Abstract
Context
Models have been developed to predict the probability that a person carries a detectable germline mutation in the BRCA1 or BRCA2 genes. Their relative performance in a clinical setting is unclear.
Objective
To compare the performance characteristics of four BRCA1/BRCA2 gene mutation prediction models: LAMBDA, based on a checklist and scores developed from data on Ashkenazi Jewish (AJ) women; BRCAPRO, a Bayesian computer program; modified Couch tables based on regression analyses; and Myriad II tables collated by Myriad Genetics Laboratories.
Design and setting
Family cancer history data were analyzed from 200 probands from the Mayo Clinic Familial Cancer Program, in a multispecialty tertiary care group practice. All probands had clinical testing for BRCA1 and BRCA2 mutations conducted in a single laboratory.
Main outcomes measures
For each model, performance was assessed by the area under the receiver operator characteristic curve (ROC) and by tests of accuracy and dispersion. Cases “missed” by one or more models (model predicted less than 10% probability of mutation when a mutation was actually found) were compared across models.
Results
All models gave similar areas under the ROC curve of 0.71 to 0.76. All models except LAMBDA substantially under-predicted the numbers of carriers. All models were too dispersed.
Conclusions
In terms of ranking, all prediction models performed reasonably well with similar performance characteristics. Model predictions were widely discrepant for some families. Review of cancer family histories by an experienced clinician continues to be vital to ensure that critical elements are not missed and that the most appropriate risk prediction figures are provided.
Keywords: Ashkenazi Jewish, Bayesian, BRCA1, BRCA2, Breast cancer, Risk prediction
Introduction
It is estimated that about 1 in 500 people in the U.S.A. carry deleterious mutations in the genes BRCA1 or BRCA2. The prevalence is higher in some ethnic groups, such as individuals of Ashkenazi Jewish (AJ) descent for whom the prevalence is about 1 in 40. In a pooled analysis of 22 studies of the relatives of 500 women with breast and ovarian cancer not selected for family history [1], the cumulative risk to age 70 years of female breast cancer was estimated to be 65% (95% confidence interval [CI] 44–78%) for BRCA1 mutation carriers and 45% (95% CI 31–56%) for BRCA2 mutation carriers. For ovarian cancer, the same cumulative risks were 39% (95% CI 18–54%) and 11% (95% CI 2.4–19%) for BRCA1 and BRCA2 mutation carriers, respectively. Other studies, particularly those performed on selected ethnic groups or on clinic-based population have reported even greater risks [2, 3]. The breast cancers that occur in BRCA1 and BRCA2 mutation carriers are on average diagnosed twenty years earlier than in the general population. There is also evidence of increased risks for carriers of cancers of the male breast, peritoneum, fallopian tube, prostate, pancreas, biliary system, stomach, and cutaneous and ocular melanoma [4–6].
These well-documented facts have led to recommendations for cancer risk management that are vastly different and more aggressive than recommendations for women with average cancer risks [7, 8]. Therefore, it is incumbent upon health care providers to consider carefully if a woman may have a high risk of being a carrier of a BRCA1 or BRCA2 mutation so that appropriate medical options can be offered.
Mutation analysis of the BRCA1 and BRCA2 genes has been clinically available since the mid-1990s. The decision to proceed with genetic testing is complex and requires discussion about how a test might impact on medical decision making, consideration of possible psychological results of testing, and addressing any cost and insurance implications. The original genetic testing guidelines proposed in 1996 by the American Society of Clinical Oncology [9] were sometimes interpreted as implying that mutation testing for BRCA genes should be offered to those individuals in whom there was a 10% or greater probability of finding a BRCA1 or BRCA2 mutation. This threshold remains a non-binding guide for genetic counseling of women considering mutation testing.
A variety of models have now been developed to predict the probability that a woman carries a detectable germline mutation in BRCA1 or BRCA2, based on her personal and family cancer history. In practice, predictions across different models are usually compared when providing a risk assessment and making decisions: a high probability would support genetic testing and/or aggressive clinical management, whereas a low probability might support a decision to forego testing. Thus, medical decisions may be directly linked to the quality of information generated by these models. In this study, we compared the performance characteristics of four mutation prediction models using families seen in the Mayo Clinic Familial Cancer Program.
Methods
Subjects
Between l996 and 2003, probands (defined as the initial consultants in the families) from 260 independent families had cancer risk assessment evaluation and counseling from the Mayo Clinic Familial Cancer Program and subsequently had clinical genetic testing for mutations in BRCA1 and BRCA2. No a priori selection criteria were required. This is a clinic-based population representing those who opted to have genetic testing after having comprehensive genetic risk assessment counseling. Thirteen families with male probands were excluded from this study because only one of the models being evaluated estimated the mutation probability for males. Eleven families were excluded because the proband had only a variant of unknown significance (VUS) in BRCA1 (n = 2) or BRCA2 (n = 9). A detailed family history of cancer diagnoses was collected by either a genetic counselor or a Familial Cancer Program study coordinator trained in genealogic collection. If possible, the pedigrees were extended to all affected and unaffected third degree relatives of the consultant. Efforts were routinely made to verify the diagnoses of breast and ovarian cancers in relatives, but this was inconsistently accomplished (less than half of cases) so risk estimates were most often reliant on family history reports alone, as is common in clinic practices. Thirty six families were excluded because several ages were unknown in a pedigree or ages at diagnosis of breast or ovarian cancer were not provided. For the purposes of this study, ductal carcinoma in situ was not considered to be invasive breast cancer, and peritoneal cancer was considered equivalent to ovarian cancer. No fallopian tube cancers were reported in these families. All consultants studied had given permission for use of their medical records for research purposes and this study was approved by the Mayo Clinic Institutional Review Board.
The final study sample consisted of 200 Caucasian probands; 30 (15%) were self-reported to be of AJ ancestry and 46 (23%) had no previous diagnosis of breast or ovarian cancer. Of the 154 with a previous diagnosis, 117 (76%) had breast cancer only, 27 (18%) had ovarian cancer only, and 10 (6%) had both breast and ovarian cancer. Forty-six probands had no type of cancer diagnosis. They were still considered to be the probands in these families.
All mutation testing for BRCA1 and BRCA2 was performed by Myriad Genetics Laboratories, Inc. and consisted of complete sequencing of both genes and, since 8/2003, testing for a large rearrangement-panel in BRCA1. Comprehensive testing was performed for all probands, affected or unaffected, Jewish and non-Jewish.
Models for mutation prediction
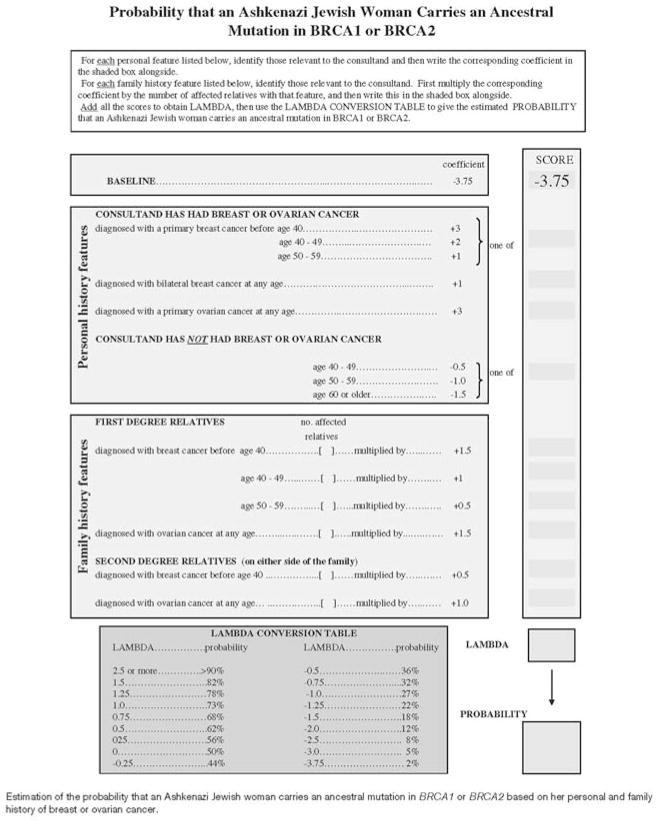
Table 1 describes the four models used to estimate the probability that the proband was a mutation carrier. The LAMBDA model [10] was developed specifically for AJ women who were tested only for the three founder mutations. It involves identifying whether the consultant and her first- and second-degree relatives had a diagnosis of breast or ovarian cancer, and if so at what age, checking appropriate boxes, summing scores and reading a probability off a prepared table. Figure 1 shows a sample worksheet for LAMBDA calculations. We applied it to both our AJ and non-AJ probands, who all had the comprehensive genetic testing, with no modification of the scoring system.
Table 1.
Description of the four models used to predict the probability that a woman caries a germline deleterious mutation in BRCA1 or BRCA2
| Model (reference) | Derivation | Input used | Comments | Output |
|---|---|---|---|---|
| LAMBDA [10] | Multiple Logistic Regression analysis of 424 AJ women in Australia and the UK with personal or family history of breast or ovarian cancer, tested for the three AJ founder mutations only | Work sheet with score based on point system on personal, FDRa, and SDRb family history of breast and ovarian cancer, including age at diagnosis, bilateral disease in proband | Women only; designed for use in AJ women | Combined probability of BRCA1 and BRCA2 for AJ women |
| Myriad II [11] | Empiric data from 10,000 women on whom BRCA sequencing was conducted | On-line risk tables, updated periodically, utilizing personal and first- and second-degree family history, for breast and ovarian cancers, grouped by those over and under age 50 years at diagnosis of breast cancer | Women only; only affected family members included; different tables for AJ and non-AJ | Combined probability for BRCA1 and BRCA2 for non- AJ or AJ women |
| BRCAPRO [12, 13] | Bayesian model assuming autosomal dominant inheritance, based on family history of FDRa and SDRb, incorporating unaffected relatives, compared with SEERc data | Cancer type and age at diagnosis in proband, FDR, and SDR entered into free computer program; only breast and ovarian cancers used in risk assessments | Men and women; allows for both AJ and non-AJ; utilizes number and ages of affected and unaffected relatives in risk assessment | Predicts individual and combined probabilities for BRCA1 and BRCA2 genes, and also provides Couch model and Myriad II model results |
| Couch [14] | Logistic regression based on 263 families of women with family history of breast cancer with or without ovarian cancer | Published tables using average age of breast cancer, and presence or absence of ovarian cancer in predicting probability of BRCA1 mutation | Women only; different tables for AJ/non-AJ; does not predict BRCA2 mutation probability | Predicts probability of deleterious BRCA1 mutation only |
FDR = first-degree relative
SDR = second-degree relative
Surveillance, Epidemiology, and End Results (SEER) program [15]
Fig. 1.
Clinic sheet from Apicella et al [10]
The second model was derived from the Myriad II tables, updated in Spring 2004 (http://www.myriad-tests.com/provider/brca-mutation-prevalence.htm) [11]. While a 2006 update has recently been posted, the data used in the 2004 tables would be more concurrent with the testing of our cohort.
The third model was the BRCAPRO computer program based on a Bayesian statistical algorithm, CancerGene 3.3.2b, University of Texas Southwestern Medical Center, Dallas, TX [12, 13].
The fourth model tested was an adaptation of the BRCA1-only prediction model based on the Couch tables (sometimes called the Penn I model) [14], and was generated by the BRCAPRO program output. We calculated and analyzed a score that we designated “Couch 1.5”, which involved multiplying the Couch table score by 1.5 and truncating at 1 to simulate a practice that is sometimes used by genetic counselors in clinical risk assessment (personal communications with author). The number derived is thought to predict the combined probability of a mutation in either BRCA1 or BRCA2, based on observations that BRCA1 mutations outnumber BRCA2 mutations in most series by a factor of about two to one.
There was adequate information to calculate an estimate for all 200 probands using the LAMBDA and Myriad II scores, for 182 using BRCAPRO, and for 170 using the Couch 1.5 score. (Note that only BRCAPRO requires information on all unaffected relatives, Couch tables require exact ages of all affected relatives, whereas the Myriad tables require only under-50/over-50 dichotomy for breast cancer.)
Statistical methods
The fits of models to the observed data were assessed in several ways. The Receiver Operating Characteristic (ROC) curve is a plot of Y = sensitivity (the proportion of subjects correctly classified when the subject is a carrier) against corresponding values of X = 1 – specificity (the proportion of subjects incorrectly classified when the subject is not a carrier). If a model has no discriminatory power the curve will not differ from the reference line Y = X, while perfect discriminatory power will result in the lines X = 0 and Y = 1. The area under the ROC curve is, therefore, a global measure of a model’s predictive performance. It can be interpreted as the probability that the model’s prediction for a randomly selected carrier (from the pool of probands) will be greater than its prediction for a randomly selected non-carrier. The greater the area, the better the model’s performance in ranking probands by carrier probability.
Each ROC curve was generated by defining a sensitivity and 1 – specificity for every number c between 0 and 1. Each c was interpreted as a cut-off to which the model’s outcome probabilities were compared, giving a binary test for which sensitivity and 1 – specificity could be calculated in the usual way. The resulting points were connected by straight lines, and the area under the curve calculated by the trapezoid rule. Standard errors and confidence intervals for the area under the ROC curve, and tests for the equality of areas under the ROC curve, were calculated using algorithms suggested by DeLong et al. [16]. Note that the area under the ROC curve depends only on the relationship between sensitivity and specificity and not on the actual values of the predicted probabilities provided their order is unchanged. That is, it is a test of the ranking of observed versus predicted probability status only.
Therefore, we also conducted two tests to assess the fit of the predicted carrier probabilities to the observed data, as described in Cox and Snell [17] and applied in Apicella et al. [10]. These tests assess the predicted carrier probabilities for systematic under- or over-estimation (i.e. accuracy) and for under- or over-dispersion. Over-dispersion typically occurs when too few of the probands with high predicted carrier probabilities are carriers (i.e. the predictions are too high at the top end) and/or too many of the probands with low predicted probabilities are carriers (i.e. the predictions are too low at the bottom end).
The degree to which a model over-estimates or is over-dispersed can be estimated (with standard errors) using logistic regression [17]. We can therefore compare the accuracy and dispersion of models by comparing their corresponding regression coefficients. We assume (conservatively) that these coefficients are uncorrelated. This likely gives an under-estimate of the variance of their difference so, for example, when testing whether two models differ in accuracy or dispersion the true P-value will be less than the one cited.
Statistical analyses were performed with STATA version 9.0 and R version 2.1.0 [18]. Following convention, all statistical tests were two-sided and statistical significance was based on a P-value of less than 0.05.
Results
Overall, 46 (23%) consultants had deleterious BRCA1 mutations, and 20 (10%) had deleterious BRCA2 mutations (total 33%). Of the 30 AJ consultants, six (20%) were carriers, one of whom had a BRCA mutation other than a founder mutation. Table 2 shows the relationship between proband age, cancer status, and mutation results.
Table 2.
Description of the study population
| Cancer status | Age group | Non-carriers | BRCA1 carriers | BRCA2 carriers | Total |
|---|---|---|---|---|---|
| Unaffected | <40 | 12 (86%) | 2 (14%) | 0 (0%) | 14 |
| 40–49 | 11 (73%) | 2 (13%) | 2 (13%) | 15 | |
| 50–59 | 6 (75%) | 2 (25%) | 0 (0%) | 8 | |
| 60+ | 7 (78%) | 2 (22%) | 0 (0%) | 9 | |
| Total unaffected | 36 (78%) | 8 (17%) | 2 (4%) | 46 | |
| Breast cancer only | <40 | 18 (53%) | 12 (35%) | 4 (12%) | 34 |
| 40–49 | 33 (72%) | 8 (17%) | 5 (11%) | 46 | |
| 50–59 | 20 (77%) | 1 (4%) | 5 (19%) | 26 | |
| 60+ | 9 (82%) | 1 (9%) | 1 (9%) | 11 | |
| Ovarian cancer only | <40 | 4 (67%) | 2 (33%) | 0 (0%) | 6 |
| 40–49 | 2 (25%) | 6 (75%) | 0 (0%) | 8 | |
| 50–59 | 2 (67%) | 1 (33%) | 0 (0%) | 3 | |
| 60+ | 8 (80%) | 1 (10%) | 1 (10%) | 10 | |
| Breast plus ovarian cancer | <40 | 1 (33%) | 2 (67%) | 0 (0%) | 3 |
| 40–49 | 1 (20%) | 2 (40%) | 2 (40%) | 5 | |
| 50–59 | 0 (0%) | 2 (100%) | 0 (0%) | 2 | |
| 60+ | 0 | 0 | 0 | 0 | |
| Total affected | 98 (64%) | 38 (25%) | 18 (12%) | 154 |
Table 3 and Fig. 2 describe the performance of the four models. The median predicted probabilities were 36% for LAMBDA, 29% for BRCAPRO, 28% for Couch 1.5 and 17% for Myriad II. LAMBDA was the only model that over-predicted carriers (by 11%). The other three models underpredicted carriers by 15% (BRCAPRO), 19% (Couch 1.5) and 48% (Myriad II). A formal test of observed versus predicted values found that the deviation was not statistically significant for LAMBDA (P = 0.3), but was for all other models; BRCAPRO (P = 0.01), Couch 1.5 (P = 0.01) and Myriad II (P < 0.001). LAMBDA gave a more accurate prediction than Myriad II (P < 0.001) and Couch 1.5 (P = 0.05).
Table 3.
Observed and expected numbers of mutation carriers as predicted by the different models, for categories based on each model’s predicted range of probability of being a carrier
| Model | Predicted probability | Number of women (%) | Numbers of carriers (%) |
||
|---|---|---|---|---|---|
| Observed | Expected | (E − O)/O (%) | |||
| LAMBDA | <10 | 53 | 11 | 3.0 (6%) | −73 |
| 10–25 | 37 | 7 | 6.7 (18%) | −4 | |
| 25–50 | 53 | 14 | 19.3 (36%) | +38 | |
| ≥50 | 57 | 34 | 42.9 (74%) | +26 | |
| Total | 200 | 66 (33%) | 72.0 (36%) | +11 | |
| BRCAPRO | <10 | 93 | 15 | 2.2 (2%) | −85 |
| 10–25 | 19 | 6 | 2.9 (15%) | −52 | |
| 25–50 | 28 | 13 | 10.2 (36%) | −10 | |
| ≥50 | 42 | 28 | 36.8 (88%) | +31 | |
| Total | 182 | 62 (34%) | 52.2 (29%) | −15 | |
| Couch 1.5 | <10 | 67 | 14 | 3.1 (5%) | −78 |
| 10–25 | 45 | 13 | 7.5 (17%) | −42 | |
| 25–50 | 26 | 10 | 9.5 (37%) | −5 | |
| ≥50 | 32 | 21 | 26.7 (83%) | +27 | |
| Total | 170 | 58 (34%) | 46.8 (28%) | −19 | |
| Myriad II | <10 | 82 | 17 | 4.9 (6%) | −71 |
| 10–25 | 84 | 24 | 13.9 (17%) | −42 | |
| 25–50 | 28 | 20 | 10.9 (39%) | −46 | |
| ≥50 | 6 | 5 | 4.4 (73%) | −12 | |
| Total | 200 | 66 (33%) | 34.0 (17%) | −48 | |
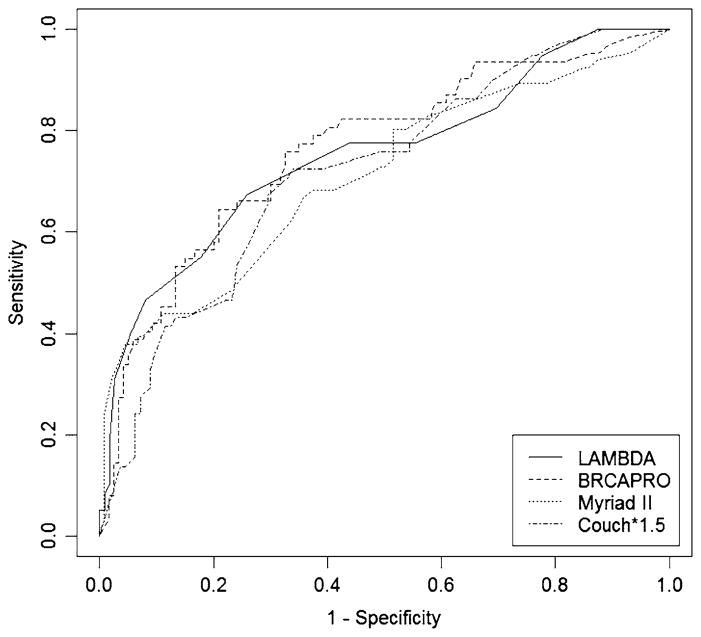
Fig. 2.
Receiver operator curves (ROCs) for the four models
A consistent feature of all models was the large under-estimation for women with low predicted carrier probabilities. For example, each model predicted that a substantial proportion of women had a carrier probability of less than 10%, yet there were at least three times as many observed carriers in this subgroup than predicted by the models. This phenomenon persisted for all models, except LAMBDA, up to the predicted probability range of 10–25%. On the other hand, all models except Myriad II over-predicted the number of carriers in the group of women with highest predicted carrier probabilities (e.g. ≥50%). A formal test found evidence of over-dispersion for all models (all P < 0.001), increasing in strength from LAMBDA to Myriad II to Couch 1.5 to BRCAPRO ( , 40, 100 and 180, respectively). LAMBDA was less dispersed than the latter two models (P < 0.02).
Table 4 shows that the under the ROC curves were similar for the four models. Ranging only from 0.76 to 0.71, BRCAPRO had the highest area under the ROC curve, but the confidence intervals illustrate that the small differences were not statistically significant (P = 0.3).
Table 4.
Areas under the ROC curves for the different models
| Model | n | Area under ROC curve (95% CI) |
|---|---|---|
| LAMBDA | 200 | 0.73 (0.66–0.79) |
| BRCAPRO | 182 | 0.76 (0.70–0.82) |
| Couch 1.5 | 170 | 0.72 (0.64–0.78) |
| Myriad II | 200 | 0.71 (0.64–0.77) |
Table 5 shows the 21 families in which one or more of the models predicted <10% chance of a mutation, but a deleterious mutation was found. In this subset of 21 families, the LAMBDA model “missed” the fewest carriers (33%), compared with BRCAPRO (71%), Couch 1.5 (61%), and Myriad II (76%).
Table 5.
The 21 women for whom a deleterious mutation was identified but one or more of the mutation-prediction models gave <10% probability which is considered here as “missing” the mutation
| Proband ID | LAMBDA | BRCAPRO | Myriad II | Couch 1.5 |
|---|---|---|---|---|
| 1 | 9.5 | 1.9 | 9.2 | NA |
| 2 | 22 | 12 | 9.2 | NA |
| 3 | 15 | 2.0 | 9.2 | NA |
| 4 | 44 | 5.9 | 23 | 55 |
| 5 | 68 | 9.3 | 23 | 55 |
| 6 | 32 | 2.3 | 6.9 | 17 |
| 7 | 32 | 1.6 | 12 | 17 |
| 8 | 9.5 | 2.4 | 12 | 16 |
| 9 | 44 | 7.5 | 16 | 7.7 |
| 10 | 6.5 | 1.4 | 9.2 | 5.4 |
| 11 | 44 | 31 | 6.9 | 2.1 |
| 12 | 56 | 98 | 6.9 | 3.2 |
| 13 | 15 | 30 | 8.4 | 3.9 |
| 14 | 15 | 44 | 8.4 | 3.9 |
| 15 | 32 | 98 | 6.9 | 27 |
| 16 | <2 | 8 | 3.1 | 3.2 |
| 17 | 9.5 | 88 | 4.3 | 19.5 |
| 18 | 15 | 1.6 | 5.7 | 7.5 |
| 19 | 15 | 5.0 | 5.7 | 4.8 |
| 20 | 9.5 | 3.0 | 2.8 | 4.8 |
| 21 | 7.0 | 1.4 | 4.2 | 3.8 |
| Total “missed” (%) | 7/21 (33%) | 15/21 (71%) | 16/21 (76%) | 11/18 (61%) |
Rows show the predicted mutation probability predicted for that case by each model. Cases 1–15 had BRCA1 mutations while cases 16–21 had BRCA2 mutations. High-risk features in some pedigrees were “missed” by all models, while in other cases models showed wide variation in risk prediction. Overall LAMBDA “missed” the fewest
Discussion
Evaluation of a human pedigree disease data for possible transmission of genetic susceptibility is complex and made all the more difficult by small family size, missing information, incomplete penetrance, variable expressivity, and environmental exposures that vary from person to person. Nevertheless, the family’s medical history remains a powerful tool for identifying individuals most likely to have a high genetically inherited risk for specific diseases. This has become all the more relevant now with the discovery of some genes which, when mutated in the germ-line, are responsible for a proportion of common diseases.
We have considered four models that derive, for a woman with a particular personal and family history of breast and ovarian cancer, the probability that she carries a germline deleterious mutation in the genes BRCA1 or BRCA2, and therefore is at substantially increased risk of breast and ovarian cancer. All models gave comparable areas under the ROC curve; i.e. they are globally similar to one another in how well they rank women according to carrier probability. All models except for LAMBDA underestimated the total number of carriers in the cohort. All models were over-dispersed; i.e. they generally predicted too high a probability for those most likely to be carriers, and/or too low a probability for those least likely to be carriers.
Our interest in assessing the LAMBDA model derived from observing how straightforward it was to use in an office setting without requiring a computer program (Fig. 2), while being more nuanced in use of age ranges than the Myriad II tables. LAMBDA was developed using data from AJ women. It is, perhaps, surprising that it performed well without any adjustment of the scoring system in families attending the Mayo Clinic, given that only 15% were of AJ descent. The LAMBDA model was less over-dispersed then the other three models. LAMBDA also was less likely to “miss” mutation carriers than all other models, which is extremely important in clinical practice where missing a carrier is of more significance than overestimating the risk of a non carrier. Further studies of this model in other non-AJ population are warranted.
The computer-based BRCAPRO model showed substantial under-estimation of risk in the lower risk groups and over-estimation in the highest risk group. It was the most demanding to implement due to the amount of data required—it is necessary to know of the existence and ages of each unaffected relative. The integration of such data using a Bayesian approach likely contributed to the model’s ability to give a higher probability to carriers in some families that were given a lower probability (i.e. more likely to be “missed”) by other models. The time required to enter data into a computer program is a deterrent to BRCAPRO becoming an office tool for any busy genetics clinic in its current form.
While the Myriad II tables were convenient to use, the quality of the data they are based on is highly dependent on the clinicians who ordered the mutation testing. Furthermore, the categorization of breast cancer cases in relatives is restrictive. Only those under 50 years at diagnosis are considered relevant. The tables do not recognize the difference between having no relatives with breast cancer after age 50 versus having multiple relatives with breast cancer after age 50. Like the other models information on third degree relatives are not incorporated. Despite these recognized issues, the tables performed similarly well in ranking carriers as did the other models.
The Couch tables, while convenient and simple to use, were created strictly for BRCA1 prediction. A rule of thumb sometimes used in the genetic counseling community (personal communications) is to multiple the “Couch” score for an individual by 1.5 so as to predict being a carrier of a mutation in either BRCA1 or BRCA2. To our knowledge this approach has never been validated. Used as intended, the “Couch” score matched BRCA1 results reasonably well (Fig. 3). Using the 1.5 multiplication rule-of-thumb gave results similar to Myriad II and BRCAPRO, with over-estimation of risk in the highest risk group and substantial under-estimation in the lower risk group.
Fig. 3.
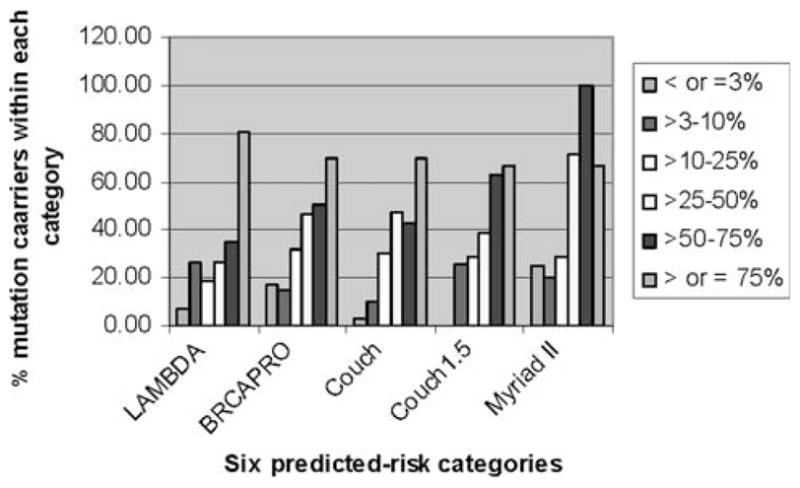
A comparison of the capacity of four BRCA-mutation-prediction models to predict deleterious mutations is shown. The color-coded bars show five prediction groups for each model and the height of each bar shows the percentage of cases in that predicted risk-group for which a mutation was actually found. For example, for the tallest bar for LAMBDA model (>75% predicted-likelihood-of-mutation), 79% of cases in that group actually had a deleterious mutation results reasonably well (Fig. 3). Using the 1.5 multiplication rule-of-thumb gave results similar to Myriad II and BRCAPRO, with over-estimation of risk in the highest risk group and substantial under-estimation in the lower risk group.
The area under the ROC curve has been used by almost every study of carrier prediction models as a test of performance, yet it only tests the ability of models to rank carriers. This test does not reveal if the models gave similar results across families. It is of great clinical relevance to know if the four models “missed” the same probands, so we reviewed the 21 women for whom a deleterious mutation was discovered (15 in BRCA1 and 6 in BRCA2) but for whom one or more of the models predicted a <10% carrier probability (Table 5). For some, such as proband 1, no model predicted a high probability. The proband was unaffected but had two paternal aunts with ovarian cancer and several third and fourth degree relatives with early onset breast cancer. None of the models take into account relatives of the consultant more distant than second degree. A genetic counselor would likely detect this pattern of cancer in the extended paternal lineage, underscoring the importance of collecting an extended pedigree and experienced clinical review. On the other hand, there were families with wide discrepancies in prediction, such as proband 12. In her family there were four women with breast cancer, one bilateral. Two of the breast cancers were diagnosed before the age of 50 years, and the others were diagnosed in the 50s and 60s. The high incidence of breast cancer in this family was interpreted by the BRCAPRO and LAMBDA models as suggesting the proband was highly likely to be a carrier, but not by the Myriad II and Couch 1.5 models. Of this set of 21 carrier probands “missed” by one or more models through using the 10% threshold, the LAMBDA model “missed” the fewest (33%), whereas BRCAPRO “missed” 71%, Couch 1.5 “missed” 61%, and Myriad II “missed” 76%.
We considered what factors related to the nature of our Mayo Clinic practice may have affected the performance characteristics of the models. Unlike some model development processes, we did not have any pre-determined inclusion criteria on who could have BRCA testing. Individuals are referred to the clinic either because of their interest or concern about their cancer risk or the concern of their personal provider. Once referred to the genetic clinic, they receive comprehensive individualized risk assessment, counseling of risks and benefits of testing including information on costs, psychological factors, and medical and surgical options for screening and risks reduction. After that discussion, individuals decide if they want to have testing. This is quite different from some centers or research registries in which predetermined family history criteria may be required to access genetic testing. As a result, we may see people who are concerned but who are actually at low risk and we may see people with more extended family histories that are not captured by simplistic family history criteria, but may be important never-the-less. For example, cancer in cousins (third degree relatives) is not included in most models. Small family size can limit eligibility for registries with pre-determined requirements and some registries might not recruit or test an unaffected family member. We thus may be ascertaining families outside the boundaries upon which the models were originally designed. We would argue, though, that understanding the performance of the models in real world populations is of considerable interest and value.
Several other groups have been evaluating risk prediction model performance [19–24]. James et al. [20] studied 257 probands from a cancer family clinic in Melbourne, Australia (27% with mutations) and compared performance of the Myriad, Couch, BRCAPRO, FHAT [25] and MANCHESTER [26] approaches. They found the best discriminator between carriers and non carriers was BRCAPRO, and this could be enhanced by incorporating pathology data. Antoniou et al. [21] studied 195 French-Canadian probands and compared predicted carrier probabilities under the BOADICEA [27] and BRCAPRO computer models. They found, as we did, that BRCAPRO over-predicted carriers with high predicted probabilities. Barcenas et al. [22] studied 472 probands (21% AJ) and compared BRCAPRO, BOADICEA, Myriad II, Manchester and Couch models. The conclusion was BOADICEA performed better than the other models for AJ families, while overall, BRCAPRO, BOADICEA, and Myriad II performed similarly with Myriad II being the easiest of these to use. Nanda et al. [23] evaluated BRCAPRO in African-American probands and found that BRCAPRO performed as well in this population as in the white and AJ families. Euhus et al. [24] studied 301 probands (42% AJ), comparing BRCAPRO with six experienced counselors. The sensitivity for identifying mutation carriers was equivalent, but BRCAPRO showed slightly superior ability to discriminate carriers from non carriers. Gerdes et al. [28] studied 267 Danish families with high-risk family histories and compared results between the Myriad tables and the Manchester model. [26] The Manchester model uses a scoring system with a maximum of 10 points for each gene, intended to reflect a >10% probability of finding a mutation in that gene. Using a 10% threshold for recommending testing, the updated Manchester model would have had 84% sensitivity and 44% specificity compared to the Myriad model which would have had 79% sensitivity and 43% specificity. Kang et al. [29] analyzed 380 pedigrees on the BRCAPRO, Manchester, Couch/Penn, and Myriad models and reported the area under the ROC as about 0.75 for all models. Simard et al. [30] studied 256 high-risk families among French-Canadians in Quebec, Canada. They compared the Myriad tables with the Manchester model and a logistic regression approach derived from the data in this study and reported that their logistic regression and the Manchester scores provided equal predictive powers and both were significantly better than the Myriad tables. Using a Manchester score of ≥18, provided a sensitivity of 86% and specificity of 82%. No published studies have reported cross-model comparisons with the LAMBDA model to date.
An important observation from our study is that LAMBDA, developed for AJ women, out-performed or at worst matched the other major tools when applied to a general clinical setting in which the vast majority (85%) were not AJ. This study suggests that the simple LAMBDA scoring system may have considerable validity outside the AJ setting, at least for ranking women in terms of their carrier probabilities. It is likely that, given an appropriately large data set, LAMBDA can be adjusted to give a better fit to non-AJ women than has been achieved here, especially if information on pathology features of tumors is available.
The results of our study reflect in part the referral and testing pattern of one institution and may not reflect model performance at other institutions. Our study has shown that BRCA1 and BRCA2 mutation probability prediction models can give misleading and discordant results in some families, especially at the extreme ends of the probability scale. It has also shown that review of cancer family histories by an experienced clinician to supplement use of multiple models to provide risk estimates would appear to be the best strategy to assure that critical elements of clinical risk assessment are not overlooked.
Abbreviations
- AJ
Ashkenazi Jewish
- CI
Confidence interval
- ROC
Receiver operating characteristic curve
- VUS
variant of unknown significance
Contributor Information
Noralane M. Lindor, Email: nlindor@mayo.edu, Department of Medical Genetics, Mayo Clinic College of Medicine, 200 First Street SW, Rochester, MN 55905, USA
Rachel A. Lindor, College of St. Benedict, St. Joseph, MN, USA
Carmel Apicella, The University of Melbourne, Melbourne, Australia.
James G. Dowty, The University of Melbourne, Melbourne, Australia
Amanda Ashley, Department of Medical Oncology, Mayo Clinic College of Medicine, Rochester, MN, USA.
Katherine Hunt, Department of Hematology/Oncology, Mayo Clinic Arizona, Scottsdale, USA.
Betty A. Mincey, Cancer Clinical Studies Unit, Mayo Clinic Jacksonville, Jacksonville, USA
Marcia Wilson, Department of Medical Genetics, Mayo Clinic College of Medicine, 200 First Street SW, Rochester, MN 44905, USA.
M. Cathie Smith, Cancer Clinical Studies Unit, Mayo Clinic Jacksonville, Jacksonville, USA.
John L. Hopper, The University of Melbourne, Melbourne, Australia
References
- 1.Antoniou A, Pharoah P, Narod S, et al. Average risk of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King M, Marks J, Mandell J, et al. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 3.Heimdal K, Maehle L, Apold J, et al. The Norwegian founder mutations in BRCA1: high penetrance confirmed in an incident cancer series and differences observed in the risk of ovarian cancer. Eur J Cancer. 2003;39(15):2205–2213. doi: 10.1016/s0959-8049(03)00548-3. [DOI] [PubMed] [Google Scholar]
- 4.Consortium TBCL. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91(15):1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 5.Thompson D, Easton D. Cancer incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002;94:1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 6.Medeiros F, Muto M, Lee Y, et al. The tubal fimbria is a preferred site for early adenocarcinoma in women with familial ovarian cancer syndrome. Am J Surg Pathol. 2006;30(2):230–236. doi: 10.1097/01.pas.0000180854.28831.77. [DOI] [PubMed] [Google Scholar]
- 7.Narod S, Offit K. Prevention and management of hereditary breast cancer. J Clin Oncol. 2005;23(8):1656–1663. doi: 10.1200/JCO.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 8.Sogaard M, Kjaer S, Gayther S. Ovarian cancer and genetic susceptibility in relation to the BRCA1 and BRCA2 genes. Occurrence, clinical importance and intervention. Acta Obstet Gynecol Scand. 2006;85(1):93–105. doi: 10.1080/00016340500324621. [DOI] [PubMed] [Google Scholar]
- 9.ASCO ASoCO. Statement of the American Society of Clinical Oncology: genetic testing for cancer susceptibility, Adopted on February 20, 1996. J Clin Oncol. 1996;14:1730–1736. doi: 10.1200/JCO.1996.14.5.1730. [DOI] [PubMed] [Google Scholar]
- 10.Apicella C, Andrews L, Hodgson S, et al. Log odds of carrying an Ancestral Mutation in BRCA1 or BRCA2 for a defined personal and family history in an Ashkenazi Jewish woman (LAMBDA) Breast Cancer Res. 2003;5(6):R206–R216. doi: 10.1186/bcr644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20(6):1480–90. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 12.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62(1):145–158. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry D, Iversen EJ, Gudbjartsson D, et al. BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J Clin Oncol. 2002;20(11):2701–2712. doi: 10.1200/JCO.2002.05.121. [DOI] [PubMed] [Google Scholar]
- 14.Couch F, DeShano M, Blackwood M, et al. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med. 1997;336(20):1409–1415. doi: 10.1056/NEJM199705153362002. [DOI] [PubMed] [Google Scholar]
- 15.National Cancer Institute. Surveillance, Epidemiology, and End Results SEER program stat database: incidence, SEER 9 Regs public-use. Version 5.2 (computer program) National Cancer Institute; Bethesda, MD: 2003. [Google Scholar]
- 16.DeLong E, DeLong D, Clarke-Pearson D. Comparing the areas under two to more correlated receiver operating curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 17.Cox D, Snell E. Analysis of binary data. 2. Chapman & Hall; London: 1989. [Google Scholar]
- 18.R Development Core Team. R: a language and environment for statistical computing. R Foundation for Stastistical Computing; Vienna, Austria: 2005. http://www.R-project.org. Cited 27 Sept 2006. [Google Scholar]
- 19.Antoniou A, Easton D. Risk prediction models for familial breast cancer. Future Oncol. 2006;2:257–274. doi: 10.2217/14796694.2.2.257. [DOI] [PubMed] [Google Scholar]
- 20.James P, Doherty R, Harris M, et al. Optimal selection of individuals for BRCA mutation testing: a comparison of available methods. J Clin Oncol. 2006;24(4):707–715. doi: 10.1200/JCO.2005.01.9737. [DOI] [PubMed] [Google Scholar]
- 21.Antoniou A, Durocher F, Smith P, et al. BRCA1 and BRCA2 mutation predictions using the BOADICEA and BRC-APRO models and penetrance estimation in high-risk French-Canadian families. Breast Cancer Res. 2005;8(1):R3. doi: 10.1186/bcr1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barcenas C, Hosain G, Arun B, et al. Assessing BRCA carrier probabilities in extended families. J Clin Oncol. 2006;24(3):354–360. doi: 10.1200/JCO.2005.02.2368. [DOI] [PubMed] [Google Scholar]
- 23.Nanda R, Schumm L, Cummings S, et al. Genetic testing in an ethnically diverse cohort of high-risk women: a comparative analysis of BRCA1 and BRCA2 mutations in American families of European and African ancestry. JAMA. 2005;294(15):1925–1933. doi: 10.1001/jama.294.15.1925. [DOI] [PubMed] [Google Scholar]
- 24.Euhus D, Smith K, Robinson L, et al. Pretest prediction of BRCA1 or BRCA2 mutation by risk counselors and the computer model BRCAPRO. J Natl Cancer Inst. 2002;94(11):844–851. doi: 10.1093/jnci/94.11.844. [DOI] [PubMed] [Google Scholar]
- 25.Gilpin C, Carson N, Hunter A. A preliminary validation of a family history assessment form to select women at risk for breast or ovarian cancer for referral to a genetics center. Clin Genet. 2000;58(4):299–308. doi: 10.1034/j.1399-0004.2000.580408.x. [DOI] [PubMed] [Google Scholar]
- 26.Evans D, Eccles D, Rahman N, et al. A new scoring system for the chances of identifying a BRCA1 AND BRCA2mutation outperforms existing models including BRCAPRO. J Med Genet. 2004;41(6):474–480. doi: 10.1136/jmg.2003.017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoniou A, Pharoah P, Smith P, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancer. Br J Cancer. 2004;91(8):1580–1590. doi: 10.1038/sj.bjc.6602175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerdes A-M, Cruger D, Thomassen M, et al. Evaluation of two different models to predict BRCA1 and BRCA2 mutations in a cohort of Danish hereditary breast and/or ovarian cancer families. Clin Genet. 2006;69:171–179. doi: 10.1111/j.1399-0004.2006.00568.x. [DOI] [PubMed] [Google Scholar]
- 29.Kang H, Williams R, Leary J, et al. Evaluation of models to predict BRCA germline mutations. Br J Cancer. 2006;95:914–920. doi: 10.1038/sj.bjc.6603358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simard J, Dumont M, Moisan A, et al. Evaluation of BRCA1 and BRCA2 mutation prevalence, risk prediction models and a multistep testing approach in French-Canadian families with high risk of breast and ovarian cancer. J Med Genet. 2007;44(2):107–121. doi: 10.1136/jmg.2006.044388. [DOI] [PMC free article] [PubMed] [Google Scholar]