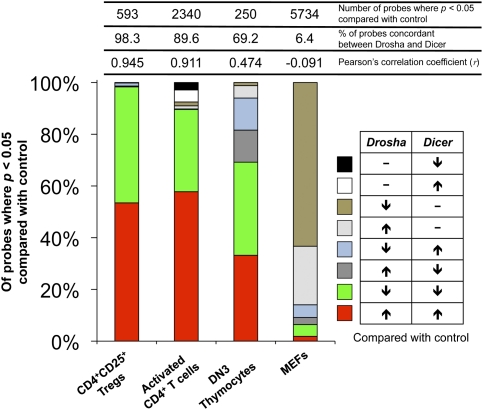
To determine if there are indeed nonredundant effects of Drosha and Dicer deficiency, microarray transcriptional profiling was performed on several cell types from the mutant mice (Fig. 2; Supplemental Tables S1–S4). Consistent with Drosha and Dicer deficiency in Tregs resulting in identical phenotypes (Chong et al. 2008), there was almost perfect correlation (Pearson's correlation coefficient = 0.945) between the transcriptional profiles of Drosha- and Dicer-deficient Tregs, with 317 probes significantly (P < 0.05) up-regulated in both and 266 probes significantly down-regulated in both. Only 10 probes were discordant between Drosha- and Dicer-deficient Tregs. Because miRNAs repress target genes at the level of mRNA and protein, direct targets are likely to be represented among the transcripts up-regulated in both mutant cells, while indirect targets may be represented among either up-regulated or down-regulated transcripts. Drosha- and Dicer-deficient CD4+ T cells also displayed very similar profiles (Pearson's correlation coefficient = 0.911). Of the probes significantly different from control CD4-cre cells, 89.6% were shared between both mutants. In DN3 thymocytes, however, 30.8% of probes significantly different from control Lck-cre cells were discordant between Drosha- and Dicer-deficient cells—a difference that was highly consistent between animals (Supplemental Fig. S4). The most extreme discordance (Pearson's correlation coefficient = −0.091) was observed in murine embryonic fibroblasts (MEFs) prepared from E14.5 embryos, with only 6.4% of the probes significantly different from control CreER cells shared between both mutants. There were many more genes up-regulated or down-regulated in Drosha-deficient than Dicer-deficient MEFs.
Figure 2.
Impact of Drosha and Dicer deficiencies on gene expression profiles. Shown is a summary of microarray gene profiles of Drosha- and Dicer-deficient CD4+CD25+ Tregs, in vitro activated CD4+ T cells, DN3 thymocytes, and MEFs compared with the appropriate control cells expressing only Cre. In Tregs and CD4+ T cells, deletion was achieved with CD4-cre; in DN3 thymocytes, deletion was achieved with Lck-cre; and in MEFs, deletion was achieved with Gt(Rosa)26SorCreER. For each genotype, cell populations to be analyzed were sorted or prepared from two (CD4 and MEFs) or three individual mice (DN3 and Treg). The data represent only those probes that were significantly different (P < 0.05 by ANOVA) in Drosha- and/or Dicer-deficient cells compared with controls. Listed above the bars are the actual numbers of probes that were significantly different in either Drosha- or Dicer-deficient cells compared with controls, the percentage of these probes that were up in both Drosha- and Dicer-deficient cells or down in both, and the Pearson's correlation coefficient calculated for the changes measured in Drosha-deficient cells compared with the changes measured in Dicer-deficient cells.
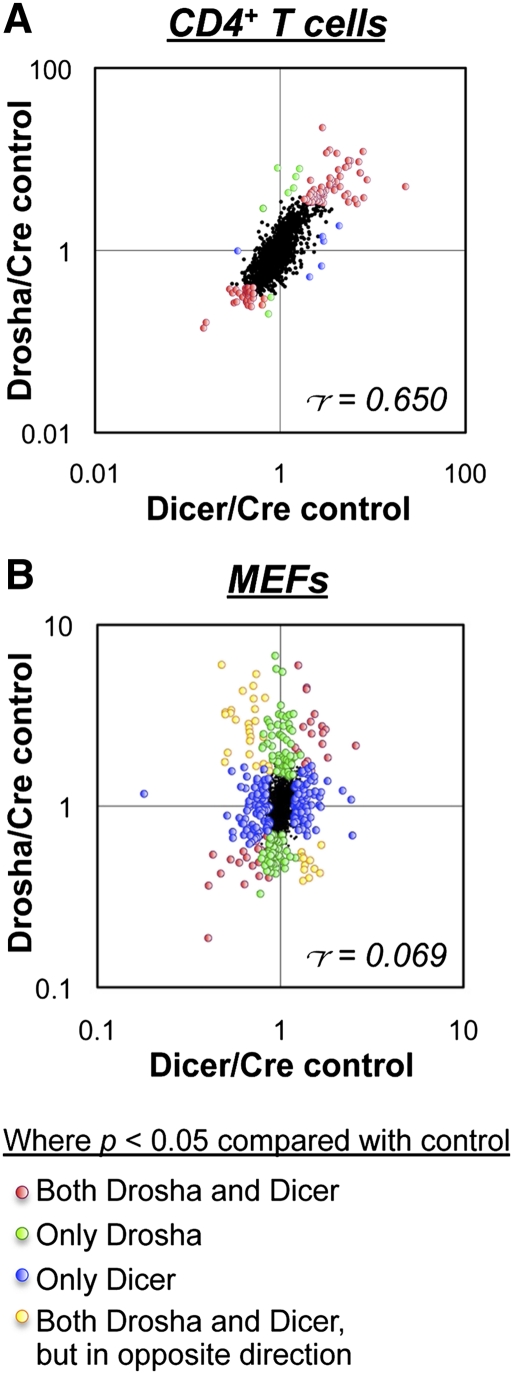
The proteomic impact of Drosha and Dicer deficiency in CD4+ T cells and MEFs was analyzed by stable isotope labeling with amino acids in culture (SILAC) (Fig. 3; Supplemental Tables S5, S6). Consistent with the transcriptional profiling data, there was poor correlation between the proteomic profiles of Drosha- and Dicer-deficient MEFs (Pearson's correlation coefficient = 0.069), whereas there was good correlation between Drosha- and Dicer-deficient CD4+ T cells (Pearson's correlation coefficient = 0.650). Thus, Drosha deficiency in some cell lineages has a greater impact than deficiency of Dicer and, presumably, absence of the canonical miRNA pathway.
Figure 3.
Impact of Drosha and Dicer deficiencies on the proteome. Shown are ratios of individual protein expression levels in in vitro activated CD4+ T cells (A) and MEFs (B) as determined by SILAC analysis. Drosha-deficient cells were cultured in L-Lys/L-Arg-deficient medium supplemented with the light isotopes L-Lys 12C614N2 and L-Arg 12C614N4, Dicer-deficient cells were cultured in medium supplemented with the intermediate isotopes L-Lys 4,4,5,5-D4 and L-Arg 13C614N4, and control cells were cultured in medium supplemented with the heavy isotopes L-Lys 13C615N2 and L-Arg 13C615N4. The labels were then inverted for repeat experiments. Labeling was performed for at least five cell divisions to label all proteins before analysis by quantitative mass spectrometry. In CD4+ T cells, deletion was achieved with CD4-cre, and in MEFs, deletion was achieved with Gt(Rosa)26SorCreER. Only data for proteins that were quantified in all three genotypes are shown. Indicated are the proteins that were found to be significantly different (P < 0.05 by ANOVA) in Drosha- and/or Dicer-deficient cells compared with controls. r = Pearson's correlation coefficient.
Such poor correlation between the transcriptional and proteomic profiles of Drosha- and Dicer-deficient MEFs was a surprise. This appeared to translate to the cellular level when we examined the ability of Oct4, Sox2, Klf4, and cMyc to reprogram Drosha- and Dicer-deficient MEFs into induced pluripotent stem cells (Takahashi et al. 2007). Although Dicer-deficient MEFs could not reprogram into pluripotent cells, they were capable of undergoing immortalization and forming colonies. In contrast, Drosha-deficient MEFs could not be immortalized (data not shown).
Overlapping phenotypes caused by Drosha and Dicer deficiency are likely to be due to loss of miRNA expression
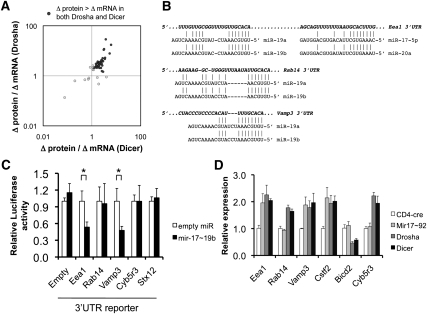
Because Drosha and Dicer deficiency resulted in both overlapping and nonoverlapping phenotypes and gene expression profiles, we wanted to determine whether the overlaps observed were indeed due to loss of miRNA expression rather than some other function of the two enzymes. In particular, we wanted to confirm that at least some of the molecular changes observed were due to loss of specific miRNAs. Because miRNAs generally affect targets more markedly at the protein than at the mRNA level (Baek et al. 2008; Selbach et al. 2008), we first searched for likely miRNA targets in CD4+ T cells by comparing protein versus mRNA derepression in Drosha- and Dicer-deficient cells (Fig. 4A; Supplemental Table S7). Those genes that were up-regulated more at the protein level in both types of mutant cells were analyzed for predicted miRNA target sites. Only sites corresponding to expressed miRNAs were analyzed further (Supplemental Table S8).
Figure 4.
Prediction of miRNA targets by measuring protein versus mRNA derepression in the absence of miRNAs. (A) Proteins that were up-regulated in both Drosha- and Dicer-deficient activated CD4+ T cells (from Fig. 3A) were analyzed for the magnitude of protein derepression versus mRNA derepression. The 3′UTRs of these genes were then analyzed for sites potentially targeted by miRNAs normally expressed in control cells. (B) Shown are predicted mir-17∼92a target sites in the 3′UTRs of Eea1, Rab14, and Vamp3, which were derepressed more at the protein than the mRNA level. (C) The entire 3′UTRs of Eea1, Rab14, and Vamp3 were inserted into a Firefly luciferase reporter and analyzed for reporter knockdown in the presence of overexpressed mir-17∼19b. (Mir-92a was left out of the expression construct because it was poorly processed.) Cyb5r3 and Stx12 3′UTRs were used as negative controls. The data represent the mean ± SEM of four experiments, normalized to Renilla luciferase as a transfection control. (*) P < 0.05 by t-test. (D) Confirmation of predicted mir-17∼92a targets in CD4+ T cells by specific deletion of the Mir17∼92a cluster. In vitro activated CD4+ T cells deficient in mir-17∼92a, Drosha, or Dicer were analyzed for expression levels of the predicted mir-17∼92a targets Eea1, Rab14, Vamp3, and Cstf2 by qRT–PCR. Analyses of Bicd2 and Cyb5r3 were included as negative controls. Bicd2, although a predicted mir-17∼92 target, was not derepressed in Drosha- or Dicer-deficient cells. The data represent the mean ± SEM of four experiments.
We first focused on up-regulated genes predicted to be targets of the polycistronic mir-17∼92a-1 cluster (Fig. 4B). The 3′ untranslated regions (UTRs) of two predicted targets—Eea1 and Vamp3—were knocked down in a luciferase reporter assay by cotransfection of the mir-17∼92a-1 primary transcript, suggesting that these are, indeed, miRNA targets (Fig. 4C). Next, we wanted to confirm that the endogenous genes are targets of mir-17∼92a-1 in vivo. Target derepression was analyzed in T cells with specific deletion of the Mir17∼92a-1 cluster (Fig. 4D). Three predicted mir-17∼92a targets—Eea1, Vamp3, and Cstf2—were derepressed in mir-17∼92a-deficient cells to the same degree as in cells defective for Drosha and Dicer, again confirming that these are targets. Reporter knockdown analysis also confirmed interactions between the Vamp3 3′UTR and miR-29b/29c, the CD28 3′UTR and miR-24, and the Stx12 3′UTR and miR-23a/23b (Supplemental Fig. S5). Thus, the overlapping phenotypes observed between Drosha- and Dicer-deficient cells appear to be due to loss of miRNAs, at least for the few targets that were validated.
Drosha regulates the expression of mRNAs containing secondary stem–loop structures by direct cleavage
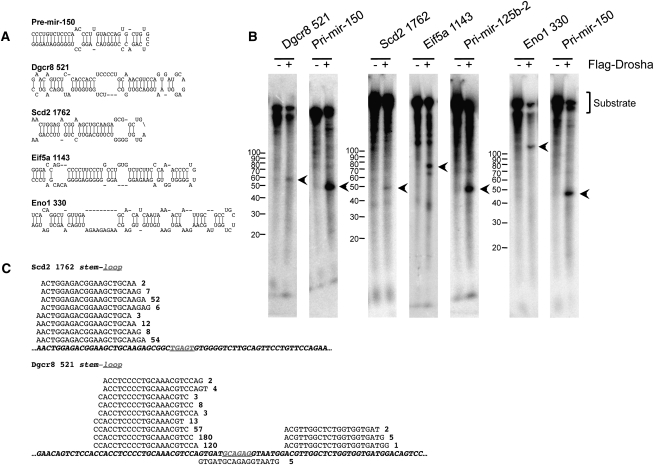
In DN3 thymocytes and MEFs, there were numerous genes derepressed by Drosha but not Dicer deficiency (Fig. 2; Supplemental Figs. S4, S6). This suggests that Drosha may be regulating these targets independently of miRNAs (or at least Dicer-dependent miRNAs). During miRNA biogenesis, Drosha normally functions to excise the pre-miRNA stem–loop from a longer primary RNA precursor. We postulated that perhaps such secondary stem–loop structures exist in other RNAs that are not miRNA precursors that may be recognized and cleaved by Drosha. Indeed, when those genes up-regulated only in Drosha-deficient DN3 thymocytes were analyzed for mRNA secondary structure using Mfold (Zuker 2003), many were found to contain putative stem–loop structures that resemble pre-miRNAs (Fig. 5A; Supplemental Fig. S7). To determine if these mRNA-embedded stem–loops can be directly recognized by Drosha, in vitro RNA cleavage assays were performed. Several mRNA-embedded stem–loops could be cleaved by immunoprecipitated Drosha, including Dgcr8, Scd2, Eif5a, and Eno1 (Fig. 5B). However, mRNA cleavage appeared less efficient compared with the processing of pri-miRNA precursors.
Figure 5.
A subset of mRNAs can be directly cleaved by Drosha. (A) Shown are the stem–loop structures found in some of the mRNAs up-regulated in Drosha- but not Dicer-deficient DN3 thymocytes. As a comparison, the stem–loop structure of pre-mir-150 is shown. (B) mRNA-embedded stem–loop structures can be cleaved by Drosha in vitro. The indicated stem–loop structures ±150 nt were in vitro transcribed with T7 RNA polymerase in the presence of α-32PUTP. These were then incubated with immunoprecipitated Flag-Drosha. As positive controls, pri-mir-150 and pri-mir-125b-2 were also cleaved. Expected cleavage products are indicated by the arrows. (C) mRNA-embedded stem–loop structures can be fully processed into small RNAs in vivo. Small RNA (with 5′ phosphate and 3′ hydroxyl termini) libraries were constructed from DN3 thymocytes, and were analyzed by deep sequencing. Shown are the small RNAs that mapped to the stem–loop structures of Scd2 1762 and Dgcr8 521. Also indicated is the number of times each RNA was sequenced.
We next looked for evidence of direct mRNA cleavage in vivo within DN3 thymocytes. We postulated that, if mRNAs are cleaved by Drosha, the excised stem–loop might be processed down to small RNAs that could be detected by deep sequencing. To this end, small RNA libraries were constructed from DN3 thymocytes for Illumina sequencing. Only small RNAs with 5′ phosphate and 3′ hydroxyl termini were cloned in order to identify RNAs processed by endonucleolytic cleavage. Not surprisingly, the majority of small RNAs present in DN3 thymocytes were found to be miRNAs. However, ∼12% of sequences were found to map to other locations within the mouse genome, including to protein-coding genes and noncoding or structural RNAs (Supplemental Fig. S8; Supplemental Table S9). Furthermore, many of the small RNAs corresponding to protein-coding transcripts mapped specifically to the predicted stem–loop structures (Fig. 5C; Supplemental Fig. S9). We also observed many other small RNAs that mapped to protein-coding mRNAs, including those for Brd2, Mbnl1, and Wipi2, which also turned out to contain stem–loop structures (Supplemental Fig. S10A,B). By qRT–PCR, we determined that these other genes were also derepressed in Drosha-deficient DN3 thymocytes (Supplemental Fig. S10C), but microarray analysis had not been sensitive enough to detect them. Thus, Drosha regulates the expression of many genes independently of miRNAs by directly cleaving secondary stem–loop structures embedded within the mRNA.
Numerous Drosha-independent miRNAs in mammalian cells
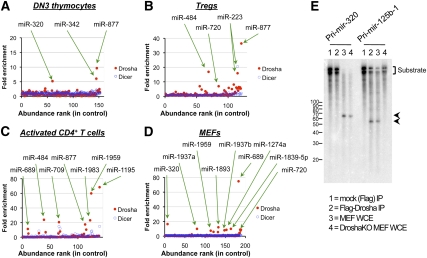
The finding that Dicer deficiency resulted in some transcriptional changes that did not overlap with Drosha deficiency was not entirely surprising, as it was assumed that Dicer would be required for the production of multiple classes of small RNAs. In particular, it had been shown in D. melanogaster and C. elegans that generation of mirtrons required Dicer and the splicing machinery, but not Drosha (Okamura et al. 2007; Ruby et al. 2007). To determine if mirtrons are also present in mammalian cells, small RNA sequencing was performed on cells in which the Rnasen or Dicer1 gene was deleted. In theory, mirtrons should be lost in Dicer-deficient but not Drosha-deficient cells, whereas canonical miRNAs should be equally affected by Drosha and Dicer deficiency. Indeed, we identified some 30 miRNAs that were enriched in Drosha-deficient but not Dicer-deficient cells (Fig. 6A–D; Supplemental Tables S10–S13), indicating that these miRNAs are generated by a Dicer-dependent, but Drosha-independent, mechanism. These miRNAs represented 1:60 miRNA molecules, and 6.9% of the miRNA species expressed across the four cell types profiled.
Figure 6.
Drosha-independent miRNAs. Small RNA libraries (with 5′ phosphate and 3′ hydroxyl termini) were constructed from Drosha-deficient, Dicer-deficient, and control DN3 thymocytes (A), CD4+CD25+ Tregs (B), activated CD4+ T cells (C), and MEFs (D); then deep sequencing was performed. Libraries were normalized to an equivalent number of reads that mapped to the mouse genome. On the X-axis is a ranking of miRNAs based on expression level in control cells, from highest to lowest. On the Y-axis is the sequencing frequency in Drosha- or Dicer-deficient cells divided by control cells. Only miRNAs expressed at >100 copies per million in control cells are shown. (E) Pri-mir-320 (Drosha-independent) and pri-mir-125b-1 (Drosha-dependent) were in vitro transcribed with T7 RNA polymerase in the presence of α-32PUTP. These were incubated with immunoprecipitated Flag-Drosha or whole-cell extracts (WCE) from control or Drosha-deficient MEFs. Whole-cell extracts were obtained from untreated RnasenF/F Gt(Rosa)26SorCreER MEFs or those that had been pulsed with 4-OH tamoxifen for 1 d, followed by culture without tamoxifen for a further 3 d. Expected cleavage products are indicated by the arrows.
We next determined if any of these Drosha-independent miRNAs originated from short introns. Only miR-877 was found to have originated from a short intron, and hence is likely to be a mirtron (Supplemental Fig. S11). The majority of Drosha-independent miRNAs appeared to be derived from long introns of protein-coding mRNAs, such as miR-342 (which is embedded in the 20.2-kb intron 3 of Evl), or from independent transcriptional units, such as miR-320. This suggests that release of the pre-miRNA stem–loop intermediate requires a processing step rather than splicing alone.
These Drosha-independent miRNAs are derived from precursors that are predicted to fold into stem–loop structures (Supplemental Fig. S12). Interestingly, these pre-miRNAs tended to form structures with extended stems of up to 65 nucleotides (nt) in length, which is longer than the typical 33-nt stems of canonical miRNAs (Han et al. 2006). To confirm that processing of these unusual miRNAs occurs through a Drosha-independent mechanism, we compared the processing of pri-mir-320 with a confirmed canonical miRNA, pri-mir-125b-1 (Fig. 6E). As expected, immunoprecipitated Drosha or whole-cell extracts from wild-type MEFs were able to cleave pri-mir-125b-1. In contrast, pri-mir-320 could not be cleaved by immunoprecipitated Drosha, but was cleaved with extracts from both wild-type or Drosha-deficient MEFs, suggesting that another enzyme in mammalian cells processes this pri-miRNA.
Discussion
Direct mRNA cleavage by Drosha as a mechanism of gene regulation
In this study, we reported the global impact of Drosha and Dicer deficiency in several cell types. Varying degrees of discordance in the cell types analyzed suggested that the enzymes also function in independent pathways in addition to canonical miRNA biogenesis. We found that Drosha is capable of directly cleaving many protein-coding mRNAs that contained secondary stem–loop structures. One of these mRNAs was Dgcr8, which encodes the dsRNA-binding partner of Drosha. Drosha-mediated cleavage of the DGCR8 mRNA in human HEK293 cells had been reported previously, and was suggested to be a mechanism of autoregulation by the microprocessor (Han et al. 2009). Further analysis in embryonic stem cells suggested that direct mRNA cleavage might be unique to the Dgcr8 mRNA (Shenoy and Blelloch 2009). However, our global analysis indicates that this is a more generalized mechanism of regulating gene expression. We showed both in vitro and in vivo that multiple mRNAs are cleaved by Drosha.
The murine Dgcr8 transcript contains two putative stem–loop structures: one in the 5′UTR (Dgcr8 200), and one in the ORF (Dgcr8 521). Although there are subtle differences in structure due to sequence variation, both stem–loops are present at the same locations in the human transcript. Interestingly, Han et al. (2009) showed that it was the 5′UTR stem–loop that was primarily processed in HEK293 cells. However, in mouse DN3 thymocytes, we found it was the ORF stem–loop that was primarily processed down to small RNAs, perhaps indicating that the activity of Drosha for the Dgcr8 mRNA may be differentially regulated in different cell types. Kadener et al. (2009) also reported a stem–loop structure within the 5′UTR of the Drosophila DGCR8 transcript. Although the structure of this stem–loop was substantially different from those in the 5′UTRs of the mouse and human transcripts, knockdown of DROSHA in S2 cells resulted in DGCR8 up-regulation, suggesting that direct DGCR8 mRNA cleavage by Drosha may also occur in the fly. Thus, this direct cleavage mechanism appears to be conserved in many species, and suggests that it must be an important mechanism of gene regulation.
Although purified Drosha could clearly cleave many mRNAs in vitro, this may not occur in all cell types in vivo. A Drosha-specific effect was obvious in DN3 thymocytes and MEFs, but was only a minor component in Tregs and activated CD4+ T cells. Despite Dgcr8 being ubiquitously expressed, mRNA levels were unaffected in Drosha-deficient Tregs. This suggests that the mRNA-cleaving activity of Drosha may vary between cell types. One possible explanation is that Drosha may have a lower affinity for mRNAs, and only when it is expressed at high levels, such as in early-stage thymocytes, are mRNAs also cleaved. A more interesting possibility is that there may be cell type-specific factors that regulate the affinity or activity of the Drosha complex for mRNAs versus pri-miRNAs. Several factors have been implicated in the modulation of Drosha-mediated pri-miRNA processing, including DEAD-box RNA helicases, Smads, and p53 (Davis and Hata 2009). It is possible that these or other cofactors, potentially cell type-specific, also regulate the activity of Drosha on mRNA substrates. Further studies will be required to clarify this issue.
Noncanonical mammalian miRNAs
Although mirtrons are clearly present in nematodes and flies (Okamura et al. 2007; Ruby et al. 2007), it has not been demonstrated whether they are also present in mammals. By deep sequencing, we showed in this study that there are numerous Drosha-independent, Dicer-dependent miRNAs in mice. However, the vast majority appeared to be derived from independent genes, or from long rather than short introns. Only miR-877 was derived from a short intron that would bypass the need for Drosha. Thus, it appears that there are very few mirtrons in mammals (at least in the cell types analyzed) that can be confirmed by cloning and sequencing. Moreover, orthologs of previously identified nematode and fly mirtrons have yet to be found in mammals (Griffiths-Jones et al. 2008).
One possible explanation for why there apparently remain so few mirtrons encoded in the genome of mammals is that introns have lengthened substantially with evolution (Lim and Burge 2001). In C. elegans and D. melanogaster, the majority of introns are ∼60 base pairs (bp) and 80 bp in length, respectively, and thus are equivalent to the size of pre-miRNA intermediates. However, in mammals, there is a broad distribution in intron lengths, from <100 bp to many kilobases.
The finding that most of the Drosha-independent miRNAs identified originated from long primary transcripts suggests that either there may be cryptic splice sites that facilitate the release of the short stem–loop structure, or there must be another enzyme that processes the transcript. Interestingly, all of the Drosha-independent miRNAs that we identified have been sequenced only in mammals or, specifically, in mice. There do not appear to be homologs of these miRNAs in C. elegans or D. melanogaster. Perhaps an enzyme distinct from Drosha that processes pri-miRNAs may have evolved later. There are some 73 proteins with demonstrated or predicted RNase activity in humans—many more than the 32 predicted in C. elegans (Ashburner et al. 2000). However, there are only two known enzymes capable of cleaving dsRNA to generate products with 5′ phosphate and 3′ hydroxyl termini: Drosha and Dicer. Further studies will be required to determine if one of the other RNase enzymes or some other protein is required for the processing of noncanonical Drosha-independent pri-miRNAs.
Our analysis of Drosha and Dicer deficiency in a variety of systems has led to the identification of novel functions of the miRNA processing machinery. Drosha-mediated mRNA cleavage, in particular, adds to an ever-increasing variety of post-transcriptional mechanisms of gene regulation. Such a variety of mechanisms highlights the importance of fine-tuning the expression of genes, rather than simply turning genes on or off. The additional activities of the two enzymes explain why, in some cell types, Drosha and Dicer deficiency did not always result in completely overlapping phenotypes. The finding in some cell types of little difference in phenotypes caused by Drosha and Dicer deficiency suggests that there may be important cell type-specific factors that modulate the activities of the miRNA machinery. Together, these data indicate that enzymes involved in miRNA pathways have additional complex mechanistic details that will need to be elucidated.
Materials and methods
Mice
RnasenF/F (Chong et al. 2008), Dicer1F/F (Harfe et al. 2005), Lck-cre (Lee et al. 2001), and CD4-cre (Wolfer et al. 2001) mice have been described previously. EIIA-cre, Gt(Rosa)26SorCreER, and Mir17∼92aF/F mice were purchased from Jackson Laboratories. RnasenΔ/+ and Dicer1Δ/+ germline null alleles were generated by breeding conditional RnasenF/F and Dicer1F/F mice with EIIA-cre transgenic mice (Jackson Laboratories) and subsequently breeding out the Cre transgene.
Microarray analyses
Total RNA was purified using Trizol (Invitrogen). Total RNA (0.1–1 μg) was labeled and hybridized to GeneChip Mouse Genome 430 2.0 arrays (Affymetrix) according to the manufacturer's protocol. Analyses were performed on cells FACS-purified from individual animals or prepared from individual embryos. Data mining was performed on the GeneSpring GX package (Agilent). One-way analyses of variance (ANOVA) were performed for statistical analyses. The *.cel raw data files can be provided on request.
SILAC
For the analysis of protein expression by SILAC, cells of each genotype were first cultured in medium deficient for L-Lys and L-Arg supplemented with 10% dialyzed FCS (Invitrogen) and different stable isotopes of L-Lys and L-Arg. “Light” medium contained natural L-Lys 12C614N2 and L-Arg 12C614N4, “medium” medium contained L-Lys 4,4,5,5-D4 (+4 Da) and L-Arg 13C614N4 (+6 Da), and “heavy” medium contained L-Lys 13C615N2 (+8 Da) and L-Arg 13C615N4 (+10 Da). Following at least five doublings in labeling media, the cells were lysed with 150 mM NaCl, 20 mM Tris (pH 7.5), 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, and protease inhibitors. Equal amounts of differently labeled lysates were mixed (based on protein concentration), separated by SDS-PAGE, digested with trypsin, and subjected to liquid chromatography–tandem mass spectrometry as described previously (Zhang et al. 2008). Protein identification and quantification were performed using Mascot and MaxQuant software as described previously (Cox and Mann 2008). One-way ANOVA were performed for statistical analyses. Please refer to the Supplemental Material for a detailed description of the SILAC method.
Luciferase reporter assays
The entire 3′UTRs of putative miRNA targets were cloned into the Firefly luciferase reporter plasmid pGL3-promoter (Promega). pri-miRNA transcripts were cloned into an MSCV retroviral vector. The Firefly luciferase reporter, the pri-miRNA retroviral vector, and a CMV-Renilla luciferase vector (as a transfection/normalization control) were cotransfected into HEK293T cells using TransIT-293 transfection reagent (Mirus). Forty-eight hours later, Firefly and Renilla luciferase reporter activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
qRT–PCRs
First strand reverse transcription was performed on total RNA with SuperScript III and oligo-dT (Invitrogen). cDNA from the equivalent of 25 ng of total RNA was then analyzed by qRT–PCR on a Roche LightCycler 480II using SybrGreen detection. All expression data were normalized to β-actin as a control. The primers used are listed in Supplemental Table S14. The measurement of mature miRNAs by qRT–PCR was performed using the NCode (Invitrogen) or TaqMan (Applied Biosystems) systems according to manufacturers' protocols, and normalized to U6 snRNA.
Small RNA deep sequencing
Small RNA libraries were constructed essentially as described (Hafner et al. 2008). In brief, 0.5 μg of total RNA was resolved on a polyacrylamide/urea gel, and the 19- to 30-nt small RNA fraction was isolated. RNA isolation and adaptor ligation were tracked by the inclusion of radiolabeled 19-nt and 30-nt synthetic oligoribonucleotides. Only small RNAs with 5′ phosphate and 3′ hydroxyl termini were cloned. To achieved this, a universal adaptor, preadenylated at the 5′ and carrying a 3′-OH-blocking group (miRNA cloning linker 1; Integrated DNA Technologies), was ligated to the 3′ end of the library using the RNA ligase 2 truncated mutant (1–249) K227Q (Hafner et al. 2008) in the absence of ATP. An Illumina-compatible adapter was then ligated to the 5′ end of the library, which subsequently allowed for deep sequencing on the Illumina GAII platform. The small RNA sequences were aligned to the mouse genome (mm9 assembly), then mapped to known miRNAs deposited in miRBase (Griffiths-Jones et al. 2008) or to other annotated genes in the University of California at Santa Cruz Genome Database (Rhead et al. 2010).
miRNA target and folding predictions
Prediction of miRNA target sites in the 3′UTRs of protein-coding mRNAs was performed using the TargetScan (Lewis et al. 2005), DIANA-microT (Maragkakis et al. 2009), and PicTar (Krek et al. 2005) databases. Folding predictions of putative RNA stem–loop structures were performed using Mfold (Zuker 2003).
In vitro RNA cleavage assays
The cleavage of RNA stem–loop structures by Drosha was analyzed essentially as described (Lee and Kim 2007). Briefly, stem–loop structures ±150 nt were cloned into the T7 promoter-containing vector pCR2.1. These were in vitro transcribed with T7 RNA polymerase in the presence of α-32PUTP. Flag-Drosha was immunoprecipitated from Flag-Drosha-transduced NIH3T3 cells with M2 anti-Flag agarose beads (Sigma), then incubated with the radiolabeled transcripts. Alternatively, whole-cell extracts prepared from RnasenF/F Gt(ROSA)26SorCreER MEFs were used to cleave radiolabeled transcripts. Drosha-deficient extracts were obtained by first inducing Rnasen deletion in the MEFs with 100 nM 4-OH tamoxifen. Following a phenol/chloroform cleanup, transcripts were run on a polyacrylamide gel to visualize cleavage products.
Accession numbers
Sequencing data have been deposited with the NCBI Gene Expression Omnibus under accession number GSE22760.
Acknowledgments
We thank Mary-Jean Sunshine for blastocyst injections; Michelle Rooks, Dick McCombie, and Diana Yang for performing the Illumina sequencing runs; Assaf Gordon for assistance with bioinformatic analyses; and Maria Ciofani and MacLean Sellars for critical reading of the manuscript. This work was supported by fellowships from the Cancer Research Institute (to M.M.W.C.) and Helen and Martin Kimmel Center for Stem Cell Biology (to M.M.W.C.), and grants from the National Institutes of Health (P30 CA106087 to T.A.N. and CA13106 to G.J.H.) the Australian National Health and Medical Research Council (637338 to M.M.W.C.), and the Howard Hughes Medical Institute (to G.J.H. and D.R.L.).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1953310.
Supplemental material is available at http://www.genesdev.org.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ 2003. Dicer is essential for mouse development. Nat Genet 35: 215–217 [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA 2007. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci 104: 18097–18102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Rasmussen JP, Rudensky AY, Littman DR 2008. The RNaseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med 205: 2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. 2010. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328: 1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT 2008. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci 105: 5614–5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hata A 2009. Regulation of microRNA biogenesis: A miRiad of mechanisms. Cell Commun Signal 7: 18 doi: 10.1186/1478-811X-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N 2008. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 doi: 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN 2004. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN 2009. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ 2005. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci 102: 10898–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT II, Nelson S, Rosbash M 2009. Genome-wide identification of targets of the drosha–pasha/DGCR8 complex. RNA 15: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. 2005. Combinatorial microRNA target predictions. Nat Genet 37: 495–500 [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim VN 2007. In vitro and in vivo assays for the activity of Drosha complex. Methods Enzymol 427: 89–106 [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15: 763–774 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB 2003. Prediction of mammalian microRNA targets. Cell 115: 787–798 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lim LP, Burge CB 2001. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci 98: 11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. 2009. DIANA-microT web server: Elucidating microRNA functions through target prediction. Nucleic Acids Res 37: W273–W276 doi: 10.1093/nar/gkp292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci 102: 12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ 2007. Critical roles for Dicer in the female germline. Genes Dev 21: 682–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L, Meister G 2007. Argonaute proteins: Mediators of RNA silencing. Mol Cell 26: 611–623 [DOI] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 38: D613–D619 doi: 10.1093/nar/gkp939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Shenoy A, Blelloch R 2009. Genomic analysis suggests that mRNA destabilization by the microprocessor is specialized for the auto-regulation of Dgcr8. PLoS ONE 4: e6971 doi: 10.1371/journal.pone.0006971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S 2007. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2: 3081–3089 [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA 2007. Maternal microRNAs are essential for mouse zygotic development. Genes Dev 21: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Wolfer A, Bakker T, Wilson A, Nicolas M, Ioannidis V, Littman DR, Lee PP, Wilson CB, Held W, MacDonald HR, et al. 2001. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat Immunol 2: 235–241 [DOI] [PubMed] [Google Scholar]
- Zhang G, Fenyo D, Neubert TA 2008. Screening for EphB signaling effectors using SILAC with a linear ion trap-orbitrap mass spectrometer. J Proteome Res 7: 4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]