Abstract
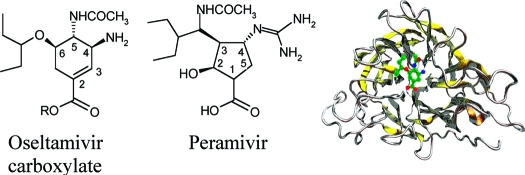
We have identified a virus, B/Perth/211/2001, with a spontaneous mutation, D197E in the neuraminidase (NA), which confers cross-resistance to all NA inhibitors. We analyzed enzyme properties of the D197 and E197 NAs and compared these to a D197N NA, known to arise after oseltamivir treatment. Zanamivir and peramivir bound slowly to the wild type NA, but binding of oseltamivir was more rapid. The D197E/N mutations resulted in faster binding of all three inhibitors. Analysis of the crystal structures of D197 and E197 NAs with and without inhibitors showed that the D197E mutation compromised the interaction of neighboring R150 with the N-acetyl group, common to the substrate sialic acid and all NA inhibitors. Although rotation of the E275 in the NA active site occurs upon binding peramivir in both the D197 and E197 NAs, this does not occur upon binding oseltamivir in the E197 NA. Lack of the E275 rotation would also account for the loss of slow binding and the partial resistance of influenza B wild type NAs to oseltamivir.
Introduction
The influenza virus neuraminidase (NA,a EC 3.2.1.18) functions in virus infection to remove sialic acid from receptors present on the surface of host cells. In the absence of NA activity, the ability of progeny virions to spread to uninfected cells is compromised. The structure of the catalytic headgroup of influenza A NA has been known since 1983,1,2 consisting of individual subunits of six-bladed β-propellers that form a boxlike tetramer with dimensions 100 Å × 100 Å × 60 Å. Structures of the influenza B/Beijing/1/873,4 and B/Lee5 NAs showed that the tetrameric head/β-propeller topology of the influenza-A NAs was conserved in influenza B NAs.
Analysis of NA crystal structures in complex with the substrate sialic acid resulted in the development of zanamivir (1)6 and oseltamivir (oseltamivir carboxylate is the active ingredient used here (2)).7 A further inhibitor, peramivir (3), is in clinical trials8,9 (Figure 1). These compounds are active against all influenza A and B viruses. While previously there have been reports of resistance from both influenza A and B viruses isolated from both immunocompromised and immunocompetent patients after treatment with 2,10−14 more recently the global spread of seasonal influenza A H1N1 strains resistant to 2(15,16) has been observed, although this appears to be unrelated to the use of 2. Furthermore, resistance to 2 is emerging in strains of the pandemic H1N1/09 viruses either with or without treatment or prophylaxis.17,18 In contrast, resistance after 1 treatment has only been reported in an immunocompromised patient infected with an influenza B strain.19 Influenza B viruses with a D197N mutation (D198 N2 numbering) have been isolated from an immunocompromised patient treated with 2(10) and arisen either spontaneously or by possible transmission from a treated patient.13 The D197N mutant NA shows decreased binding to both 1 and 2, thus demonstrating the importance of D197 in the influenza B NAs for tight binding of the NA inhibitors. Unlike other residues that confer resistance, D197 is not absolutely conserved across influenza A and B NAs, as analyses of NA sequences in the databases show that wild type influenza A N7 and N9 subtype NAs have N197. Residue D197 does not interact directly with substrate or inhibitor in the NA but engages in a salt bridge interaction with R150 (R152 N2 numbering), which forms a hydrogen bond with the N-acetyl group of sialic acid and the NA inhibitors. An influenza B virus with an R150K NA mutation was isolated after prolonged treatment of an immunocompromised child with 1(19) with a significant impact on enzyme activity and cross-resistance to other NA inhibitors, clearly demonstrating the importance of interactions of the R150 with the substrate and inhibitors.
Figure 1.
Neuraminidase inhibitors used in the study.
The B/Perth/211/2001 (B/Perth) virus was isolated from an infant with no history of treatment with or contact with NA inhibitors. The sample contained both wild type and mutant viruses with a D197E mutation in the NA. The mutant NA had reduced sensitivity to 1, 2,and 3.20 When expressed in insect cells, recombinant B/Perth wild type and mutant NAs had properties similar to those of the virus associated NAs. As we were unable to culture these viruses in eggs, we describe here the use of this recombinant NA for structural studies.
The clinical effectiveness of 2 against influenza B infection in children is reported to be less than against influenza A.21,22 However, until now, data on the structure of an influenza B NA with oseltamivir bound have not been available. We present here structures of B/Perth/211/2001 NAs with D (B/Perth D) or E (B/Perth E) at position 197 in the apo form and in complex with 3 and provide an insight into the mechanism of resistance of mutations at D197. Furthermore, the structure of the B/Perth E complex with 2 is presented, and from these data we propose the mechanism of reduced binding of 2 in wild type influenza B NAs.
Results
Protein Purification
We wished to determine the impacts that the mutations at D197 had on the structure and function of the mutant proteins in order to understand the mechanisms of resistance to all NA inhibitors. We previously reported that the native B/Perth wild type D197 and mutant E197 NAs expressed in insect cells had similar resistance profiles as the influenza virus associated NAs.20 However, the recombinant NAs had no tags to facilitate purification. Hence, we developed conditions for cleavage of the membrane anchor and stalk regions and for purification of the NA from the insect cells. Acetone fixation of the cells enabled us to store them with no effect on the enzyme function. Digestion with either pronase or trypsin showed that optimal cleavage was obtained with trypsin at 2 mg/mL, between residues K69 and G70 in the stalk, comparable to cleavage of NA heads from other influenza B viruses.3,23
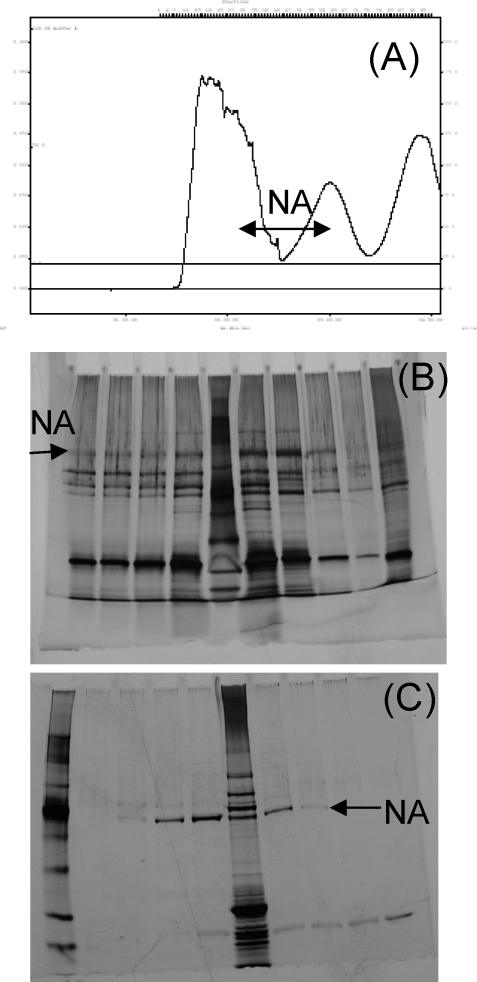
After separation of the digests by Superose-12 and lentil lectin affinity chromatography, PAGE analysis showed a single band corresponding to the NA (Figure 2). Yields of purified protein were approximately 100 μg/L.
Figure 2.
Purification of recombinant B/Perth NA heads by trypsin digestion of SF21 cells expressing the full length B/Perth NA: (A) gel filtration profile of crude extract; (B) silver stained PAGE of gel filtration fractions; (C) samples after lentil lectin affinity chromatography.
Electron Microscopy
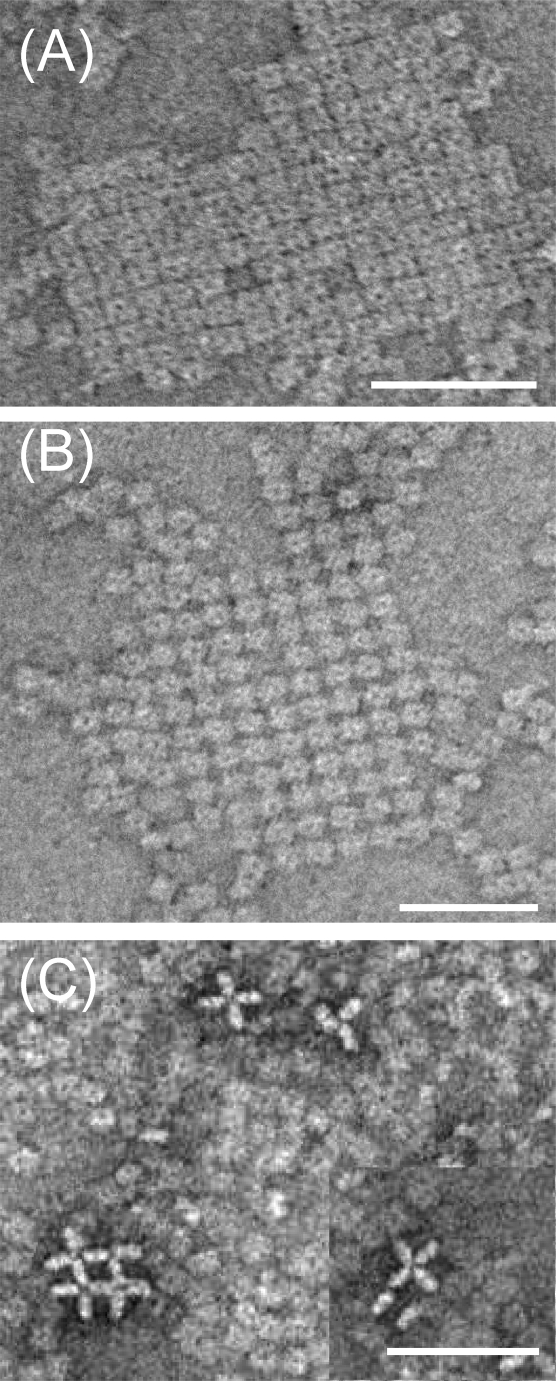
Since this was the first time we had attempted to use recombinant NA for structural studies, we used electron microscopy, as previously reported,24 to examine the integrity of NA heads. The B/Perth D heads were clearly tetrameric but interestingly spontaneously formed two-dimensional arrays on the carbon substrate. Three different patterns were observed (Figure 3). The most common was a simple square tessellation, and the second form consisted also of orthogonal rows and columns but with the square NA heads rotated through 45°, forming a “hound’s-tooth” pattern. The third and least common form had NA heads oriented on their sides or edges, arranged in an open square lattice. These first two forms are a rare example of actually seeing how the protein packs in crystalline arrays, corresponding to crystallographic arrangements subsequently detected by X-ray crystallography.
Figure 3.
Electron micrographs of recombinant B/Perth D NA heads. Samples were negatively stained with uranyl acetate and show two-dimensional arrays of NA heads: (A) simple square tessellation; (B) “hound’s-tooth” pattern of orthogonal rows and columns but with the square NA heads rotated through 45°; (C) an open square lattice with NA heads oriented on their sides or edges. Scale bars, 50 nm.
Enzyme Studies
We have previously shown that the B/Perth E197 NA and the B/Yamagata N197 NA demonstrate cross-resistance to the NA inhibitors 1, 2, and 3.20,25,26 Some influenza NAs demonstrate time dependent binding of the NA inhibitors often called slow binding.27 Many NAs with a mutation in the active site are reported to have lost this property, binding the inhibitors rapidly.27−29 Since D197 does not directly interact with substrate or inhibitor, we therefore were interested in whether the D197E or D197N mutations affected the enzyme function or the rate of binding of the inhibitors.
Comparisons of the activity of the purified recombinant B/Perth D197 and E197 NA proteins demonstrated that the specific activity of the mutant E197 NA was approximately 70% of that of the wild type D197 enzyme.
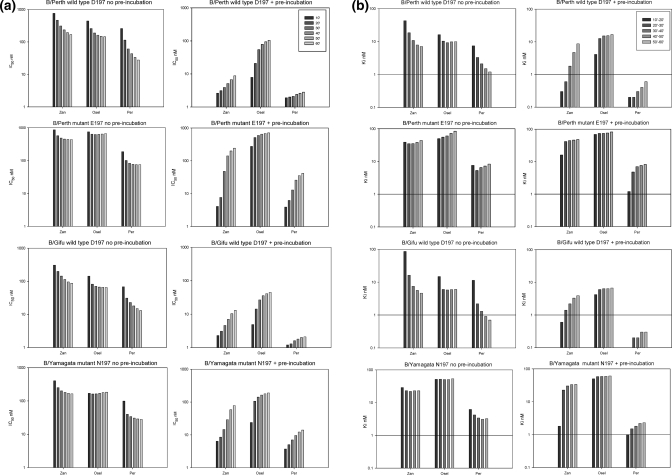
For comparing values for the Km, Ki, and IC50, we used detergent extracts of each of the four influenza viruses grown in cell culture, since we did not have recombinant NA from either the B/Gifu or B/Yamagata viruses. The B/Perth wild type D197, mutant E197, and the wild type D197 B/Gifu NAs had similar Km values for MUNANA of 12.4 ± 4.2, 12.1 ± 3.7, and 10.5 ± 1.4 μM, respectively. Although the D197N mutation has less impact on IC50 than the E197 (Table 1), it had a slightly higher Km of 18.8 ± 4.5 μM.
Table 1. Comparison of IC50 Values of Wild Type and Mutant NAs after 60 min in MUNANA Based Enzyme Inhibition Assay with or without Preincubation with Inhibitorsa.
| IC50, nM |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D197 Perth |
E197 Perth |
D197 Gifu |
N197 Yamagata |
|||||||||
| Zan | Osel | Per | Zan | Osel | Per | Zan | Osel | Per | Zan | Osel | Per | |
| no prec | 167.4 | 144.2 | 27.8 | 433.8 | 659.7 | 75.2 | 87.0 | 65.0 | 13.0 | 163.9 | 182.5 | 28.1 |
| fold resistanceb | 2.6 | 4.6 | 2.7 | 1.9 | 2.8 | 2.2 | ||||||
| prec | 8.8 | 104.4 | 2.8 | 242.0 | 708.0 | 41.5 | 13.0 | 45.0 | 2.1 | 77.8 | 188.4 | 14.0 |
| fold resistanceb | 27.5 | 6.8 | 17.3 | 6.0 | 4.2 | 6.7 | ||||||
| no pre/prec | 19 | 1.4 | 9.9 | 1.8 | 0.90 | 1.8 | 6.7 | 1.4 | 6.2 | 2.1 | 1.0 | 2.0 |
Results are the mean of duplicates.
Fold resistance is the ratio of the mutant to the wild type IC50 values.
No pre or pre is the fold differences in the final IC50 values with no preincubation with inhibitor compared to 30 min of preincubation.
We carried out two different experiments for each virus/drug combination to study the rate of inhibitor binding. The first experiment had no preincubation with the inhibitors, which enabled us to examine the rate of association of the drug with the NA, and the second assay had a 30 min preincubation, which enabled us to examine whether any further association or dissociation of the NA−inhibitor complex occurred upon addition of substrate. Both used a 60 min reaction with substrate. Others have used preincubation times from 10 min to 2 h,6,29,30 and it is not clear what impact these preincubation times, or the subsequent reaction times with substrate, may have on the calculated IC50. Since the IC50 is also known to vary with substrate concentration, others have calculated Ki values in addition to the IC50, since Ki values are meant to be a more invariant measure of affinity for an inhibitor independent of substrate concentration. We therefore compared IC50 and Ki values at each 10 min interval between 10 and 60 min either with no preincubation or after preincubation with the inhibitors.
When the curves for the total FU versus elapsed time were compared, there were three types of curves. The first type of curve (Figure 4A) seen after preincubation with 1 or 3 in the wild type D197 NAs showed a gradual increase in the rate of reaction, indicating slow dissociation of inhibitor. The second type of curve where there was no preincubation of inhibitor, with 1 and 3 in the wild type NAs, showed a gradually decreasing rate of reaction (Figure 4B), indicating slow association of the inhibitor. The third type of curve reached a constant rate during the reaction (Figure 4C) and was seen with 2 in the D197 NAs and with all inhibitors with the mutant NAs, indicating that rapid equilibrium had been reached.
Figure 4.
Neuraminidase activity changes in the presence of inhibitor using a MUNANA based fluorescent assay. MUNANA was added to detergent treated wild type and mutant viruses either after a 30 min preincubation or without incubation with inhibitors ranging from 10 000 to 0.1 nM. Activity was monitored for 60 min after addition of substrate: (A) B/Perth D activity in 1 after 30 min of preincubation showing an increase in rate with time, corresponding to slow dissociation; (B) B/Perth D activity in 1 without preincubation, showing a decrease in rate with time, corresponding to slow association of 1; (C) B/Yamagata (D197N) activity in 1 without preincubation, showing a constant rate, corresponding to a rapid association of 1.
The effects these changing rates have on the IC50 and Ki values are shown in Figure 5. For both wild type D197 NAs without preincubation there was a gradual decrease in IC50 corresponding to a slow association of both 1 and 3. After preincubation with these inhibitors there was a gradual increase in IC50, indicating slow dissociation. In contrast there was rapid association of 2 without preincubation with little change in IC50 after the first 20 min. There was also rapid dissociation of 2 after preincubation. Although the initial IC50 values were not that much higher than for 1, there was more than a 10-fold change over the 60 min reaction time. Because of the slow association of 3 and 1 in the wild type NAs, the final 60 min IC50 values with no preincubation were still 6- to 19-fold higher than with preincubation (Table 1), demonstrating that the inhibitor binding had not yet reached equilibrium. In contrast, there was less than a 2-fold difference between the 60 min no preincubation and preincubation IC50 values for 2 in both wild type NAs.
Figure 5.
Effect of incubation times after addition of substrate and inhibitor on (a) IC50 and (b) Ki values. IC50 values were calculated after each 10 min interval as the drug concentration causing 50% inhibition compared to the uninhibited control. Ki values were calculated based on the slope between consecutive 10 min time intervals. Final IC50 values after 60 min for 1 and 2 in wild type B/Perth and B/Gifu NAs were much higher without preincubation compared to 30 min preincubation, reflecting the slow binding of these two inhibitors. There was much less change in IC50 values for the binding of 2 in the wild type and 1, 2, and 3 in the mutant NAs, demonstrating loss of slow binding. Conversely 2 dissociated rapidly from the wild type NA, in contrast to slow dissociation of 1 and 3, and 1, 2, and 3 all dissociated more rapidly from the mutant B/Perth E and B/Yamagata NAs. (b) Ki values also changed with time, reflecting the differences in the slow or fast binding or dissociation of the inhibitors, with less change seen with 2 in the wild type and 1, 2, and 3 in the mutant NAs.
When the IC50 values are compared without preincubation for all inhibitors for both mutant NAs, the graphs show there is much less change in IC50 after the first 10 min. 2 also appears to bind even more rapidly to both mutant NAs than to the wild type D197 NAs. There is also faster dissociation of 1 and 3 after the NAs are preincubated with inhibitor, compared to the wild type NAs. For all inhibitors for both mutant NAs the ratios after 60 min of the no preincubation to preincubation are all within 2-fold, indicating much more rapid equilibration of all inhibitors compared to the wild type NAs.
The slow binding of the 1 and 3 in the wild type D197 NAs also resulted in the Ki values continuing to change over the 60 min period, whereas for 2 in the wild type and all the inhibitors with the mutant NAs the Ki values stabilized much more quickly after the first 10−20 min period, as seen in Figure 5 and Tables 1 and 2. Therefore, the reaction time affected both the Ki and IC50 values. This means that the “fold resistance”, often used to describe how resistant the isolates are, will vary significantly during the course of the reaction, depending on whether the inhibitors are faster or slower binding or dissociating compared to the wild type.
Table 2. Comparison of Ki Values of Wild Type and Mutant NAs after 60 min in a MUNANA Based Enzyme Inhibition Assay with or without Preincubation with Inhibitorsa.
|
Ki, nM |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D197 Perth |
E197 Perth |
D197 Gifu |
N197 Yamagata |
|||||||||
| Zan | Osel | Per | Zan | Osel | Per | Zan | Osel | Per | Zan | Osel | Per | |
| no pre | 7.1 | 9.9 | 1.2 | 43.8 | 84.0 | 8.3 | 4.6 | 6.0 | 0.7 | 22.8 | 53.4 | 3.2 |
| fold resistanceb | 6.2 | 8.5 | 6.9 | 5.0 | 8.9 | 4.6 | ||||||
| pre | 8.7 | 16.6 | 0.6 | 48.0 | 82.0 | 8.2 | 3.9 | 6.7 | 0.3 | 33.5 | 60.7 | 2.3 |
| fold resistanceb | 5.5 | 4.9 | 13.7 | 8.6 | 9.1 | 7.7 | ||||||
Results are the mean of duplicates.
Fold resistance is the ratio of the mutant to the wild type Ki values.
Crystallography
The apo form of B/Perth D crystallized in space group I4 and was solved by molecular replacement using A/tern/Australia/G7OC/75 N9 NA (PDB code 7NN9) followed by automatic rebuilding using PHENIX; the R and Rfree were 33% and 39%, respectively. Several cycles of model building and refinement gave a high quality final model (Table 3). Crystal contacts between subunits in the C-direction are formed entirely by carbohydrate-mediated interactions. A carbohydrate chain composed of GlcNAc and mannoside residues was appended to residue N284, consistent with known patterns of N-linked oligosaccharides in insect cell lines.31 The asymmetric unit contained two monomers, each adjacent to a 4-fold crystallographic axis. Thus, two tetramers are generated from each of the two monomers. The packing of tetramers in layers observed in this crystal form appears similar to that observed in electron microscopy (Figure 3A). The subunit structure is similar to previously determined influenza B NA structures. It adopts the classic β-propeller arrangement with six four-stranded β-sheets. The B/Perth D structure superimposes with a rmsd of 0.32 Å (over 388 Cα atoms) and 0.28 Å (over 385 Cα atoms) with B/Lee/40 (PDB 1INV) and B/Beijing/1/87 (PDB code 1NSB) NAs, respectively, illustrating the high degree of structural conservation in influenza B NAs. The active-site residues of all three strains are identical. The active site of B/Perth D contained water molecules and a sulfate group bound between the guanidinium moieties of R116, R292, and R374.
Table 3. X-ray Dataa.
| B/Perth D | peramivir B/Perth D |
B/Perth E | peramivir B/Perth E |
oseltamivir B/Perth E |
|
|---|---|---|---|---|---|
| X-ray source | AS-PX-1b | AS-PX-1b | AS-PX-1b | AS-PX-1b | PF-17Ab |
| space group | I4 | I4 | P1 | P1 | P1 |
| unit cell | |||||
| a, Å | 87.6 | 88.7 | 111.2 | 111.96 | 111.8 |
| b, Å | 87.6 | 88.7 | 123.7 | 124.87 | 123.8 |
| c, Å | 197.2 | 207.5 | 123.8 | 125.25 | 124.0 |
| α, deg | 90.0 | 90.0 | 90.0 | 90.0 | 90.0 |
| β, deg | 90.0 | 90.0 | 90.2 | 92.1 | 90.2 |
| γ, deg | 90.0 | 90.0 | 90.2 | 91.2 | 90.1 |
| resolution range (Å) | 26.7−2.20 (2.32−2.20) | 40.7−2.00 (2.11−2.00) | 62−2.2 (2.32−2.20) | 63−2.6 (2.74−2.6) | 87−2.6 (2.74−2.6) |
| Rmerge c (%) | 12.2 (43.8) | 12.8 (33.5) | 10.2 (36.5) | 17.6 (40.6) | 10.4 (35.8) |
| ⟨I/σ(I)⟩ | 11.6 (2.7) | 10.1 (3.8) | 8.1 (2.6) | 5.9 (2.9) | 6.0 (1.9) |
| unique observations | 34306 (4204) | 47163 (7141) | 316772 (45355) | 201957 (28682) | 199227 (29004) |
| completeness | 91.5 (77.6) | 87.6 (90.9) | 94.6 (92.7) | 96.9 (94.3) | 97.4 (96.7) |
| multiplicity | 4.5 (2.9) | 4.6 (4.1) | 2.0 (1.8) | 3.6 (3.2) | 2.0 (2.0) |
| R, Rfree d | 18.2, 23.9 | 17.1, 23.7 | 19.1, 22.0 | 19.8, 21.4 | 20.7, 22.8 |
| Root-Mean-Square Deviations from Ideal Geometry | |||||
| bond lengths (Å) | 0.014 | 0.014 | 0.017 | 0.016 | 0.016 |
| bond angles (deg) | 1.560 | 1.625 | 1.513 | 1.560 | 1.610 |
| torsion angle (deg) | 7.942 | 7.638 | 7.11 | 7.061 | 7.00 |
| chiral volume (Å3) | 0.091 | 0.115 | 0.104 | 0.102 | 0.104 |
| planar group (Å) | 0.006 | 0.006 | 0.007 | 0.006 | 0.007 |
Numbers in parentheses correspond to the highest resolution bin.
AS-MX1 is beamline MX-1 at the Australian Synchrotron. PF-17A is beamline 17A at the Photon Factory, Tsukuba, Japan.
Rmerge = ∑hkl∑i|I(hkl)i − ⟨I(hkl)⟩|/∑hkl∑iI(hkl)i.
R =∑hkl|Fo(hkl) − Fc(hkl)|/∑hkl|Fo(hkl)|, where Fo and Fc are the observed and calculated structure factors, respectively.
For the B/Perth E crystals, although the unit cell suggested tetragonal symmetry, good merging statistics were obtained only in space group P1. The B/Perth E structure was solved by molecular replacement using tetramers of B/Perth D as a search model. MOLREP selected a radius of integration of 61 Å, and four tetramers were found with peak heights of 21.4, 18.9, 18.0, and 15.3 σ. Solutions for all four monomers were found in the translation function, resulting in an initial model with R and Rfree factors of 31.2% and 31.1%. Refinement proceeded using tight NCS restraints. The 16-fold NCS-averaged maps were used for model building in COOT. Refmac detected pseudomerohedral twinning, and corrections were applied accordingly. The structure of B/Perth E is highly similar to the B/Perth D structure in spite of the different crystal form. The tetramers form layers that resemble the “hound’s-tooth” pattern described above from electron microscopy (Figure 3B). A sulfate group is bound between the guanidinium moieties of R116, R292, and R374, and a yttrium ion of partial occupancy is bound adjacent to the sulfate group. These groups are displaced by 2 or 3 upon soaking with those inhibitors (see below).
Crystals of B/Perth D soaked with 3 adopted similar packing to the apoenzyme. 3 was included in the model at a late stage of refinement, and the final model is of high quality (Table 3).
Soaking inhibitors into crystals of B/Perth E proved challenging, with relatively weak data being obtained for crystals of this isozyme in the presence of 3 and 2 (Table 3). Nevertheless, the structure refinement and map interpretation were aided by 15 noncrystallographic symmetry (NCS) restraints. Averaged electron density maps allowed for clear and unambiguous interpretation of the structures including, where present, inhibitors. Refinement of the B/Perth E complex with 3 commenced using the apo-form of this mutant as the starting model (R = 40.5%, Rfree = 40.4%). 3 was included in the model at a late stage of refinement. Similarly, refinement of the B/Perth E 2 complex commenced using B/Perth E apo structure as the starting model (R = 34.1%, Rfree = 35.3%). For both complexes tight NCS restraints were used throughout. Pseudomerohedral twinning was detected in both cases and corrected in Refmac. Final model statistics for all models are in Table 3.
Electron density for all inhibitor complexes is unambiguous. 3 binds in a similar fashion to related inhibitors observed in previously determined B/Beijing and B/Lee structures. The carboxylic acid group lies in the pocket formed by R292, R374, and R116. The guanidinium group is buried in a pocket formed by E149 and E117. The sec-pentyl moiety is stacked against the E275-Cβ group (E276 N2 numbering) (Figure 6B). Upon inhibitor binding, E275 must rotate away from the inhibitor in a manner analogous to that described previously for B/Beijing NA in complex with dihydropyranphenethylpropylcarboxamide.32 This inhibitor has an ethyl moiety that corresponds to part of the sec-pentyl group of 3.
Figure 6.
Comparisons of the active sites of B/Perth wild type and mutant NAs uncomplexed and with bound inhibitors (A, B) B/Perth wild type D and (C, D, E) B/Perth mutant E structures. Apo (A, C) and 3-bound (B, D) forms are shown. B/Perth E in complex with 2 is shown (E). (F) A model of the D197N mutant based on the wild-type B/Perth structure is shown. Active-site residues are shown in stick form and the backbone in cartoon form. Arrow shows rotation of the E275 upon binding of 3.
Surprisingly, rotation of E275 is not observed in the B/Perth E complex with 2, which does not form any hydrophobic contacts with E275. Instead, the sec-pentyl group makes less favorable contacts with the charged portions of R223, E275, and R292 (Figure 6E). In this structure, there is only partial rotation of E275 away from the active site and hence only partial insertion of one arm of the sec-pentyl moiety into the resulting hydrophobic cleft (Figure 6D).
The D197E mutation in B/Perth affects the way the carboxylic acid group of this residue engages with R150. In the structure of B/Perth D determined in the absence of inhibitor, the carboxylic acid group of D197 engages side-on with the guanidinium group of R150 as seen in most influenza B NA structures. In the B/Perth E apo structure, the guanidinium group of R150 is rotated to engage in a stacking interaction with the carboxylic acid moiety of E197. Furthermore, the guanidinium group has rotated 180° so that the Nη1-atom is now pointing away from the active site (Figure 6C). In the structure of B/Perth E with 3, R150 has rotated toward the active site relative to its position in the apo structure and engages in a hydrogen bond with the N-acetyl oxygen atom via the Nε-atom. The distances of the R150 to N-acetyl hydrogen bonds are longer in B/Perth E compared with P/Perth D: 3.4 Å versus 2.7 Å, respectively. In the complex of B/Perth E with 2, R150 is in the conformation observed in B/Perth D, with atom Nη1 engaging in a hydrogen bond with the inhibitor N-acetyl oxygen atom (2.6 Å). While the distance is not significantly different from the equivalent distance in the 3 complex, the R150 guanidinium group and N-acetyl group are no longer coplanar, indicating a geometrically less favorable and hence weakened interaction.
Inhibition with 2,3-Difluoro KDN (4)
As an additional way of demonstrating that the reduced binding of the inhibitors in the D197E and D197N NAs was due to altered interactions with the N-acetyl group of the sugar ring, we compared inhibition of all four NAs with 2,3-difluoro-2-keto-3-deoxy-d-glycero-d-galactononulosonic acid 4.33 Although it is only a weak inhibitor, it has no N-acetyl group; hence, values should be similar for wild type and mutant NAs if this interaction can no longer occur.
There was no resistance to 4 with the mutant NAs compared to the D197 wild type NA. In fact the IC50 for each mutant was less than for the wild type pair, B/Perth E197 NA 19.4 ± 1.7 μM compared to the wild type 37.7 ± 1.7 μM and the B/Yamagata N197 NA 41.6 ± 0.4 μM compared to the B/Gifu wild type of 134 ± 17 μM, respectively. This confirmed that decreased sensitivity was due solely to altered interactions with the N-acetyl group.
Discussion and Conclusions
We have used structural and functional studies here to gain an understanding of the mechanism of resistance to the NA inhibitors of influenza B viruses with mutations at residue 197. Equally important, our studies provide insights into why influenza B wild type NAs have reduced binding of 2 compared to influenza A NAs.
We demonstrate that although D197 does not interact directly with substrate or inhibitors, mutations of D197E and D197N in influenza B alter binding of substrate and all three NA inhibitors 1, 2, and 3, as shown by decreased specific activity and increased Ki and IC50 values. The D197E mutation also conferred greater resistance than the D197N mutation. We also demonstrate using a modified approach to the enzyme inhibition assay that the reaction time can significantly affect both the Ki and IC50 differently for wild type and mutant NAs, depending on whether the inhibitor is fast or slow binding. Others6 have also reported variation in Ki over time, due to the time dependent slow binding of 1. They observed a 10-fold decrease in Ki over the course of their reaction, with a 10 min preincubation and 15 min reaction time after addition of substrate. Because of the shorter preincubation time in their case, 1 was obviously still binding, rather than in our case where after a 30 min preincubation we start to see dissociation. Thus, despite Ki values being thought to be more consistent, because of the variation in the methods used by different laboratories6,34,35 and the impact of fast and slow binding with time of incubation, the Ki values would appear to be more suitable as relative values within a laboratory for comparing enzyme properties of wild type and mutant NAs rather than as absolute values that different laboratories can directly compare.
We have also shown that the end point IC50 experiment can be adapted to “IC50 kinetics” experiments by simply monitoring changes in IC50 with time without the addition of stop solution. Comparison of the changes in IC50 with preincubation or no preincubation with inhibitor shows that this approach can provide additional information about impacts of mutations on the rates of inhibitor binding and dissociation, compared to just a single end point IC50. This is an assay that can be carried out in any laboratory with a modern fluorimeter, without the need for any additional equipment. Laboratories globally compare IC50 values, and we are striving to understand parameters that can affect the IC50 values to enable a better comparison of IC50 values from different laboratories. Clearly both the preincubation and incubation times are critical.
Structural analysis indicates that the effect of the D to E mutation at position 197 was to destabilize the interaction with R150 and to reduce the stability of this crucial inhibitor-binding residue. It appears that the D197 to E mutant prefers a conformation in which the guanidinium group of R150 is rotated and moved slightly out of the active site but can rotate back so as to engage with an inhibitor. However, this interaction is less stable than in the D197 form. All clinically approved inhibitors possess an N-acetyl group that interacts with R150 or its equivalent and are potentially susceptible to this mechanism of drug resistance. This is consistent with the cross-resistance seen to 1, 2, and 3. Lack of resistance to another inhibitor without the N-acetyl group, 4, also confirmed that resistance was due to altered binding to the N-acetyl group.
The mechanism of resistance in B/Perth E is different from that observed in N1 and N9 NAs to date.27,35−37 In both the N1 NA with an H274Y mutation and the N9 NA with an R292K mutation the altered binding is due to altered interactions of the E276 (N2 numbering) with the isopentyl ether group on 2. The influence of these mutations on the binding of 1 is less pronounced, as the side chain of E276 is able to maintain favorable interactions with the glycerol side chain. In contrast, reduced binding of 1 with an E119G mutant NA is partly due to reduced interactions with the guanidinium group at the 4 position on the sugar ring.27 The reduced binding in the D197 mutant NAs is due to altered interactions with yet another part of the ring, the N-acetyl group, which is common to all inhibitors and substrate. The viability of the D197N mutation demonstrates that a salt bridge between this residue and R150 is not essential for function. We anticipate that the effect of the D197N mutation in B NAs is to weaken the interaction of this residue with the R150 guanidinium group through the elimination of the salt bridge interaction of D197 and R150. N197 could still interact through a hydrogen bond between the Oε1 atom and the Nε or Nη2 groups of R150. This will have a similar effect to the D197E mutation in that it affords more flexibility to the R150 side chain and weakens the interaction with the N-acetyl group of the inhibitors (modeled in Figure 6F).
While NAIs are described as time dependent slow binding inhibitors, we have shown here that both D197E and D197N lead to loss of slow binding of 1, 2, and 3. Loss of slow binding is generally associated with mutations in the NA active site, leading to NA inhibitor resistance.27 One proposed mechanism of slow binding is due to the need for the rotation of the E27530 in inhibitors with the modified glycerol side chain.29,38 A proposed mechanism for the slow binding of 1 is the slow release of a water molecule by the guanidinium group.39 This would also apply to 3 which shares this group. However, we see here a loss of slow binding in the NAs with mutations at D197, remote from the position occupied by the guanidinium groups in both 3 and 1, and rotation of the E275 still occurs upon binding 3. Thus, slow binding of the NA inhibitors is clearly affected by more than interactions in either the vicinity of the 4′-guanidino group or the rotation of the E275.
Although in the E197 mutant NA the binding of 1 was reduced by nearly 30-fold compared to about 7-fold reduction for 2, the overall IC50 for 2 was higher. This is due to the lower sensitivity, or higher IC50, of the wild type B/Perth NA for 2(40) before the additional mutation.13 As the levels of 1 delivered to the upper respiratory tract after 10 mg doses are estimated to be up to 10 000 nmol/L,41 this would still be more than 50-fold higher than the IC50 of the E197 enzyme. In contrast the plasma levels of 2 are estimated to range from 400 to 1200 nmol/L,42 and levels in saliva are estimated to be less than 5% of plasma levels.43 Thus, with a potential level in the upper respiratory tract of only 20−60 nM,44 efficacy of 2 against a similar D197E mutant strain could be significantly reduced.
In addition to loss of slow binding of the mutant NAs to 1, 2, and 3, our enzyme analyses show here that there is a loss of slow binding of 2 to the wild type B/Perth and B/Gifu D197 enzymes compared to binding of 1 and 3, as well as faster dissociation of 2. This is consistent with the lower sensitivity or partial resistance of wild type influenza B strains to 2 in enzyme assays compared to influenza A strains, especially in the MUNANA assay where the IC50 values are around 12−70 nM (compared to the NA-Star assay, ∼2−11 nM 40) and compared to an IC50 of around 0.5−2 nM for influenza A strains in both assays. The loss of slow binding is consistent with the observations of Baum and colleagues,29 although Kati et al. had described 2 to be also slow binding in influenza B viruses.30
Consistent with our enzyme observations, we also importantly present structural evidence to explain the partial resistance of influenza B NAs to 2. Upon binding 2, residue E275 of B/Perth NA fails to rotate to allow binding of the sec-pentyl moiety to the aliphatic portion of this residue as observed in the equivalent residue (E276) in N1 and N9 NAs.36 Rotation of this residue is necessary for high affinity binding of 2, and failure to occur is consistent with resistance to 2, seen in other mutant NAs.35,36 While we were unable to obtain the structure of B/Perth D with 2, a previous publication also describes lack of full rotation of the E275 in the B/Lee wild type NA.45 It may be a general feature of type B NAs that this part of the active site is more rigid and E275 is less able to rotate to accommodate hydrophobic groups, although rotation does occur upon binding 3. The floor of the active site of B-type NAs has been described as being more sterically crowded than for A-type enzymes, indicating that residues in type-B NA might be tightly constrained to the observed positions in the uncomplexed enzyme.32 The observations presented here appear to confirm this view.
We conclude that the rotation of residue E275 needed for high affinity binding of 2 does not occur in the current strains of influenza B wild type NAs, and this would correlate with the loss of slowing binding of 2, the higher IC50 values seen especially in the MUNANA assay, and possible decreased clinical efficacy of 2 in children.21,22
Experimental Section
Viruses and Inhibitors
Isolation of the B/Perth viruses with wild type D197 NA and mutant E197 NA has been previously described.20 We also wanted to investigate how the D197N mutation affected the NA enzyme properties. For this purpose we obtained the B/Yamagata/186/05 virus with a D197N mutation in the NA,46 but as this has several other NA sequence differences compared to the B/Perth, we obtained a control for this virus NA which only had a single amino acid difference in the stalk region, B/Gifu/11/2005. Both viruses were obtained from the NIID, Tokyo, Japan. All virus stocks were serially plaque purified in MDCK cells.
1, 2, and 3 were synthesized by GSK, (Stevenage, U.K.). 2-Keto-3-deoxy-d-glycero-d-galactononulosonic acid (4) (2,3, difluoro KDN)33 was provided by Dr. Andrew Watts (University of Bath, U.K.) (Figure 1). This is a sialic acid based inhibitor but lacks the N-acetyl group. Serial log10 dilutions of inhibitors were prepared in water for inhibition assays ranging from 0.01 to 10 000 nM for 1, 2, and 3 and from 0.01 to 10 000 μM for 4.
Protein Expression and Purification
The full length B/Perth wild type D197 and mutant E197 NAs were expressed in Sf21 insect cells as previously described.20 An amount of 4 L of cells at (1−2) × 106 cells/mL was infected with a multiplicity of infection of 1.5 plaque forming units per cell and were harvested at day 4, when about 30% cell death had occurred. The cells were recovered by centrifugation and resuspended in TBS buffer at 40× concentration. An equal volume of acetone was added to fix the cells, and these were stored on ice until required.
After removal of the acetone and being washed three times, the cells were resuspended in Tris-buffered saline to a density of 1 × 108 cells/mL. Cells were digested with trypsin (Worthington, TPCK) or Pronase (Calbiochem) at concentrations from 0.1 to 2 mg/mL for 2 h at 37 °C to remove the NA. The cells were centrifuged at 14 000 rpm for 10 min. NA was recovered in the supernatant, and the cell pellet was resuspended in TBS and redigested. The pooled supernatants were concentrated in an Amicon 8050 stirred cell concentrator using a Pall 30K MWCO polysulfone filter. The concentrated NA was separated from other proteins in the trypsin digest by gel filtration using Superose 12 in TBS.47 Activity of the fractions was determined by the 4-methylumbelliferyl-N-acetylneuraminic acid (MUNANA, Sigma-Aldrich) fluorescent enzyme assay.48,49 The active fractions were collected, analyzed by SDS−PAGE, and concentrated down again. These were further purified by running several times over a Lentil Lectin Sepharose 4B column (Amersham Biosciences) and eluting with 100 mM methyl-α-d-mannopyranoside in 20 mM Tris-Cl, pH 7.4, 0.5 M NaCl (Lancaster).
The activity of the fractions was again checked by the MUNANA assay, and the purity was checked by SDS−PAGE and silver staining. The purest active fractions of both NAs were concentrated to 4.5 mg/mL for crystallization trials.
Electron Microscopy
Purified B/Perth D NA was examined by negative staining as follows. The protein was diluted to 0.2 mg/mL in TBS. Then 300-mesh copper grids were coated with a thin carbon film and glow-discharged in nitrogen for 30 s. The 5−10 μL aliquots of the sample were pipetted onto the grids, and after 1 min of adsorption time, excess solution was drawn off using Whatman 541 filter paper. The grids were washed with 5 μL of TBS, and the grid was then stained with 2% uranyl acetate and was air-dried. The grids were examined in a Tecnai 12 transmission electron microscope (FEI, Eindhoven, The Netherlands) at an operating voltage of 120 kV, and images were recorded using a Megaview III CCD camera and AnalySIS camera control software (Olympus).
Enzyme Studies
(i) Specific Activity. Activities of purified samples of recombinant B/Perth D197 and E197 NAs were titrated in the MUNANA based NA enzyme assay.48 Relative specific activity was calculated based on units of fluorescence per microgram of protein.
(ii) Km and Ki.Km and Ki values were calculated using viruses solubilized by the addition of CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) to a final volume of 1%. Each of the extracts was titrated in the MUNANA based enzyme assay to determine similar final values of fluorescent units without the addition of stop solution, since this enabled continual monitoring of the reactions. The rate of hydrolysis of MUNANA was measured at substrate concentrations ranging from 6.25 to 200 μM, with readings taken every minute in a Victor 2 (Wallac) or BMG FLUOstar Optima reader. Experiments were carried out in duplicate and repeated four times. The maximum slope for each reaction was determined by comparing the slopes over different overlapping time intervals. Initial velocities of the reactions were then calculated by measuring the maximum slopes plotted as a function of substrate concentrations. The Michaelis Menten constant, Km, which represents the affinity of the enzyme for substrate, was calculated using a nonlinear regression function in GraphPad Prism.
While the NA inhibitors are competitive inhibitors, some wild type influenza NAs are described as time dependent or slow binders of the inhibitors, and some mutant NAs have lost this slow binding property. We followed the kinetics of inhibitor binding two ways, with preincubation of inhibitor for 30 min or without any preincubation, followed by 60 min of incubation with the MUNANA substrate for both assays. All experiments were carried out in duplicate. We used a constant substrate concentration of 100 μM MUNANA and inhibitor concentrations ranging from 10000 to 0.01 nM. Fluorescent readings were taken every minute in a BMG FLUOstar Optima reader for 60 min. Graphs of concentration of inhibitor versus % enzyme inhibition compared to the control were plotted for each 10 min data set. The IC50 was calculated as the concentration of inhibitor resulting in a 50% reduction in enzyme activity compared to the control for each 10 min time point. By use of the Km and the substrate concentration, the Ki can also be calculated using nonlinear regression and one-site competitive binding, using the equation of Cheng and Prusoff50Ki = IC50/(1 + [substrate]/Km). As we had calculated the Km, we then calculated Ki values in Graph Pad Prism using this method for each 10 min interval. Experiments were carried out in duplicate.
Crystallization and X-ray Data Collection
All crystals were grown at the Bio21 Collaborative Crystallization Centre (www.csiro.au/c3). Either a Phoenix (Art Robbins Industries) or a Mosquito (TTP) dispensing robot was used to set up sitting drops in 96-well SD-2 plates (IDEX Corp.). Plates were stored at 281 K in a Gallery 700 incubator and imaged with a Minstrel HT imaging system (Rigaku). A single commercial screen (The JCSG+ Suite from Qiagen) was set up initially to determine if the B/Perth D protein concentration was appropriate for crystallization trials, using 0.2 μL droplets consisting of 50% protein solution mixed with 50% reservoir solution, and equilibrated against a reservoir of 50 μL. Small (<50 μm) crystals grew from PEG-based conditions in the JCSG+ Suite trials, and subsequent crystallization trials were set up using The PACT Suites (Qiagen), as well as from screens designed around the hits in these two commercial screens. X-ray data for flash-cooled crystals were collected at the Australian Synchrotron beamline MX-1 using the Blu-ice software51 or Photon Factory beamline 17A. MOSFLM52 was used for processing all diffraction images. SCALA53 was used to scale all diffraction data.
B/Perth D protein (4.5 mg/mL) was mixed with reservoir solution in a 1:1 ratio (drop volume 0.2 μL). X-ray data were measured from crystals grown at 8 °C in 0.2 M Na2SO4, 20% w/v PEG 3350, 0.1 M bis-Tris propane, pH 6.5. These crystals were transferred to mother liquor with 10% v/v ethylene glycol and 10% v/v glycerol added just prior to flash-cooling to 100 K. Remaining crystals were used for soaking inhibitors (1, 2, and 3). A successful soak of 3 was performed by seeding some solid compound into a drop containing crystals and allowing the sample to equilibrate for 10 days prior to flash-cooling to 100 K. The well solution in this case was 0.2 M NaNO3, 20% w/v PEG3350, 0.1 M Bis-Tris propane, pH 6.5. We were not able to obtain crystals containing either 1 or 2 with the D197 protein.
B/Perth E (4.5 mg/mL) was used to grow crystals under conditions similar to those described above. The best crystals for diffraction experiments were grown in 12−17% w/v PEG 3350, 0.2−0.3 M Na2SO4, and 5 mM YCl3. The complex of B/Perth E with 3 was obtained by placement of the compound directly into the drop as described above for B/Perth D. The complex of B/Perth E with 2 was obtained by adding the compound (5 mM) to the cryoprotectant (the same as for B/Perth D) prior to flash cooling.
Structure Solution and Refinement
Structures were solved by molecular replacement using PHENIX54 and MOLREP.55 All structures were manually rebuilt using COOT56 and refined in REFMAC.57 The weighting of X-ray and geometric parameters in refinement and the type of NCS restraints were based on their effects on Rfree cross-validation.
Acknowledgments
We thank Dr. Ross Fernley and Pat Pilling for assistance with protein purification and crystallization, Peter Schmidt for helpful discussions, the technical staff of the Bio21 C3 Centre for help with crystallization, the Australian Synchrotron for access to MX-1, and the Photon Factory (Japan) for access to beamline 17A. This access was supported by the Australian Synchrotron Research Program which is funded by the Commonwealth of Australia under the MNRF Program.
This work was supported by Grant RO1A1062721 from NIAID and grants from GSK U.K. and Australia. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Australian synchrotron, NIH, NIAID, or GSK.
Coordinates and structure factors have been deposited in the PDB under accession numbers 3K36 (B/Perth D), 3K37 (B/Perth D peramivir 3 complex), 3K38 (B/Perth E), 3K39 (B/Perth/E 3 complex), and 3K3A (B/Perth E oseltamivir 2 complex).
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: FU, fluorescent unit; MUNANA, 4-methylumbelliferyl-N-acetylneuraminic acid; NA, neuraminidase; KDN, 2,3-difluoro-2-keto-3-deoxy-d-glycero-d-galactononulosonic acid.
References
- Varghese J. N.; Laver W. G.; Colman P. M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 1983, 303, 35–40. [DOI] [PubMed] [Google Scholar]
- Colman P. M.; Varghese J. N.; Laver W. G. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 1983, 303, 41–44. [DOI] [PubMed] [Google Scholar]
- Burmeister W. P.; Daniels R. S.; Dayan S.; Gagnon J.; Cusack S.; Ruigrok R. W. Sequence and crystallization of influenza virus B/Beijing/1/87 neuraminidase. Virology 1991, 180, 266–272. [DOI] [PubMed] [Google Scholar]
- Burmeister W. P.; Ruigrok R. W.; Cusack S. The 2.2 Å resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J. 1992, 11, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janakiraman M. N.; White C. L.; Laver W. G.; Air G. M.; Luo M. Structure of influenza virus neuraminidase B/Lee/40 complexed with sialic acid and a dehydro analog at 1.8-Å resolution: implications for the catalytic mechanism. Biochemistry (Moscow) 1994, 33, 8172–8179. [DOI] [PubMed] [Google Scholar]
- von Itzstein M.; Wu W. Y.; Kok G. B.; Pegg M. S.; Dyason J. C.; Jin B.; Van Phan T.; Smythe M. L.; White H. F.; Oliver S. W.; Colman P. M.; Varghese J. N.; Ryan D. M.; Woods J. M.; Bethell R. C.; Hotham V. J.; Cameron J. M.; Penn C. R. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 1993, 363, 418–423. [DOI] [PubMed] [Google Scholar]
- Kim C. U.; Lew W.; Williams M. A.; Liu H.; Zhang L.; Swaminathan S.; Bischofberger N.; Chen. M. S.; Mendel D. B.; Tai C. Y.; Laver W. G.; Stevens R. C. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site:design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 1997, 119, 681–690. [DOI] [PubMed] [Google Scholar]
- Babu Y. S.; Chand P.; Bantia S.; Kotian P.; Dehghani A.; El-Kattan Y.; Lin T. H.; Hutchison T. L.; Elliott A. J.; Parker C. D.; Ananth S. L.; Horn L. L.; Laver G. W.; Montgomery J. A. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J. Med. Chem. 2000, 43, 3482–3486. [DOI] [PubMed] [Google Scholar]
- BioCryst Peramivir (Neuraminidase Inhibitor). http://www.biocryst.com/peramivir.htm (accessed March 10, 2009).
- Gubareva L. V. Molecular mechanisms of influenza virus resistance to neuraminidase inhibitors. Virus Res. 2004, 103, 199–203. [DOI] [PubMed] [Google Scholar]
- de Jong M. D.; Tran T. T.; Truong H. K.; Vo M. H.; Smith G. J.; Nguyen V. C.; Bach V. C.; Phan T. Q.; Do Q. H.; Guan Y.; Peiris J. S.; Tran T. H.; Farrar J. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 2005, 353, 2667–2672. [DOI] [PubMed] [Google Scholar]
- Ward P.; Small I.; Smith J.; Suter P.; Dutkowski R. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic. J. Antimicrob. Chemother. 2005, 55(Suppl. 1), i5–i21. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S.; Sugaya N.; Ito M.; Yamazaki M.; Ichikawa M.; Kimura K.; Kiso M.; Shimizu H.; Kawakami C.; Koike K.; Mitamura K.; Kawaoka Y. Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. JAMA, J. Am. Med. Assoc. 2007, 297, 1435–1442. [DOI] [PubMed] [Google Scholar]
- Stephenson I.; Democratis J.; Lackenby A.; McNally T.; Smith J.; Pareek M.; Ellis J.; Bermingham A.; Nicholson K.; Zambon M. Neuraminidase inhibitor resistance after oseltamivir treatment of acute influenza A and B in children. Clin. Infect. Dis. 2009, 48, 289–296. [DOI] [PubMed] [Google Scholar]
- Hurt A. C.; Ernest J.; Deng Y. M.; Iannello P.; Besselaar T. G.; Birch C.; Buchy P.; Chittaganpitch M.; Chiu S. C.; Dwyer D.; Guigon A.; Harrower B.; Kei I. P.; Kok T.; Lin C.; McPhie K.; Mohd A.; Olveda R.; Panayotou T.; Rawlinson W.; Scott L.; Smith D.; D’Souza H.; Komadina N.; Shaw R.; Kelso A.; Barr I. G. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res. 2009, 83, 90–93. [DOI] [PubMed] [Google Scholar]
- Meijer A.; Lackenby A.; Hungnes O.; Lina B.; van-der-Werf S.; Schweiger B.; Opp M.; Paget J.; van-de-Kassteele J.; Hay A.; Zambon M. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007−08 season. Emerging Infect. Dis. 2009, 15, 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Dwyer D. E.; Blyth C. C.; Soedjono M.; Shi H.; Kesson A.; Ratnamohan M.; McPhie K.; Cunningham A. L.; Saksena N. K. Detection of the rapid emergence of the H275Y mutation associated with oseltamivir resistance in severe pandemic influenza virus A/H1N1 09 infections. Antiviral Res. 2010, 87, 16–21. [DOI] [PubMed] [Google Scholar]
- Le Q. M.; Wertheim H. F.; Tran N. D.; van Doorn H. R.; Nguyen T. H.; Horby P. A community cluster of oseltamivir-resistant cases of 2009 H1N1 influenza. N. Engl. J. Med. 2010, 362, 86–87. [DOI] [PubMed] [Google Scholar]
- Gubareva L. V.; Matrosovich M. N.; Brenner M. K.; Bethell R. C.; Webster R. G. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 1998, 178, 1257–1262. [DOI] [PubMed] [Google Scholar]
- Hurt A. C.; Iannello P.; Jachno K.; Komadina N.; Hampson A. W.; Barr I. G.; McKimm-Breschkin J. L. Neuraminidase inhibitor-resistant and -sensitive influenza B viruses isolated from an untreated human patient. Antimicrob. Agents Chemother. 2006, 50, 1872–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya N.; Mitamura K.; Yamazaki M.; Tamura D.; Ichikawa M.; Kimura K.; Kawakami C.; Kiso M.; Ito M.; Hatakeyama S.; Kawaoka Y. Lower clinical effectiveness of oseltamivir against influenza B contrasted with influenza A infection in children. Clin. Infect. Dis. 2007, 44, 197–202. [DOI] [PubMed] [Google Scholar]
- Sugaya N.; Tamura D.; Yamazaki M.; Ichikawa M.; Kawakami C.; Kawaoka Y.; Mitamura K. Comparison of the clinical effectiveness of oseltamivir and zanamivir against influenza virus infection in children. Clin. Infect. Dis. 2008, 47, 339–345. [DOI] [PubMed] [Google Scholar]
- Burmeister W. P.; Baudin F.; Cusack S.; Ruigrok R. W. Comparison of structure and sequence of influenza B/Yamagata and B/Beijing neuraminidases shows a conserved “head” but much greater variability in the “stalk” and NB protein. Virology 1993, 192, 683–686. [DOI] [PubMed] [Google Scholar]
- Sahasrabudhe A.; Lawrence L.; Epa V. C.; Varghese J. N.; Colman P. M.; McKimm-Breschkin J. L. Substrate, inhibitor, or antibody stabilizes the Glu 119 Gly mutant influenza virus neuraminidase. Virology 1998, 247, 14–21. [DOI] [PubMed] [Google Scholar]
- Hurt A. C.; McKimm-Breschkin J. L.; McDonald M.; Barr I. G.; Komadina N.; Hampson A. W. Identification of a human influenza type B strain with reduced sensitivity to neuraminidase inhibitor drugs. Virus Res. 2004, 103, 205–211. [DOI] [PubMed] [Google Scholar]
- Tashiro M.; McKimm-Breschkin J. L.; Saito T.; Klimov A.; Macken C.; Zambon M.; Hayden F. G. Surveillance for neuraminidase-inhibitor-resistant influenza viruses in Japan, 1996−2007. Antiviral Ther. 2009, 14, 751–761. [DOI] [PubMed] [Google Scholar]
- Blick T. J.; Tiong T.; Sahasrabudhe A.; Varghese J. N.; Colman P. M.; Hart G. J.; Bethell R. C.; McKimm-Breschkin J. L. Generation and characterization of an influenza virus neuraminidase variant with decreased sensitivity to the neuraminidase-specific inhibitor 4-guanidino-Neu5Ac2en. Virology 1995, 214, 475–484. [DOI] [PubMed] [Google Scholar]
- McKimm-Breschkin J. L.; Sahasrabudhe A.; Blick T. J.; McDonald M.; Colman P. M.; Hart G. J.; Bethell R. C.; Varghese J. N. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J. Virol. 1998, 72, 2456–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum E. Z.; Wagaman P. C.; Ly L.; Turchi I.; Le J.; Bucher D.; Bush K. A point mutation in influenza B neuraminidase confers resistance to peramivir and loss of slow binding. Antiviral Res. 2003, 59, 13–22. [DOI] [PubMed] [Google Scholar]
- Kati W. M.; Saldivar A. S.; Mohamadi F.; Sham H. L.; Laver W. G.; Kohlbrenner W. E. GS4071 is a slow-binding inhibitor of influenza neuraminidase from both A and B strains. Biochem. Biophys. Res. Commun. 1998, 244, 408–413. [DOI] [PubMed] [Google Scholar]
- Kubelka V.; Altmann F.; Kornfeld G.; Marz L. Structures of the N-linked oligosaccharides of the membrane glycoproteins from three lepidopteran cell lines (Sf-21, IZD-Mb-0503, Bm-N). Arch. Biochem. Biophys. 1994, 308, 148–157. [DOI] [PubMed] [Google Scholar]
- Taylor N. R.; Cleasby A.; Singh O.; Skarzynski T.; Wonacott A. J.; Smith P. W.; Sollis S. L.; Howes P. D.; Cherry P. C.; Bethell R.; Colman P.; Varghese J. Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B. J. Med. Chem. 1998, 41, 798–807. [DOI] [PubMed] [Google Scholar]
- Watts A. G.; Oppezzo P.; Withers S. G.; Alzari P. M.; Buschiazzo A. Structural and kinetic analysis of two covalent sialosyl-enzyme intermediates on Trypanosoma rangeli sialidase. J. Biol. Chem. 2006, 281, 4149–4155. [DOI] [PubMed] [Google Scholar]
- Hart G. J.; Bethell R. C. 2,3-Didehydro-2,4-dideoxy-4-guanidino-N-acetyl-d-neuraminic acid (4-guanidino-Neu5Ac2en) is a slow-binding inhibitor of sialidase from both influenza A virus and influenza B virus. Biochem. Mol. Biol. Int. 1995, 36, 695–703. [PubMed] [Google Scholar]
- Collins P. J.; Haire L. F.; Lin Y. P.; Liu J.; Russell R. J.; Walker P. A.; Skehel J. J.; Martin S. R.; Hay A. J.; Gamblin S. J. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 2008, 453, 1258–1261. [DOI] [PubMed] [Google Scholar]
- Varghese J. N.; Smith P. W.; Sollis S. L.; Blick T. J.; Sahasrabudhe A.; McKimm-Breschkin J. L.; Colman P. M. Drug design against a shifting target: a structural basis for resistance to inhibitors in a variant of influenza virus neuraminidase. Structure 1998, 6, 735–746. [DOI] [PubMed] [Google Scholar]
- Smith B. J.; McKimm-Breschkin J. L.; McDonald M.; Fernley R. T.; Varghese J. N.; Colman P. M. Structural studies of the resistance of influenza virus neuramindase to inhibitors. J. Med. Chem. 2002, 45, 2207–2212. [DOI] [PubMed] [Google Scholar]
- Wang M. Z.; Tai C. Y.; Mendel D. B. Mechanism by which mutations at His274 alter sensitivity of influenza A virus N1 neuraminidase to oseltamivir carboxylate and zanamivir. Antimicrob. Agents Chemother. 2002, 46, 3809–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg M. S.; von Itzstein M. Slow-binding inhibition of sialidase from influenza virus. Biochem. Mol. Biol. Int. 1994, 32, 851–858. [PubMed] [Google Scholar]
- McKimm-Breschkin J.; Trivedi T.; Hampson A.; Hay A.; Klimov A.; Tashiro M.; Hayden F.; Zambon M. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob. Agents Chemother. 2003, 47, 2264–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass L. M.; Brown J.; Pickford M.; Fayinka S.; Newman S. P.; Johansson C. J.; Bye A. Pharmacoscintigraphic evaluation of lung deposition of inhaled zanamivir in healthy volunteers. Clin. Pharmacokinet. 1999, 36(Suppl. 1), 21–31. [DOI] [PubMed] [Google Scholar]
- Oo C.; Barrett J.; Hill G.; Mann J.; Dorr A.; Dutkowski R.; Ward P. Pharmacokinetics and dosage recommendations for an oseltamivir oral suspension for the treatment of influenza in children. Paediatr. Drugs 2001, 3, 229–236; erratum appears in Paediatr. Drugs2001, 3 (4), 246. [DOI] [PubMed] [Google Scholar]
- Wattanagoon Y.; Stepniewska K.; Lindegardh N.; Pukrittayakamee S.; Silachamroon U.; Piyaphanee W.; Singtoroj T.; Hanpithakpong W.; Davies G.; Tarning J.; Pongtavornpinyo W.; Fukuda C.; Singhasivanon P.; Day N. P.; White N. J. Pharmacokinetics of high-dose oseltamivir in healthy volunteers. Antimicrob. Agents Chemother. 2009, 53, 945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.; Roy S.; Rayner C.; Amer A.; Howard D.; Smith J. R.; Evans T. G. A randomized, crossover study to evaluate the pharmacokinetics of amantadine and oseltamivir administered alone and in combination. PLoS One 2007, 2, e1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew W.; Chen X.; Kim C. U. Discovery and development of GS 4104 (oseltamivir): an orally active influenza neuraminidase inhibitor. Curr. Med. Chem. 2000, 7, 663–672. [DOI] [PubMed] [Google Scholar]
- Neuraminidase Inhibitor Susceptibility Network (NISN). Monitoring of neuraminidase inhibitor resistance among clinical influenza virus isolates in Japan during the 2003−2006 influenza seasons. Wkly. Epidemiol. Rec. 2007, 27, 149−150. [PubMed] [Google Scholar]
- McKimm-Breschkin J. L.; Caldwell J. B.; Guthrie R. E.; Kortt A. A. A new method for the purification of the influenza A virus neuraminidase. J. Virol. Methods 1991, 32, 121–124. [DOI] [PubMed] [Google Scholar]
- Potier M.; Mameli L.; Belisle M.; Dallaire L.; Melancon S. B. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal. Biochem. 1979, 94, 287–296. [DOI] [PubMed] [Google Scholar]
- McKimm-Breschkin J. L.; Blick T. J.; Sahasrabudhe A.; Tiong T.; Marshall D.; Hart G. J.; Bethell R. C.; Penn C. R. Generation and characterization of variants of NWS/G70C influenza virus after in vitro passage in 4-amino-Neu5Ac2en and 4-guanidino-Neu5Ac2en. Antimicrob. Agents Chemother. 1996, 40, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.; Prusoff W. H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50% inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [DOI] [PubMed] [Google Scholar]
- McPhillips T. M.; McPhillips S. E.; Chiu H. J.; Cohen A. E.; Deacon A. M.; Ellis P. J.; Garman E.; Gonzalez A.; Sauter N. K.; Phizackerley R. P.; Soltis S. M.; Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 2002, 9, 401–406. [DOI] [PubMed] [Google Scholar]
- Leslie A. W. G.Recent changes to MOSFLM package for processing film and image plate data. Joint CCP4 ESF-EAMCB Newslett. Protein Crystallogr. 1992, 26. [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006, 62, 72–82. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Grosse-Kunstleve R. W.; Hung L. W.; Ioerger T. R.; McCoy A. J.; Moriarty N. W.; Read R. J.; Sacchettini J. C.; Sauter N. K.; Terwilliger T. C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002, 58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- Vagin A.; Teplyakov A. MOLREP:an automated program for molecular replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar]
- Emsley P.; Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997, 53, 240–255. [DOI] [PubMed] [Google Scholar]