Abstract
Context
Approximately 60% of families that meet the Amsterdam-I criteria (AC-I) for hereditary nonpolyposis colorectal cancer (HNPCC) have a hereditary abnormality in a DNA mismatch repair (MMR) gene. Cancer incidence in AC-I families with MMR gene mutations is reported to be very high, but cancer incidence for individuals in AC-I families with no evidence of an MMR defect is unknown.
Objective
To determine if cancer risks in AC-I families with no apparent deficiency in DNA MMR are different from cancer risks in AC-I families with DNA MMR abnormalities.
Design, Setting, and Participants
Identification (1997–2001) of 161 AC-I pedigrees from multiple population- and clinic-based sources in North America and Germany, with families grouped into those with (group A) or without (group B) MMR deficiency by tumor testing. A total of 3422 relatives were included in the analyses.
Main Outcome Measures
Cancer incidence in groups A and B (excluding the 3 affected members used to define each pedigree as AC-I) and computed age- and sex-adjusted standardized incidence ratios (SIRs) using Surveillance, Epidemiology, and End Results data.
Results
Group A families from both population- and clinic-based series showed increased incidence of the HNPCC-related cancers. Group B families showed increased incidence only for colorectal cancer (SIR, 2.3; 95% confidence interval, 1.7–3.0) and to a lesser extent than group A (SIR, 6.1; 95% confidence interval, 5.2–7.2) (P<.001).
Conclusions
Families who fulfill AC-I criteria but who have no evidence of a DNA MMR defect do not share the same cancer incidence as families with HNPCC-Lynch syndrome (ie, hereditary MMR deficiency). Relatives in such families have a lower incidence of colorectal cancer than those in families with HNPCC-Lynch syndrome, and incidence may not be increased for other cancers. These families should not be described or counseled as having HNPCC-Lynch syndrome. To facilitate distinguishing these entities, the designation of “familial colorectal cancer type X” is suggested to describe this type of familial aggregation of colorectal cancer.
Hereditary nonpolyposis colorectal cancer (HNPCC) is a dominantly inherited syndrome characterized by significantly increased risks for colon cancer as well as for cancers of the endometrium, stomach, small intestine, hepatobiliary system, kidney, ureter, and ovary.1,2 Most studies have not reported increased risks for lung, breast, or prostate cancers in HNPCC kindreds. Many experts currently use the term HNPCC synonymously with a hereditary DNA mismatch repair (MMR) gene deficiency, and studies of cancer risks in the syndrome have generally focused on families with MMR deficiency. Based on fairly consistent cancer risks in studies of various designs, clinical screening guidelines have been proposed, focusing especially on screening for cancers of the colon and endometrium. 3–6 However, in the broader clinical realm and in some current medical literature, the term HNPCC continues to be based on pedigree criteria, typically the strict Amsterdam I criteria (AC-I),7 which have 3 elements: (1) there are at least 3 relatives with histologically verified colorectal cancer (1 a first-degree relative of the other 2), and familial adenomatous polyposis should be excluded; (2) at least 2 successive generations should be affected; and (3) 1 of the relatives’ colorectal cancers should be diagnosed before age 50 years.
About half of the families with AC-I pedigrees have no evidence of a heritable DNA MMR defect, either by gene sequencing or tumor phenotyping for microsatellite instability (MSI), the hallmark of MMR deficiency.8 For this large group of families, cancer risks have not been studied, and appropriate screening guidelines are unknown.
In the present study, we identified families fulfilling the strict AC-I criteria and analyzed the family history for cancer incidences, stratified by evidence of a DNA MMR defect.
METHODS
Classification of AC-I Families
Families fulfilling AC-I criteria were collected from 11 sources (1997–2001), distinguishing between population-based and clinic-based ascertainment. The majority of families (139 of 161) came from the Colon Cooperative Family Registry, a National Cancer Institute–supported consortium established in 1997 to create a multinational comprehensive collaborative infrastructure for interdisciplinary studies in the genetic epidemiology of colorectal cancer (detailed information about the registry is available at http://epi.grants.cancer.gov/CFR/). Population-based registries were used to identify eligible cases at the Fred Hutchinson Cancer Research Center, the University of Hawaii Cancer Research Center, and Cancer Care Ontario. Sites that included both population- and clinic-based collections were the Mayo Clinic, University of Southern California Consortium, and Australia. All families identified as fulfilling the AC-I criteria were eligible for recruitment at each Colon Cooperative Family Registry site. Additional clinic-based families were identified at the Mayo Clinic, at the University of California, San Francisco, and in Düsseldorf, Germany.
The study was approved by the institutional review board of each institution. After providing written consent to participate in research, AC-I families provided a family history, and enrollment of additional relatives began. Efforts were made to verify reported cancer diagnoses by use of multiple interviews of family members and by review of medical records, death certificates, pathology reports, and tumor tissues. Colorectal tumor blocks were tested for MSI using 10 microsatellite loci (4 mononucleotide markers [BAT25, BAT26, BAT40, BAT34C4], 5 dinucleotide markers [D5S346, D17S250, ACTC, D18S55, D10S197], and MYCL). Tumors were classified as microsatellite instability–high (MSI-H) if more than 30% of markers demonstrated instability, as microsatellite instability–low (MSI-L) if 30%or less demonstrated instability, and as microsatellite stable (MSS) if no marker exhibited instability. If MSI status was not available, a surrogate for MSI-H was documentation of a definite deleterious mutation in either the hMLH1 or hMSH2 genes. Immunohistochemical testing for MMR proteins of MLH1, MSH2, and MSH6 was conducted on all tumors with a high or low level of tumor MSI.9
As required by the AC-I criteria,7 each family contained a “triad” of family members with colorectal cancer. In many families, there were multiple potential triads. To maintain a consistent reference point, we assigned the core triad to be the one that included the earliest-generation member (eg, a grandparent-parent-child triad would be chosen over a parent and 2 offspring). This rule, while arbitrary, was implemented to ensure consistency of triad assignment among pedigrees.
Pedigrees were classified into 2 groups: those with MSI-H, reflecting DNA MMR deficiency (group A), and those with MSI-L or MSS, reflecting intact DNA MMR capacity (group B). Group A corresponds to hereditary MMR deficiency. We included only families for whom we had a high level of confidence in the assignment to the 2 groups; ambiguous families were excluded. Consequently: (1) An MSI test result for at least 1 of the defining triad members was available for every pedigree included in the analysis. (2) If the 1 tumor that was used to define the MSI classification of the pedigree was MSI-H and that person was younger than 60 years at the time of diagnosis, this pedigree was classified as group A. (3) Pedigrees were excluded from the study if the only MSI analysis performed for the entire pedigree was in a person older than 60 years and was MSI-H, because of the high probability with advancing age that these were due to epigenetic hMLH1 silencing rather than to germline MMR mutation. (4) Absence of expression of hMLH1 or hMSH2 demonstrated by immunohistochemical testing in a tumor was an acceptable surrogate for missing MSI data because of the demonstrated excellent correlation between immunohistochemical and MSI analyses.9 The same age-related rules were used for immunohistochemistry as for MSI, per point 3 above. (5) Normal expression of hMLH1 or hMSH2 demonstrated by immunohistochemical testing was not used to infer MSI status in the absence of actual MSI data. (6) If multiple triad members with colorectal cancer had MSI testing but results were discordant, then that pedigree was excluded. However, if multiple members with colorectal cancer were MSI-H with consistent immunohistochemistry results showing loss of expression of a particular MMR protein between different family members’ tumors, or if there was a known germline mutation in the triad member, we included this family as MSI-H, even if there was a relative with an MSS tumor outside the triad. We interpreted this as an apparent sporadic phenocopy.
Overall, only 12 of 173 pedigrees (7%) were excluded because of our inability to assign with confidence to groups A or B.
Relatives in the 2 groups of families were included in the cancer-incidence evaluation analysis. To provide the most conservative interpretation of risks, all AC-I–defining triad members were excluded from analysis. Relatives related by blood to all defining triad members were included in the risk calculations. “Primary-zone relatives” (ie, those at 50% mendelian risk of being a carrier of a dominant gene carried by each triad member) were defined as first-degree relatives of any member of the defining triad. “Secondary-zone relatives” (ie, those at 25% mendelian risk of being a carrier of a dominant gene carried by each triad member) were defined as second-degree relatives of any member of the defining triad who was not in the primary zone. Primary- and secondary-zone relatives so defined were mutually exclusive groups.
Statistical Methods
The incidence of cancer in the AC-I families was calculated as the ratio of observed cases to the number of person-years at risk. Person-years were calculated from age 20 years until the earliest cancer diagnosis or death. All cancers, except nonmelanoma skin cancers, were recorded.
The standardized incidence ratios (SIRs) of each cancer among members were calculated as the ratio of the observed to the expected numbers of cases. The latter was calculated as the sum of the products of the number of person-years for each 5-year age/sex group and the corresponding age/sex-specific incidence rates from the Surveillance, Epidemiology, and End Result (SEER) database. 10,11 Because individuals in groups A and B were blood relatives, the SEs in the confidence intervals (CIs) were underestimated using a standard analytical procedure. To correctly estimate the SEs, a bootstrap analysis using 1000 replicates12 was run on all cancers, and 95% CIs were reported from the bootstrap results.
All analyses were stratified by MSI status (group A [MSI-H] vs group B [MSI-L/MSS]). In addition, analyses were conducted separately in the population- and clinic-based groups, and in the primary and secondary zones. P values for the difference in cancer incidence between the MSI-H and the MSI-L/MSS groups were computed.13 Statistical analyses were performed using SAS version 8 (SAS Institute Inc, Cary, NC); P<.05 was used to determine statistical significance.
RESULTS
After exclusion of the reference triad of each pedigree, 3422 relatives (1680 women, 1742 men) were included in the analyses, 1657 in the primary zone and 1765 in the secondary zone. Of these, 518 (15.1%) reported being diagnosed with cancer, including 438 with 1 tumor, 62 with 2 primary tumors, 17 with 3, and 1 with 4, all of which were counted. There were 90 group A AC-I pedigrees (46 population-based) with MSI-H, and 71 group B pedigrees (46 population-based) with either MSI-L (n=11) or MSS (n=60). None of the group B cases had loss of expression of MSH6 demonstrated by immunohistochemical testing.
TABLE 1 shows that group A relatives had greatly increased incidences for cancers of the colorectum and uterus (endometrium), stomach, urinary tract (kidney/ureter), ovary, and small intestine. There was also evidence for increased incidences for cancers of the pancreas and liver, but no increases for cancers of the breast, lung, prostate, or cervix, or for melanoma. In contrast, group B showed increased incidences only for colorectal cancer, and the SIR was less than half of that seen in group A.
Table 1.
Standardized Incidence Ratios Comparing First- and Second-Degree Relatives of Group A vs Group B
| Group A (MSI-H) (n = 1855)* |
Group B (MSI-L/MSS) (n = 1567)* |
||||
|---|---|---|---|---|---|
| Tumor Site | No. of Tumors (Men/Women) |
SIR (95% CI)† | No. of Tumors (Men/Women) |
SIR (95% CI)† |
P Value‡ |
| Colorectum | 182 (94/88) | 6.1 (5.2–7.2)§ | 55 (23/32) | 2.3 (1.7–3.0)§ | <.001 |
| Uterus | 41 (0/41) | 4.1 (2.9–5.6)§ | 6 (0/6) | 0.8 (0.3–1.6) | <.001 |
| Stomach | 21 (14/7) | 4.6 (2.7–6.6)§ | 5 (1/4) | 1.4 (0.3–2.8) | .008 |
| Breast | 21 (0/21) | 0.5 (0.3–0.7) | 13 (0/13) | 0.4 (0.2–0.6) | .59 |
| Kidney | 15 (6/9) | 2.6 (1.4–4.0)§ | 4 (2/2) | 0.9 (0.2–1.8) | .04 |
| Lung | 12 (9/3) | 0.3 (0.2–0.5) | 7 (3/4) | 0.2 (0.1–0.4) | .55 |
| Ovary | 12 (0/12) | 2.0 (1.0–3.2)§ | 7 (0/7) | 1.5 (0.4–2.9) | .60 |
| Hematopoietic | 11 (5/6) | 0.5 (0.2–0.8) | 5 (3/2) | 0.3 (0.1–0.6) | .29 |
| Pancreas | 10 (6/4) | 1.7 (0.7–2.8) | 2 (1/1) | 0.4 (0.0–1.2) | .05 |
| Prostate | 10 (10/0) | 0.3 (0.1–0.6) | 9 (9/0) | 0.4 (0.1–0.6) | .79 |
| Small intestine | 6 (3/3) | 7.6 (2.5–13.9)§ | 1 (0/1) | 1.6 (0.0–5.1) | .10 |
| Cervix | 6 (0/6) | 0.3 (0.1–0.5) | 5 (0/5) | 0.3 (0.1–0.6) | .96 |
| Bladder | 5 (2/3) | 0.4 (0.1–0.9) | 1 (1/0) | 0.1 (0.0–0.3) | .17 |
| Liver | 4 (0/4) | 2.4 (0.6–5.0) | 1 (1/0) | 0.8 (0.0–2.3) | .28 |
| Ureter | 4 (2/2) | 9.0 (2.0–18.3)§ | 1 (1/0) | 2.9 (0.0–9.2) | .29 |
| Brain | 3 (0/3) | 0.7 (0.0–1.7) | 2 (2/0) | 0.6 (0.0–1.4) | .84 |
| Head/neck | 3 (3/0) | 0.3 (0.0–0.6) | 3 (2/1) | 0.3 (0.0–0.8) | .79 |
| Melanoma | 2 (0/2) | 0.2 (0.0–0.4) | 1 (1/0) | 0.1 (0.0–0.3) | .71 |
Abbreviations: CI, confidence interval; MSI-H, microsatellite instability–high; MSI-L, microsatellite instability–low; MSS, microsatellite stable; SIR, standardized incidence ratio.
Number of first- and second-degree relatives of triad members fulfilling Amsterdam-I criteria.
SIRs and bootstrapped 95% CIs of specific cancers, by DNA mismatch repair status, combining population- and clinic-based families.
Computed by comparing the SIR of the DNA mismatch repair groups (group A vs group B).
Statistically significant estimate compared with the Surveillance, Epidemiology, and End Results population.9
TABLE 2 presents results stratified by method of ascertainment. The SIRs in the clinic-based families in group A were greater than for the population-based families for colorectal cancer (9.6 [95% CI, 7.5–12.3] vs 4.3 [95% CI, 3.4–5.3], respectively) and for uterine cancer (5.4 [95% CI, 3.1–7.9] vs 3.4 [95% CI, 1.9–4.8]). In group B, the SIRs for colorectal cancer were greater in the clinic-based families than in the population-based families (3.1 [95% CI, 1.9–4.3] vs 2.0 [95% CI, 1.3–2.7]).
Table 2.
Standardized Incidence Ratios of Group A vs Group B, Showing Analysis of Population-Based vs Clinic-Based Families
| Group A (MSI-H) SIR (95% CI)* | Group B (MSI-L/MSS) SIR (95% CI)* | P Value† | ||||
|---|---|---|---|---|---|---|
| Tumor Site | Population-Based (n = 1052)‡ |
Clinic-Based (n = 803)‡ |
Population-Based (n = 976)‡ |
Clinic-Based (n = 591)‡ |
Population-Based or Unselected |
Clinic- Based |
| Colorectum | 4.3 (3.4–5.3)§ | 9.6 (7.5–12.3)§ | 2.0 (1.3–2.7)§ | 3.1 (1.9–4.3)§ | <.001 | <.001 |
| Uterus | 3.4 (1.9–4.8)§ | 5.4 (3.1–7.9)§ | 0.6 (0.0–1.5) | 1.1 (0.0–2.6) | .001 | .002 |
| Stomach | 3.3 (1.4–5.4)§ | 7.1 (3.1–11.7)§ | 1.7 (0.4–3.5) | 0.8 (0.0–2.6) | .25 | .007 |
| Breast | 0.4 (0.2–0.6) | 0.6 (0.3–1.0) | 0.5 (0.2–0.8) | 0.2 (0.0–0.5) | .68 | .15 |
| Kidney | 1.8 (0.5–3.3) | 4.1 (1.5–7.1)§ | 1.0 (0.0–2.2) | 0.6 (0.0–2.1) | .36 | .03 |
| Lung | 0.2 (0.0–0.4) | 0.6 (0.2–1.1) | 0.3 (0.1–0.5) | 0.2 (0.0–0.5) | .69 | .19 |
| Ovary | 1.6 (0.5–3.1) | 2.6 (0.8–5.1) | 1.4 (0.3–3.0) | 1.8 (0.0–4.0) | .84 | .60 |
| Hematopoietic | 0.7 (0.2–1.1) | 0.3 (0.0–0.7) | 0.2 (0.0–0.5) | 0.5 (0.0–1.1) | .07 | .47 |
| Pancreas | 2.1 (0.8–3.8) | 1.0 (0.0–2.7) | 0.3 (0.0–1.1) | 0.6 (0.0–2.1) | .04 | .74 |
| Prostate | 0.3 (0.1–0.6) | 0.3 (0.0–0.7) | 0.1 (0.0–0.3) | 1.0 (0.3–1.8) | .17 | .09 |
| Small intestine | 9.7 (1.9–19.3)§ | 3.6 (0.0–12.0) | 2.5 (0.0–7.8) | NA‖ | .17 | .45 |
| Cervix | 0.2 (0.0–0.4) | 0.5 (0.0–0.9) | 0.1 (0.0–0.3) | 0.6 (0.1–1.3) | .67 | .65 |
| Bladder | 0.3 (0.0–0.7) | 0.8 (0.0–1.8) | NA‖ | 0.3 (0.0–1.1) | .21 | .46 |
| Liver | 0.9 (0.0–3.2) | 5.3 (0.0–11.7) | 1.1 (0.0–3.7) | NA‖ | .85 | .10 |
| Ureter | 10.0 (0.0–22.9) | 6.8 (0.0–21.8) | NA‖ | 8.7 (0.0–30.0) | .11 | .85 |
| Brain | 0.7 (0.0–1.9) | 0.6 (0.0–2.1) | NA‖ | 1.7 (0.0–4.2) | .19 | .43 |
| Head/neck | 0.4 (0.0–0.9) | NA‖ | 0.2 (0.0–0.5) | 0.7 (0.0–1.8) | .44 | .11 |
| Melanoma | 0.2 (0.0–0.6) | NA‖ | NA‖ | 0.3 (0.0–0.8) | .19 | .29 |
Abbreviations: CI, confidence interval; MSI-H, microsatellite instability–high; MSI-L, microsatellite instability–low; MSS, microsatellite stable; NA, not available; SIR, standardized incidence ratio.
SIRs and bootstrapped 95% CIs for cancers in first- and second-degree relatives of triad members fulfilling Amsterdam-I criteria, by microsatellite instability status as well as ascertainment mode.
Computed by comparing the SIR of the DNA mismatch repair groups (group A vs group B).
Number of first- and second-degree relatives, excluding triad members, included in the analysis.
Statistically significant estimate compared with the Surveillance, Epidemiology, and End Results population.9
Zero cancer events, so no bootstrap 95% CI was generated.
TABLE 3 shows that primary-zone relatives had greater SIRs overall than secondary-zone relatives. In group A, the SIRs for cancers of the colorectum, uterus, stomach, kidney, ovary, and small intestine were all significantly elevated in primary-zone relatives. In group B, this was true only for colorectal cancer among primary-zone relatives (2.7 [95% CI, 1.9–3.4]).
Table 3.
Standardized Incidence Ratios of Group A vs Group B Showing Analysis of Primary- vs Secondary-Zone Relatives
| Group A (MSI-H) SIR (95% CI)* | Group B (MSI-L/MSS) SIR (95% CI)* | P Value† | ||||
|---|---|---|---|---|---|---|
| Tumor Site | Primary Zone (n = 870)‡ |
Secondary Zone (n = 985)‡ |
Primary Zone (n = 787)‡ |
Secondary Zone (n = 780)‡ |
Primary Zone |
Secondary Zone |
| Colorectum | 6.1 (4.9–7.5)§ | 6.1 (4.6–7.9)§ | 2.7 (1.9–3.4)§ | 1.1 (0.4–2.1) | <.001 | <.001 |
| Uterus | 5.0 (3.4–7.1)§ | 2.5 (0.9–4.3) | 0.9 (0.2–1.7) | 0.5 (0.0–1.8) | <.001 | .09 |
| Stomach | 5.9 (3.2–9.0)§ | 2.4 (0.5–5.3) | 1.4 (0.3–3.0) | 1.2 (0.0–4.0) | .004 | .58 |
| Breast | 0.6 (0.3–0.9) | 0.3 (0.1–0.6) | 0.4 (0.2–0.6) | 0.4 (0.0–0.9) | .30 | .59 |
| Kidney | 3.1 (1.4–4.9)§ | 1.8 (0.4–3.9) | 1.2 (0.3–2.5) | NA‖ | .08 | .11 |
| Lung | 0.4 (0.2–0.7) | 0.2 (0.0–0.5) | 0.3 (0.1–0.5) | 0.2 (0.0–0.6) | .50 | .81 |
| Ovary | 2.9 (1.3–4.7)§ | 0.4 (0.0–1.4) | 1.5 (0.3–3.0) | 1.6 (0.0–3.9) | .22 | .28 |
| Hematopoietic | 0.7 (0.3–1.2) | 0.2 (0.0–0.6) | 0.3 (0.1–0.7) | 0.2 (0.0–0.6) | .20 | .96 |
| Pancreas | 2.1 (0.8–3.6) | 1.0 (0.0–2.5) | 0.6 (0.0–1.5) | NA‖ | .07 | .38 |
| Prostate | 0.3 (0.1–0.6) | 0.4 (0.1–0.8) | 0.4 (0.1–0.6) | 0.4 (0.0–1.2) | .80 | .77 |
| Small intestine | 8.2 (1.9–17.6)§ | 6.7 (0.0–16.6) | 2.2 (0.0–6.8) | NA‖ | .20 | .33 |
| Cervix | 0.4 (0.1–0.7) | 0.2 (0.0–0.5) | 0.3 (0.0–0.7) | 0.3 (0.0–0.7) | .86 | .75 |
| Bladder | 0.4 (0.0–0.9) | 0.5 (0.0–1.3) | 0.1 (0.0–0.4) | NA‖ | .34 | .37 |
| Liver | 3.8 (0.9–8.0) | NA‖ | 1.0 (0.0–3.1) | NA‖ | .19 | NA¶ |
| Ureter | 7.0 (0.0–18.7) | 12.6 (0.0–33.1) | 3.6 (0.0–11.5) | NA‖ | .60 | .41 |
| Brain | 1.2 (0.0–2.8) | NA‖ | 0.4 (0.0–1.3) | 0.9 (0.0–2.8) | .65 | .24 |
| Head/neck | 0.4 (0.0–1.1) | NA‖ | 0.3 (0.0–0.8) | 0.4 (0.0–1.4) | .69 | .21 |
| Melanoma | 0.3 (0.0–0.7) | NA‖ | 0.1 (0.0–0.4) | NA‖ | .61 | NA¶ |
Abbreviations: CI, confidence interval; MSI-H, microsatellite instability–high; MSI-L, microsatellite instability–low; MSS, microsatellite stable; NA, not available; SIR, standardized incidence ratio.
SIRS and bootstrapped 95% CIs for cancers in primary-zone and secondary-zone relatives (see text) of triad members fulfilling Amsterdam-I criteria, by microsatellite instability status.
Computed by comparing the SIR of the DNA mismatch repair groups (group A vs group B).
Number of relatives included in the analysis.
Statistically significant estimates compared with the Surveillance, Epidemiology, and End Results population.9
Zero cancer events, so no bootstrap 95% CI was generated.
Zero cancer events in both comparison groups, so no P value was calculated.
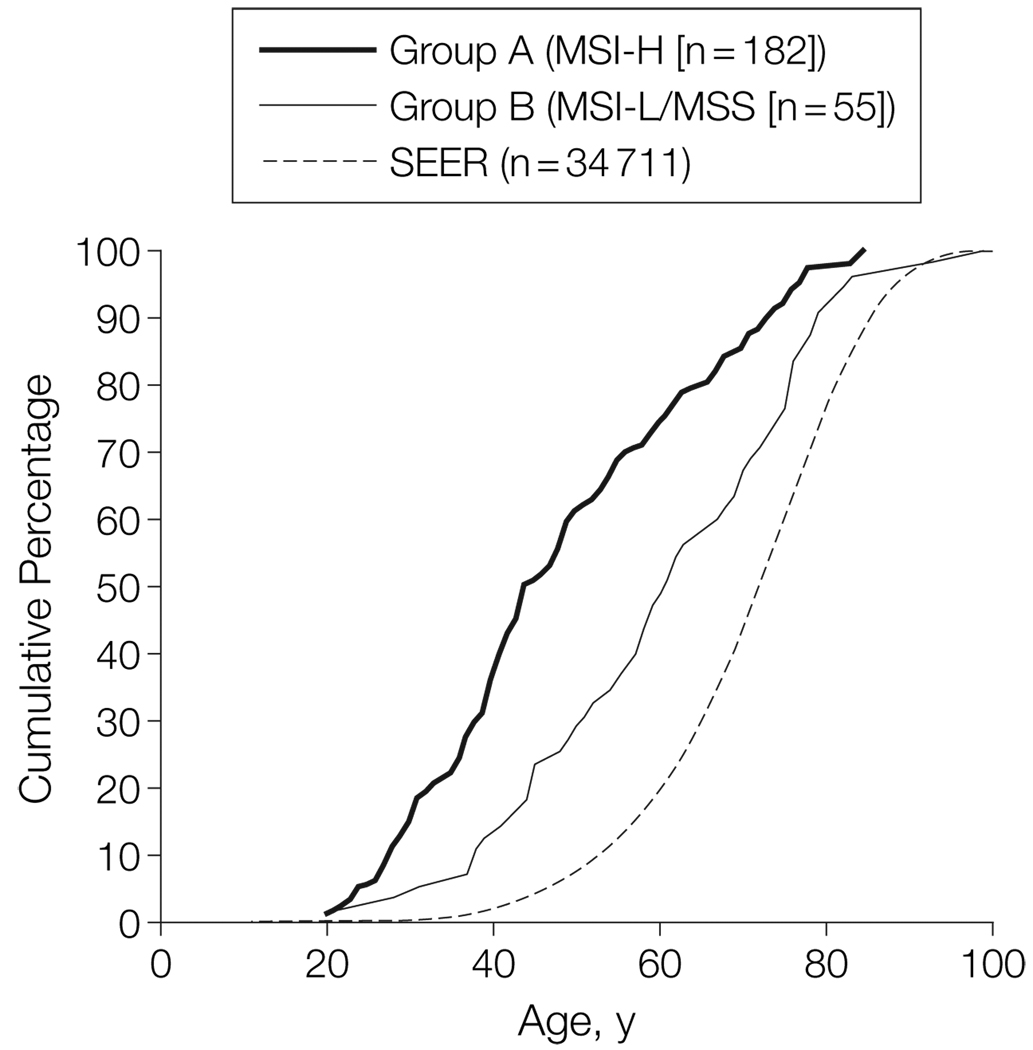
The cumulative age-of-onset curves for colorectal cancer among all relatives in groups A and B, compared with the distribution of colorectal cancer diagnoses in the SEER data, are shown in FIGURE 1. The curves are clearly distinguishable, indicating that relatives in AC-I families (groups A and B) tend to develop colorectal cancer at a younger age than the general (ie, SEER) population and that group A relatives tend to develop colorectal cancer at a younger age (mean, 48.7 years) than group B relatives (mean, 60.7 years).
Figure 1.
Cumulative Age of Onset of Colorectal Cancer in Primary- and Secondary-Zone Relatives in Families Fulfilling Amsterdam-I Criteria and Distribution of Colorectal Cancer Diagnoses in the SEER Registry9
Curves exclude the 3 affected family members used to define the Amsterdam-I criteria. MSI-H indicates microsatellite instability–high; MSI-L, microsatellite instability–low; MSS, microsatellite stable; SEER, Surveillance, Epidemiology, and End Results.
COMMENT
We have documented that families fulfilling the stringent AC-I criteria7 without evidence of MMR deficiency have a distinctly different pattern of cancer incidences than AC-I families that do have MMR deficiency. The group A families, with presumed hereditary DNA MMR deficiency, showed cancer incidences that are typical of what has been previously reported: very high SIRs for cancers of the colorectum, endometrium, stomach, small intestine, and ureter but no increases in incidence of cancer of the breast, lung, prostate, or other sites. This is consistent with the majority of literature on hereditary MMR deficiency. In the absence of evidence of MMR deficiency, there was only a modest increase in the incidence of colorectal cancer and no increase in the risk of other malignancies. The literature regarding cancer risks in AC-I families without MMR deficiency has been very limited but has hinted that there could be differences from the AC-I families with MMR deficiency. 14,15 Thus, in counseling such families, clinicians can now provide them with more accurate and lower-risk information, using these new data in combination with the specific family history.
We hypothesize that the AC-I families with no MMR defects are a heterogeneous group comprised of (1) some cancer aggregation occurring by chance alone, (2) some aggregation related to shared lifestyle factors, and (3) some yet-to-be-defined genetic syndromes. Group B families with particularly strong or unusual histories are best counseled based on a customized assessment of the pedigree, but such families should not automatically be triaged to HNPCC screening algorithms, as is current practice.
It is notable that the mean age at diagnosis of colorectal cancer in group A relatives (48.7 years) was substantially lower than that for group B relatives (60.7 years), further underscoring the inappropriateness of standard HNPCC screening guidelines for many group B families. Published cancer screening guidelines for HNPCC, based on expert opinion and some observational studies, suggest that annual colonoscopy should begin between ages 20 to 25 years and that annual endometrial cancer screening by transvaginal ultrasound or endometrial aspirate should begin at ages 25 to 35 years.3–6 In addition, consideration of prophylactic subtotal colectomy and hysterectomy is suggested. These guidelines do seem appropriate for those families with hereditary DNA MMR defects, based on abundant and consistent data. However, these guidelines do not seem indicated for the group B families described in this study, ie, those with AC-I pedigree but no demonstrable defect in DNA MMR capacity. It is our opinion that for group B families it may be reasonable to offer colorectal cancer screening initiated 5 to 10 years prior to the age of earliest colorectal cancer diagnosis in these families, with frequency determined by initial findings but no less often than every 5 years. Aggressive endometrial cancer screening in group B families is not supported by our data. In families that meet AC-I criteria but in whom tumor MSI testing or genetic testing is not feasible or informative, screening recommendations should default to those used for families with hereditary DNA MMR.
One limitation of this study is the possibility of underreporting or misreporting of cancers. Efforts were made to verify reported cancers, but this was not always feasible. It is important to note, however, that this deficiency would affect the group A families to the same extent as the group B families. The fact that the group A families revealed a profile of incidences for cancers that are extremely consistent with studies of families with hereditary MMR gene mutations suggests that underreporting or misreporting is unlikely to explain the differences between group A and group B.
With such emphasis in this study on distinguishing between families with and without MSI-H, it is important to consider how clinical risk-assessment triage based on tumor MSI/immunohistochemical testing could be misleading. First, phenocopies can happen in any family (eg, an MSS tumor arising as a sporadic event in a group A family). Second, sporadic MSI-H tumors due to promoter methylation of hMLH1 can arise at any age, and tumor testing cannot distinguish this finding from MSI-H related to germline mutation ofhMLH1.16 Third, there are genes involved in the DNA MMR pathway whose mutation only inconsistently results in an MSI-H tumor (hMSH6, hPMS2).17,18 Additional laboratory testing can be useful in resolving these uncertainties. Use of immunohistochemistry for these gene products can be of great help in defining genetic causality. Testing tumors from multiple affected family members is also a very powerful way to clarify genetic etiology in AC-I families. Genetic testing is available forhMLH1,hMSH2, andhMSH6. All these perspectives underscore the imperative for individualizing cancer risk assessment for AC-I families in whom no DNA MMR defect can be found.
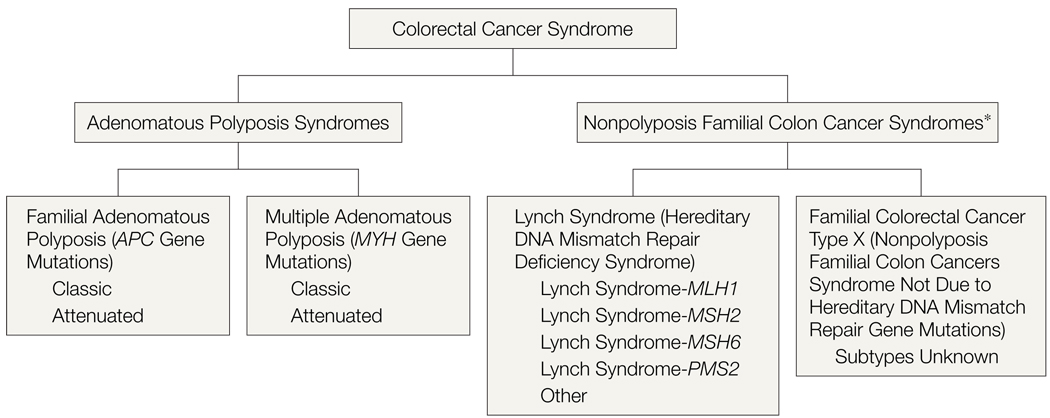
The use of HNPCC as a label needs to be refined or made obsolete. The term HNPCC encompasses considerable heterogeneity and has come to mean different entities to different people. We prefer the term “Lynch syndrome” or “HNPCC-Lynch syndrome” to specify those individuals or families with germline mutations in the DNA MMR genes. As diagnostic and clinical studies are refined, we envision variants described as Lynch syndrome-MLH1, Lynch syndrome-MSH2, and so on (FIGURE 2). It may be reasonable to introduce a term for families similar to our group B families, who have a clustering of colorectal cancer but in whose tumors no DNA MMR gene defect is evident. We suggest the term “familial colorectal cancer type X.” This term does not define this group as having hereditary colorectal cancer (which usually implies single-gene etiology), and it acknowledges our lack of understanding of the etiology (thus the “X”). As additional single-gene disorders are discovered in this group, the remaining families with unexplained cancer etiology can retain the familial colorectal cancer type X designation until the etiology is explained. Regardless of what term is eventually adopted, it is essential that the term HNPCC not be used without clearly defining it, to acknowledge that families with Lynch syndrome (hereditary DNA MMR deficiency) and those with familial colorectal cancer type X are not equivalent entities.
Figure 2.
Categories of Colorectal Cancer Syndromes
Schematic showing the 2 categories of colorectal cancer syndromes, illustrating that nonpolyposis disorders are heterogeneous but based on tumor biology can be distinguished as those having defective mismatch repair (Lynch syndrome; group A) and those with proficient mismatch repair (group B in this study, called here familial colorectal cancer type X). Diagram excludes syndromes characterized by hamartomatous/hyperplastic polyposis.
*Defined by any number of pedigree and/or laboratory criteria, including but not limited to the Amsterdam criteria. Hereditary nonpolyposis colon cancer syndrome is the term that has traditionally been used in this context, encompassing those entities that have emerged as distinguishable clinical entities (ie, Lynch syndrome and familial colorectal cancer type X).
Acknowledgment
We thank the many families who have participated in research studies that facilitated this specific study. We also thank the following individuals for their assistance in this study: Kristin Lee, John Hake, Hema Vankayala, MD, Robert Vierkant, MAS, and Duncan Thomas, PhD.
Funding/Support: This work was supported by the National Cancer Institute, National Institutes of Health, under RFA CA-95-011 and through cooperative agreements with the members of the Colon Cancer Family Registry and principal investigators.
Role of the Sponsors: The National Cancer Institute and the Colon Cancer Family Registry had no role in the design and conduct of the study; in the collection, analysis, and interpretation of the data; or in the preparation, review, or approval of the manuscript.
Footnotes
Reprints/E-prints reprints@ama-assn.org
Author Contributions: Dr Lindor had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Lindor, Rabe, Petersen, Goldberg, de Andrade, Boardman.
Acquisition of data: Lindor, Rabe, Petersen, Baron, Gallinger, Aronson, Hopper, Jass, LeMarchand, Grove, Potter, Newcomb, Terdiman, Conrad, Moslein, Goldberg, Anton-Culver, Thibodeau, Boardman.
Analysis and interpretation of data: Lindor, Rabe, Petersen, Haile, Casey, Baron, Bapat, Hopper, LeMarchand, Potter, Newcomb, Goldberg, Ziogas, de Andrade, Siegmund, Seminara.
Drafting of the manuscript: Lindor, Rabe, Petersen, Baron, Hopper, Moslein, Anton-Culver.
Critical revision of the manuscript for important intellectual content: Lindor, Rabe, Petersen, Haile, Casey, Baron, Gallinger, Bapat, Aronson, Hopper, Jass, LeMarchand, Grove, Potter, Newcomb, Terdiman, Conrad, Moslein, Goldberg, Ziogas, de Andrade, Siegmund, Thibodeau, Boardman, Seminara.
Statistical analysis: Lindor, Rabe, Haile, Ziogas, de Andrade, Siegmund.
Obtained funding: Lindor, Petersen, Gallinger, Hopper, Jass, LeMarchand, Grove, Potter, Newcomb, Seminara.
Administrative, technical, or material support: Lindor, Petersen, Casey, Hopper, Jass, Goldberg, Anton-Culver, Thibodeau, Boardman.
Study supervision: Lindor, Petersen, Hopper, Moslein.
Financial Disclosures: None reported.
Publisher's Disclaimer: Disclaimer: The content of this article does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Cooperative Family Registries, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government or the Cooperative Family Registry. Collaborating centers include the Australian Colorectal Cancer Family Registry (UO1 CA097735), the USC Familial Colorectal Neoplasia Collaborative Group (UO1 CA074799), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (UO1 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (UO1 CA074783), Seattle Colorectal Cancer Family Registry (UO1 CA074794), University of Hawaii Colorectal Cancer Family Registry (UO1 CA074806), and University of California, Irvine Informatics Center (UO1 CA078296).
REFERENCES
- 1.Watson P, Lynch H. Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer. 1993;71:677–685. doi: 10.1002/1097-0142(19930201)71:3<677::aid-cncr2820710305>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 2.Aarnio M, Sankila R, Pukkala E, et al. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer. 1999;81:214–218. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 3.Burke W, Petersen G, Lynch P, et al. Cancer Genetics Studies Consortium. Recommendations for follow-up care of individuals with an inherited predisposition to cancer, I: hereditary nonpolyposis colon cancer. JAMA. 1997;277:915–919. [PubMed] [Google Scholar]
- 4.Jarvinen HJ, Aarnio M. Surveillance on mutation carriers of DNA mismatch repair genes. Ann Chir Gynaecol. 2000;89:207–210. [PubMed] [Google Scholar]
- 5.Smith RA, Von Eschenbach AC, Wender R, et al. American Cancer Society. American Cancer Society guidelines for the early detection of cancer, 2004. CA Cancer J Clin. 2004;54:41–52. doi: 10.3322/canjclin.54.1.41. [DOI] [PubMed] [Google Scholar]
- 6.National Comprehensive Cancer Network. [Accessed March 11, 2005];National Comprehensive Cancer Network Practice Guidelines in Oncology. 2004 Available at: http://www.nccn.org/professionals/physician_gls/PDF/colorectal_screening.pdf.
- 7.Vasen HF, Mecklin J-P, Meera Khan P, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 8.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 9.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 10.Bergstalh EJ, Offord KP, Kosanke JL, Augustine GA. PERSONYRS: A SAS Procedure for Person-Year Analyses. Technical Report Series, No. 31. Cary, NC: SAS Institute Inc; 1986. [Google Scholar]
- 11.National Cancer Institute. Surveillance, Epidemiology, and End Results (SEER) Program SEER*Stat Database: Incidence—SEER 9 Regs Public-Use. Bethesda, Md: National Cancer Institute; 2003. Apr, [computer program] Version 5.2. based on the November 2002 submission. [Google Scholar]
- 12.Efron B. Regional Conference Series in Applied Mathematics, No. 38. Philadelphia, Pa: SIAM; 1982. The Jackknife, the Bootstrap and Other Resampling Plans. [Google Scholar]
- 13.Cox DR. Some simple approximate tests for Poisson variates. Biometrika. 1953;40:354–360. [Google Scholar]
- 14.Jass JR, Cottier DS, Jeevaratnam P, et al. Diagnostic use of microsatellite instability in hereditary non-polyposis colorectal cancer. Lancet. 1995;346:1200–1201. doi: 10.1016/s0140-6736(95)92902-9. [DOI] [PubMed] [Google Scholar]
- 15.Scott RJ, McPhillips M, Meldrum CJ, et al. Hereditary nonpolyposis colorectal cancer in 95 families: differences and similarities between mutation-positive and mutation-negative kindreds. Am J Hum Genet. 2001;68:118–127. doi: 10.1086/316942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakar S, Burgart LJ, Thibodeau SN, et al. Frequency of loss of hMLH1 expression in colorectal carcinoma increases with advancing age. Cancer. 2003;97:1421–1427. doi: 10.1002/cncr.11206. [DOI] [PubMed] [Google Scholar]
- 17.Buttin BM, Powell MA, Mutch DG, et al. Penetrance and expressivity of MSH6 germline mutations in seven kindreds not ascertained by family history. Am J Hum Genet. 2004;74:1262–1269. doi: 10.1086/421332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucci-Cordisco E, Rovella V, Carrara S, et al. Mutations of the “minor” mismatch repair gene MSH6 in typical and atypical hereditary nonpolyposis colorectal cancer. Fam Cancer. 2001;1:93–99. doi: 10.1023/a:1013872914474. [DOI] [PubMed] [Google Scholar]