Abstract
Yeast prions provide a powerful model system for examining prion formation and propagation in vivo. Yeast prion formation is driven primarily by amino acid composition, not by primary amino acid sequence. However, although yeast prion domains are consistently glutamine/asparagine-rich, they otherwise vary significantly in their compositions. Therefore, elucidating the exact compositional requirements for yeast prion formation has proven challenging. We have developed an in vivo method that allows for estimation of the prion propensity of each amino acid within the context of a yeast prion domain.1 Using these values, we are able to predict the prion-propensity of various glutamine/asparagine-rich yeast domains. These results provide insight into the basis for yeast prion formation, and may aid in the discovery of additional novel prion domains. Additionally, we examined whether amino acid composition could drive interactions between heterologous glutamine/asparagine-rich proteins.2 Although inefficient interactions between yeast prion domains have previously been observed, we found that one prion protein, Ure2, is able to interact with compositionally similar domains with unprecedented efficiency. This observation, combined with the growing number of yeast prions, suggests that a broad network of interactions between heterologous glutamine/asparagine-rich proteins may affect yeast prion formation.
Key words: yeast, prion, amyloid, Ure2, Sup35
The highly-studied yeast prion proteins Sup35, Ure2 and Rnq1, which form the [PSI+], [URE3] and [PIN+] prions, respectively, each contain a glutamine/asparagine (Q/N) rich domain that drives prion formation. 3 Prion formation is thought to involve conversion of the soluble proteins into an insoluble amyloid form.3 Four additional yeast prion proteins containing Q/N-rich prion-forming domains have recently been discovered, and domains with similar Q/N-content are over-represented in various eukaryotic genomes.4–8 This suggests that numerous other prion proteins may remain to be discovered, yet predicting which of these Q/N-rich domains is likely to form prions has proven difficult.
Randomizing the order of the amino acids in either the Sup35 or Ure2 prion domains does not block prion formation, demonstrating that prion formation is driven primarily by amino acid composition, not primary sequence.9,10 We therefore explored two related questions. First, can amino acid composition be used to accurately predict prion propensity?1 Second, to what extent can compositionally similar Q/N-rich proteins interact during prion formation?2
Surprisingly, although composition drives yeast prion formation, compositional similarity to known prion domains is a poor predictor of prion propensity. In addition to high Q/N content, yeast prion domains show an under-representation of both charged and hydrophobic residues (Table 1), yet neither the number of charged residues nor hydrophobic residues differs significantly between Q/N-rich domains with and without prion-like activity.1,11 Alberti et al. recently identified the 100 yeast domains with greatest compositional similarity to the known prion domains, and tested each domain in four different assays for prion-like activity.4 This remarkable effort revealed numerous new potential prion domains; however, there was very little correlation between a domain’s compositional similarity to known prion domains and its prion-like activity.1,4 The most likely explanation for this unexpected finding is that because the yeast prion domains are likely not optimized for maximum prion propensity, some compositional deviations will increase prion propensity and some will decrease it. However, algorithms that predict prion propensity based on compositional similarity to known prion domains are predicated on the assumption that all compositional changes will reduce prion propensity. Instead, accurate prediction of prion propensity requires understanding how changes in amino acid composition affect prion propensity.
Table 1.
Prion propensity, order propensity and prevalence for each amino acid
| Amino acid | Prion propensitya | Prevalence in prion domainsb | Hydrophobicityc |
| Phenylalanine | 0.84 | 2.5 | 0.81 |
| Isoleucine | 0.81 | 1.1 | 1.0 |
| Valine | 0.81 | 1.5 | 0.97 |
| Tyrosine | 0.78 | 7.8 | 0.36 |
| Methionine | 0.67 | 1.7 | 0.71 |
| Tryptophan | 0.67 | 0 | 0.40 |
| Cysteine | 0.42 | 0 | 0.78 |
| Serine | 0.13 | 10.0 | 0.41 |
| Asparagine | 0.080 | 23.5 | 0.11 |
| Glutamine | 0.069 | 22.0 | 0.11 |
| Glycine | −0.039 | 12.9 | 0.46 |
| Leucine | −0.040 | 2.3 | 0.92 |
| Threonine | −0.12 | 2.1 | 0.42 |
| Histidine | −0.28 | 0.9 | 0.14 |
| Alanine | −0.40 | 3.9 | 0.70 |
| Arganine | −0.41 | 2.5 | 0.00 |
| Glutamic Acid | −0.61 | 1.5 | 0.11 |
| Proline | −1.20 | 1.7 | 0.32 |
| Aspartic Acid | −1.28 | 1.3 | 0.11 |
| Lysine | −1.58 | 0.7 | 0.07 |
Experimentally determined prion propensity from ref. 1. Prion propensities were calculated as the natural log of the fold over/under-representation of the amino acid among prion-forming clones relative to the naïve library.
The average percent prevalence among the Sup35, rnq1 and Ure2 prion domains.
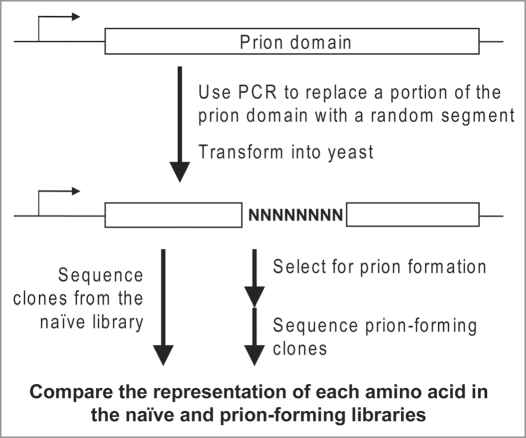
We therefore developed the first method to quantify the prion propensity of each amino acid within the context of a Q/N-rich prion domain (Fig. 1). We utilized Sup35-27, a scrambled version of Sup35 that forms prions with high efficiency. Using an oligonucleotide-based mutagenesis approach, we replaced eight consecutive codons within the SUP35-27 DNA sequence with (NNB)8, where N represents any of the four nucleotides and B represents any nucleotide except adenine. This generated a library of sequences in which all 20 amino acids should be present at each position within the mutated region. We then selected for the subset of proteins that maintained the ability to form prions. By comparing the naïve library to the prion-forming subset, we were able to determine the degree to which each amino acid was over- or under-represented among the prion-forming clones. These numbers were then used to generate a scale ranking the prion-propensity of all 20 amino acids (Table 1). Consistent with previous bioinformatic and mutational analysis, we found that charged residues and prolines were strongly under-represented among the prion-forming clones.1,11,12 By contrast, hydrophobic residues were strongly overrepresented, and no detectable bias was seen for or against glutamines and asparagines.1 Similar results were seen at a second position, demonstrating that these biases were not an artifact of local interactions.
Figure 1.
Mutagenesis method. Oligonucleotides were designed with a degenerate region, flanked by regions complementary to SUP35-27. These degenerate oligonucleotides were used to PCR amplify SUP35-27, generating a library of sequences in which eight codons were randomly mutated. Using plasmid shuffling, the library was used to replace wild-type Sup35 as the sole copy of Sup35 in the cell. Mutants were selected for prion formation by plating on medium lacking adenine. Isolates from the naïve library and the prion-forming subset were sequenced to determine which amino acids were over/under-represented among the prion-forming isolates.
Many of these results can be rationalized based on the proposed structure of Sup35-27 amyloid fibrils. Amyloid fibrils are β-sheet-rich structures in which the β-strands are predominantly oriented perpendicular to the long axis of the fibril. Solid state NMR indicates that for Sup35-27, the β-strands adopt an inregister parallel alignment.13 This inregister alignment should strongly disfavor charged residues due to electrostatic repulsion. Proline residues are known β-sheet breakers, so it is not surprising that they would also be disfavored. Hydrophobic residues have been proposed to drive formation of other inregister parallel amyloid structures, so their high prion-propensities in our assay should not be a complete surprise.14
Other aspects of our results are less easily explained based on proposed structures. X-ray diffraction studies of small fragments from Sup35 indicated that hydrogen bonding of in-register glutamines and asparagines stabilizes prion fibers, analogous to the polar zippers that have been proposed for poly-Q.15,16 However, we saw no bias in favor of glutamines and asparagines. This lack of bias in favor of glutamines and asparagines was particularly surprising based on the compositions of known yeast prion domains. Although high Q/N content is not an absolute requirement for a protein to act as a prion in yeast, Q/N residues are consistently over-represented in yeast prion domains (Table 1).11 Likewise, although we observed a strong bias in favor of hydrophobic residues, and although tyrosines have been implicated in prion formation and propagation, hydrophobic residues in general are strongly under-represented among yeast prion domains (Table 1).1,11,17,18
This raises the question, why is there such a large discrepancy between the residues that most strongly promote prion formation and those that are actually present in yeast prion domains? The simplest explanation is that the composition of yeast prion domains may reflect functions of the domains other than prion formation. Bioinformatic analysis can reveal what compositional features are present in prion domain, but not why they are present. Nevertheless, it seems surprising that yeast prion domains could form prions when they are largely lacking in the most strongly prion-promoting residues. We propose that native state intrinsic disorder is essential to yeast prion formation, and can help explain the ability of yeast prion domains to form prions despite their relative lack of strongly prion-prone amino acids. The yeast prion domains are intrinsically disordered, and for Sup35, this structural flexibility seems to be important for prion formation.19 The yeast prion domains are biased towards residues that balance intrinsic disorder and prion-propensity. For most proteins, amyloid formation must compete with native-state structure; in the absence of this competition, yeast prion domains are able to form prions despite relatively modest prion propensities. Although hydrophobic residues have high prion propensity, large numbers of hydrophobic residues would reduce intrinsic disorder by promoting hydrophobic collapse, potentially creating a stable fold that would compete with amyloid formation.
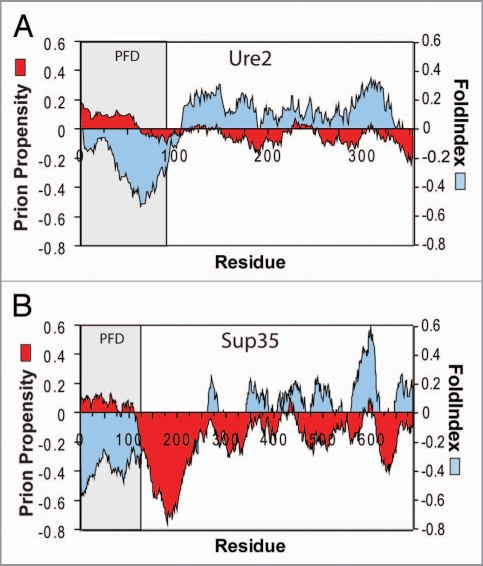
A significant question was whether mutagenesis results from short eightresidue segments could be used to predict prion propensities of entire prion domains. To test this, we scanned various proteins with a 41-amino-acid window size, calculating for each window prion propensity as the sum of the experimentally determined prion propensities for each amino acid in the window, and the order propensity using FoldIndex.20 FoldIndex is a simple algorithm for identifying disordered regions based on hydrophobicity (using the Kyte/Doolittle scale) and net charge.20,21 Strikingly, regions with similar predicted prion propensity to the yeast prion domains are common in non-prion proteins; however, these regions are consistently predicted to be ordered.1 By contrast, for Sup35 and Ure2, the prion domains are easily distinguishable as regions with positive prion propensity and negative FoldIndex order propensity (Fig. 2), supporting a role for intrinsic disorder in facilitating prion formation. Although the prion domains for the other yeast prion proteins are not as clearly defined, similar disordered and prion-prone regions are found in all of the yeast prion domains except that of Cyc8. By contrast, while Q/N-rich domains with low prion-like activity are consistently predicted to be disordered, they generally have low predicted prion propensity. By scoring proteins based on the predicted prion propensity of the most prion-prone region, we were able to distinguish with greater than 90% accuracy between Q/N-rich domains with and without prion-like activity. This is the first time that such prediction accuracy has been achieved for Q/N-rich domains.
Figure 2.
Prion propensity maps. Ure2p (A) and Sup35 (B) were scanned using a window size of 41 amino acids, calculating for each window the average order propensity using FoldIndex and prion propensity as the sum of the experimentally determined prion propensities for each amino acid across the window. The prion domain (PFD) is shaded.
These results provide insight into the differences between Q/N-rich and non-Q/N-rich amyloid proteins. Amyloid formation by non-Q/N-rich proteins is thought to be driven by short peptide stretches.22 Therefore, most amyloid prediction algorithms are designed to look for local regions of high prion propensity. However, our algorithm uses a relatively large 41-amino-acid window size. When we used a smaller window size, our algorithm lost the ability to distinguish between Q/N-rich domains with and without prion-like activity. Thus, the yeast prion domains are characterized by extended disordered regions of modest prion propensity, not local regions of high prion propensity. This explains why many amyloid prediction algorithms underestimate the amyloid propensity of the polar yeast prion domains.23
An obvious question is whether mutagenesis data from a single protein can really be used to make such broad claims about the basis for prion formation by all Q/N-rich yeast prion proteins. Similarly, FoldIndex is a relatively simple disorder-prediction algorithm that may not be specifically optimized for Q/N-rich proteins. However, these concerns are somewhat allayed by the accurate predictions that our algorithm, when combined with FoldIndex, is able to make. The fact that mutagenesis data from a single Q/N-rich protein allows for accurate prediction of a broad range of other Q/N-rich proteins suggests that the basic mechanisms of prion formation are similar for the various Q/N-rich prion domains. There is little doubt that a broader data set derived from multiple yeast prion domains could improve prediction accuracy; likewise, other disorder-prediction algorithms might be better suited for analyzing yeast prions. Nevertheless, the prediction accuracy that we have achieved suggests that while such changes might modestly improve our prediction accuracy, they are unlikely to fundamentally change our general conclusions about yeast prion domains.
Although our results provide an exciting first step towards understanding the sequence requirements for prion formation, numerous questions remain unanswered. For Sup35, separate regions of the prion domain are required for prion formation and prion propagation.24 Therefore, future experiments will be needed to dissect the distinct sequence requirements for prion formation versus propagation. Likewise, although numerous domains have been identified that can drive prion-like activity when inserted in the place of the Sup35 prion domain,4,25 flanking sequences outside of the Sup35 prion domain affect prion propagation.26 Therefore, there is no guarantee that such domains will be able to act as prions in their native contexts. Understanding the role of flanking sequences will be critical for predicting prion propensity of domains within their native contexts. Similarly, expression levels and patterns and cellular localization likely influence prion formation and propagation.
The effects of primary sequence on prion propensity are also still unclear. The dominant effect of amino acid composition on prion formation has made it difficult to distinguish the more subtle effects of primary sequence. By delineating the compositional requirements for prion formation, our work should make it easier to identify primary sequence elements that affect prion propensity. Specifically, examination of outlier proteins not accurately predicted by our composition-based algorithm may reveal primary sequence features that promote or inhibit prion formation.
One such aspect of primary sequence is already incorporated into our algorithm. In examining the 100 Q/N-rich proteins studied by Alberti et al. we discovered a subtle, but statistically significant bias in the distribution of proline residues.1,4 When prolines were present in Q/N-rich domains with prion-like activity, they tended to occur in clusters, while the prolines were more likely to be dispersed in domains without prion-like activity. This is not surprising, as prolines are known β-sheet breakers. If multiple prolines are present in single cluster, they will disrupt the β-sheet structure at a single location; by contrast, the same number of prolines dispersed throughout a sequence will result in multiple locations where the β-sheet structure is disrupted.
Another potential example of the effects of primary sequence is seen in Cyc8, the only yeast prion protein incorrectly predicted by our algorithm not to form prions. Interestingly, the prion domain contains an imperfect (QA)32 repeat. Patterns of alternating polar and non-polar residues are thought to promote amyloid formation, although alanine has not generally been included among the non-polar residues when considering such patterns.27 Therefore, this primary sequence element may explain why our algorithm, which predominantly considers amino acid composition, predicts Cyc8 to have relatively low prion propensity.
The high level of prediction accuracy achieved by our algorithm should also facilitate the identification of new prion proteins. The large number of yeast prion proteins, combined with the prevalence of Q/N-rich domains in eukaryotic genomes, suggests that similar prion-like structural conversions may be common in other organisms. However, only two prion proteins have been identified outside of Saccharomyces cerevisiae—the mammalian protein PrP and Podospora anserine protein HET-s—neither of which is Q/N-rich. Identification of new prion proteins has been hindered by the lack of accurate prediction algorithms. In yeast, genetic screens and compositional homology searches have led to the identification of novel prions, but with relatively low success rates.4–8 Understanding the compositional requirements for prion formation should improve this success rate, allowing for more targeted testing of potential prion proteins.
As the list of Q/N-rich yeast prion proteins continues to grow, an obvious question will be, how do interactions between heterologous prion proteins affect prion formation and propagation? If amino acid composition is the predominant factor determining prion-like activity, can amino acid composition also drive interactions between heterologous prion proteins? Various interactions have been observed among the yeast prion proteins. Under normal cellular conditions, prion formation by Sup35 requires the presence of [PIN+].28 In vitro and in vivo evidence suggests that Rnq1 aggregates can act as imperfect templates for seeding Sup35 aggregation.29 Overexpression of various Q/N-rich proteins can substitute for [PIN+], allowing [PSI+] formation in cells lacking [PIN+]; this suggests that Sup35 can interact with a broad range of Q/N-rich proteins.30,31 [PIN+] also promotes [URE3] formation, while [PSI+] inhibits [URE3] formation.32,33 However, each of these interactions is quite inefficient; for example, Rnq1 aggregates are at least 50-fold less efficient than Sup35 aggregates at seeding Sup35 amyloid formation.29
Therefore, we were surprised to discover that [URE3] prion formation can be stimulated with high efficiency by a variety of compositionally similar domains.2 Overexpression of the prion domain of Ure2 significantly increases [URE3] formation. Surprisingly, overexpression of each of the scrambled versions of Ure2 stimulates wild-type [URE3] formation with comparable efficiency.2 In vitro, amyloid aggregates formed by the scrambled Ure2s were able to efficiently seed amyloid aggregation by wild-type Ure2, suggesting that wild-type Ure2 is able to directly interact with scrambled Ure2 amyloid aggregates. There were limits to Ure2’s promiscuity, as overexpression of scrambled Sup35 prion domains did not promote [URE3] formation.
To determine whether Ure2 could similarly interact with naturally occurring yeast proteins, we performed a genomic search to identify the five yeast protein fragments with greatest compositional similarity to the Ure2 prion domain.2 Four of these five fragments were able to stimulate wild-type [URE3] formation. The efficiency of this cross-seeding was unprecedented; one of the fragments, from Sap30, was able to stimulate [URE3] formation three-fold more efficiently than the wild-type Ure2 prion domain. These results raise the possibility that such interactions could be physiologically relevant, acting as regulators of either beneficial or deleterious amyloid formation. Interestingly, not all prion proteins appear to be as promiscuous as Ure2. Prion formation by Sup35 was not significantly affected by overexpression of scrambled versions of Sup35. Defining the sequence features that allow Ure2 to interact with compositionally similar domains, and defining the limits of Ure2’s promiscuity, will be critical for determining the extent to which similar interactions affect the normal physiology of yeast prions and mammalian amyloid diseases.
Although it is clear that Ure2 is able to promiscuously interact with a broad range of compositionally similar proteins, our results say nothing about why such interactions occur. These interactions may have evolved to positively or negatively regulate prion formation. Alternatively, such prion-promoting interactions may simply be a byproduct of the natural function of the Ure2 prion domain. Although the exact function of the Ure2 prion domain is unknown, regions of intrinsic disorder domains are often used to recognize multiple binding partners.34 Promiscuous prion-promoting interactions may simply be an unintended consequence of such activity. Therefore, future studies will be needed to determine the function, if any, of Ure2’s promiscuity. Likewise, although our mutagenesis results clearly indicate that the compositions of the yeast prion domains reflect a balance of intrinsic disorder and prion propensity, our studies say nothing about why the yeast prion domains have evolved this balance. These compositional characteristics may have evolved for a reason unrelated to prion formation. Alternatively, the yeast prion domains may have specifically been optimized for the purpose of prion formation.
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/12190
References
- 1.Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30:319–332. doi: 10.1128/MCB.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross CD, McCarty BM, Hamilton M, Ben-Hur A, Ross ED. A promiscuous prion: Efficient induction of [URE3] prion formation by heterologous prion domains. Genetics. 2009;183:929–940. doi: 10.1534/genetics.109.109322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wickner RB, Shewmaker F, Kryndushkin D, Edskes HK. Protein inheritance (prions) based on parallel in-register beta-sheet amyloid structures. Bioessays. 2008;30:955–964. doi: 10.1002/bies.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemecek J, Nakayashiki T, Wickner RB. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci USA. 2009;106:1892–1896. doi: 10.1073/pnas.0812470106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross ED, Baxa U, Wickner RB. Scrambled prion domains form prions and amyloid. Mol Cell Biol. 2004;24:7206–7213. doi: 10.1128/MCB.24.16.7206-7213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci USA. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 13.Shewmaker F, Ross ED, Tycko R, Wickner RB. Amyloids of shuffled prion domains that form prions have a parallel in-register beta-sheet structure. Biochemistry. 2008;47:4000–4007. doi: 10.1021/bi7024589. [DOI] [PubMed] [Google Scholar]
- 14.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, et al. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Nat Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandrov IM, Vishnevskaya AB, Ter-Avanesyan MD, Kushnirov VV. Appearance and propagation of polyglutamine-based amyloids in yeast: tyrosine residues enable polymer fragmentation. J Biol Chem. 2008;283:15185–15192. doi: 10.1074/jbc.M802071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohhashi Y, Ito K, Toyama BH, Weissman JS, Tanaka M. Differences in prion strain conformations result from non-native interactions in a nucleus. Nature. 2010;6:225–230. doi: 10.1038/nchembio.306. Epub 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheibel T, Lindquist SL. The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat Struct Biol. 2001;8:958–962. doi: 10.1038/nsb1101-958. [DOI] [PubMed] [Google Scholar]
- 20.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, et al. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 21.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 22.Esteras-Chopo A, Serrano L, de la Paz ML. The amyloid stretch hypothesis: Recruiting proteins toward the dark side. Proc Natl Acad Sci USA. 2005;102:16672–16677. doi: 10.1073/pnas.0505905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linding R, Schymkowitz J, Rousseau F, Diella F, Serrano L. A comparative study of the relationship between protein structure and [beta]-aggregation in globular and intrinsically disordered proteins. J Mol Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 24.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 26.Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+] Proc Natl Acad Sci USA. 2002;99:16446–16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West MW, Wang W, Patterson J, Mancias JD, Beasley JR, Hecht MH. De novo amyloid proteins from designed combinatorial libraries. Proc Natl Acad Sci USA. 1999;96:11211–11216. doi: 10.1073/pnas.96.20.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, et al. Dependence and independence of [PSI(+)] and [PIN(+)]: a twoprion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 31.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 32.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]