Abstract
Alu DNA elements were long considered to be of no biological significance and thus have been only poorly defined. However, in the past Alu DNA elements with well-defined nucleotide sequences have been suspected to contribute to disease, but the role of Alu DNA element transcripts has rarely been investigated. For the first time, we determined in a real-time approach Alu DNA element transcription in buffy coat cells isolated from the blood of humans suffering from sporadic Creutzfeldt-Jakob disease (sCJD) and other neurodegenerative disorders. The reverse transcribed Alu transcripts were amplified and their cDNA sequences were aligned to genomic regions best fitted to database genomic Alu DNA element sequences deposited in the UCSC and NCBI data bases. Our cloned Alu RNA/cDNA sequences were widely distributed in the human genome and preferably belonged to the “young” Alu Y family. We also observed that some RNA/cDNA clones could be aligned to several chromosomes because of the same degree of identity and score to resident genomic Alu DNA elements. These elements, called paralogues, have purportedly been recently generated by retrotransposition. Along with cases of sCJD we also included cases of dementia and Alzheimer disease (AD). Each group revealed a divergent pattern of transcribed Alu elements. Chromosome 2 was the most preferred site in sCJD cases, besides chromosome 17; in AD cases chromosome 11 was overrepresented whereas chromosomes 2, 3 and 17 were preferred active Alu loci in controls. Chromosomes 2, 12 and 17 gave rise to Alu transcripts in dementia cases. The detection of putative Alu paralogues widely differed depending on the disease. A detailed data search revealed that some cloned Alu transcripts originated from RNA polymerase III transcription since the genomic sites of their Alu elements were found between genes. Other Alu DNA elements could be located close to or within coding regions of genes. In general, our observations suggest that identification and genomic localization of active Alu DNA elements could be further developed as a surrogate marker for differential gene expression in disease. A sufficient number of cases are necessary for statistical significance before Alu DNA elements can be considered useful to differentiate neurodegenerative diseases from controls.
Key words: transcription of Alu DNA elements, chromosomal patterns of active Alu DNA elements, sporadic CJD, differential gene expression in neurodegeneration
Introduction
Short interspersed elements (SINE) represent a major part of noncoding sequences within the human genome. For quite some time such non-coding, repetitive DNA elements were disparagingly denominated as junk DNA. That view has dramatically changed in recent years since these elements have been found to modulate the transcriptome and proteome. Particularly Alu DNA elements have been ascribed certain functions in cells. The biology of transposable elements has received heightened interest because retrotransposons have been found to shape the genome and thus gene expression programs.1 Gene expression can be altered when Alu DNA elements are retrotransposed into or close to exons and introns of coding gene sequences.2 Alu DNA element transcripts are known to interfere with translation by modifying an initiation factor and transcription by interfering with RNA polymerase II complexes.3–8 Neither the role of Alu DNA elements, in particular their sequences such as the internal poly A tails, nor their non-coding transcripts have been sufficiently analyzed as to their association with disease. Recent reports have related functions to specific sequence motifs of resident Alu DNA elements and their transcription, which is key for retrotransposition of Alu DNA elements in the genome. Transcribed Alu DNA elements are designated “active” elements. If Alu DNA elements undergo mutations, then biological consequences such as retrotranspostion can be altered. The presence of active Alu retrotransposons has rarely been investigated but recently Bennett et al.9 began to look for active, i.e., transcribed, Alu DNA elements. They began with bioinformatics and then examined distinct cloned Alu elements that (1) could be transcribed in cell culture and (2) whose transposition could be monitored. Our experimental approach was the other way round: In a real-time experimental approach with human cells, we isolated Alu transcripts, prepared cDNA clones and focused on extensive sequence alignments. Recent publications reported on sequence criteria for retrotransposition or as targets for microRNAs.10–12 Another experimental approach in line with our study analyzed the expression patterns of DNA elements for non-coding RNAs in various tissues.1,13 From this standpoint, we present for the first time data on active or expressed (i.e., transcribed into RNA) Alu DNA elements in healthy controls and in humans with neurodegenerative disorders. Our findings demonstrate distinct Alu transcription patterns in healthy and diseased humans.
This study may provide another experimental approach for monitoring gene expression in cells by establishing transcription profiles of active Alu elements indicative of differential gene expression in neurodegenerative diseases. Future analyses of genome sequences flanking the expressed, i.e., active Alu DNA elements may reveal chromosomal regions, including genes with embedded Alu sequences, which were transcriptionally activated and which may be related to pathogenic consequences such as inflammation or cell death.
Results
Isolation of RNA with stringent protocols to eliminate genomic DNA.
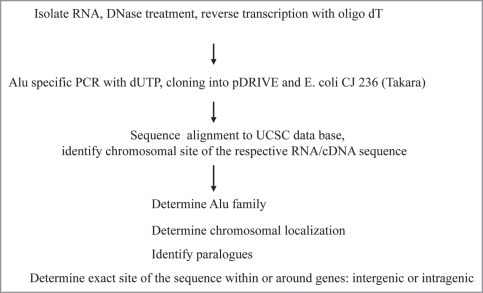
The experimental strategy is outlined in Figure 1. Total RNA was isolated from buffy coat and treated with DNase to eliminate genomic DNA. Since we were critical of established protocols, we improved our procedure thoroughly to remove Alu DNA contaminating cellular RNA. Each RNA preparation was subjected to an agarose gel electrophoresis before and after treatment with DNase to visualize loss of cellular DNA but most importantly to prove that RNA was not completely degraded. At an advanced stage of our Alu RNA isolation and oligo dT-directed cDNA syntheses, we switched to a dUTP PCR step, cloning cDNA into pDRIVE plasmid (Qiagen) before transfection into UNG negative E. coli strain CJ 236 (Takara). This strategy was superior to standard procedures since we could avoid carry-over and did not experience degradation of U-containing plasmid DNA by uracyl glycosidase. These modifications in the protocols ensured reliable results on Alu DNA element transcription. Additional RT PCR protocols omitting template or RT as well as testing RNA preparations with β-actin primers were used to verify loss of genomic DNA.
Figure 1.
Experimental approach to identify transcribed “active” Alu DNA elements and their chromosomal location. A stringent protocol was developed to isolate Alu RNA and to prepare cDNA sequences in order to search of the active, i.e., expressed/transcribed Alu DNA elements within the human genome.
Alu RNA/cDNA sequence alignment to human chromosomes.
The Alu-specific cDNA sequences (277 bps) were submitted and 257 bps of each clone were aligned to chromosomes using the UCSC and NCBI data bases. Those genomic sites with highest nucleotide similarity were used to define the genomic loci of active Alu elements. One example of an aligned sequence is shown in Figure 2 (data base search and determination of genomic locations are provided in Suppl. Figs. 1–4).
Figure 2.
Cloning of Alu RNA/cDNA and its sequence for alignment. The amplicon was 277 bps in size and the sequence for alignment 257 bps in size. The Eco RI cloning sites and the primer sites are in bold and the 5′ and 3′ sites for alignments are underlined. The clone shown: sequence 884 clone no. 1 sCJD and sequence obtained with M13 reverse primer.
Chromosomal localization of active Alu DNA elements.
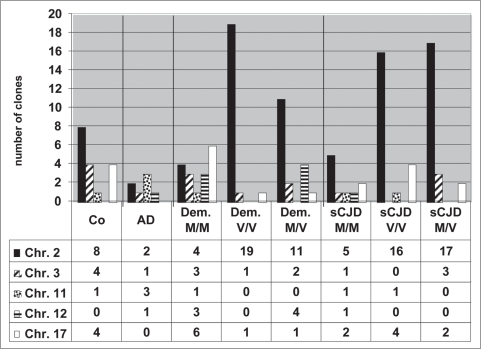
We found chromosome 2 to be the location of many active Alu DNA elements. A broad distribution of active Alu elements was found and is presented in a histogram (Fig. 3). To a certain degree distinct patterns of active Alu elements could be, for example, linked to health status. AD cases demonstrated active Alu elements particularly on chromosome 11, or sCJD M/V cases showed active Alu elements on chromosome 2 and 3.
Figure 3.
Frequency of active Alu DNA elements on chromosomes of healthy and diseased humans. The numbers of RNA/cDNA clones targeting individual chromosomes are given. Clearly, chromosome 2 is the chromosome where transcribed = active Alu DNA elements preferentially reside.
Since our study probably cannot produce sufficient statistics, this analysis should serve as a first attempt to correlate chromosomal patterns of active Alu DNA elements with neurodegenerative diseases. With regard to molecular biology, the Alu element expression patterns may be used as indicator for differential transcriptional activation of the genome. The data base search that aligned cDNA sequences to identify the best matched chromosomal Alu sequence as a site for the respective active Alu element is shown in the Supplementary Figure 2. Identity and score put one chromosomal Alu DNA element on top and this chromosome was then taken for interpretation. Some RNA/cDNA clones matched perfectly to more than one chromosome. The results revealed that one cloned Alu Ya5 sequence from a control could be localised on chromosomes 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, 13, 15 and 19. This “multilocalization” of some individual RNA/cDNA clones may point to recent transposition(s) and implied that our RNA/cDNA sequences identified Alu DNA element paralogues. Identity and score of one clone were shared with several identical chromosomal sequences. However, we were not able to distinguish whether one chromosomal site was preferably transcribed or whether each of the multiple Alu sites gave rise to Alu transcripts. A detailed look at the Alu transcripts revealed that multilocalization occurred to a large extent with Alu Y family transcripts, particularly Alu Ya5 and, to a lesser extent, Alu Yb8 and a8 (Suppl. Fig. 3).
A first survey on paralogues and their chromosomal distribution showed that a “diagnostic” flow chart could be made (data not shown). For example, chromosome 15 seemed to be a preferred location for Alu DNA element paralogues, whereas chromosome 21 was never found to harbor an Alu paralogue. However, chromosomes 8 and 17 were the only chromosomes for paralogues in sCJD and dementia cases, respectively. In AD cases we found chromosomes 13 and 14 to be loci for paralogues, whereas no cDNA clone from sCJD or dementia could be located on chromosomes 13 and 14. Moreover, the cDNA clones from AD cases, i.e., paralogues, were never found on chromosomes 17–22. Since the number of cases was not sufficient for statistical analysis, one would need more cases to substantiate diagnosis by paralogue patterns as a supportive or predictive diagnostic tool.
Classification of RNA/cDNA clones into Alu family/subfamily.
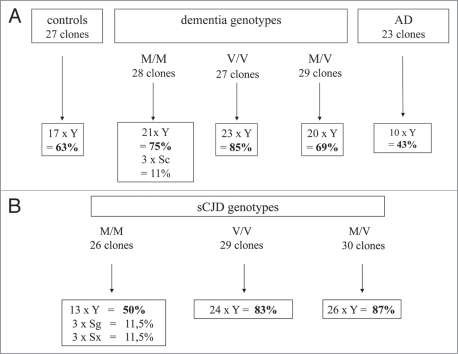
According to the UCSC data base we mostly identified Alu Y species, but Alu Sg and Alu Sx were also isolated. These real-time results confirmed that evolutionary “young” but also “intermediate” Alu elements are transcribed in human peripheral blood cells. Independent of the health status, most cases and controls have Alu transcripts preferably belonging to the Alu Y family (Fig. 4A and B). By chance, we could further characterize dementia and sCJD cases since their prion protein gene types (PRNP) were known. The M/M, V/V and M/V correspond to homozygous methionine or valine or heterozygous methione genotypes, respectively. Dominance of Alu Y transcripts may be explained since these “younger” Alu elements are supposed to be active in comparison to the evolutionarily older Alu members. We found certain nucleotides preserved in our RNA/cDNA clones. One example is the conserved G as part of the UGU/TGT motif around position 24/25 in a prototype Alu structure (unpublished; data not shown). This G has been shown to be responsible for retrotransposition of murine B1 elements,10 which may point to “functional” retrotransposable Alu transcripts isolated in our real-time approach. Our RNA/cDNA clones contained the sequence motif “GCACUU” that has been predicted by bioinformatics as a target for micro RNA (unpublished; data not shown).12 We mention this observation on Alu transcripts since these short sequence motifs have gained heightened interest in molecular biology.
Figure 4.
Frequency of Alu Y transcripts in buffy coat cells of healthy humans and cases with neurodegenerative diseases. Numbers of Alu Y RNA/cDNA clones obtained from healthy controls, dementia and Alzheimer disease cases are shown in (A) and sporadic CJD are shown in (B). The prion protein genotypes are either homozygous or heterozygous for methionine or valine: M/M, V/V or M/V.
Intergenic and intragenic localization of active Alu DNA elements.
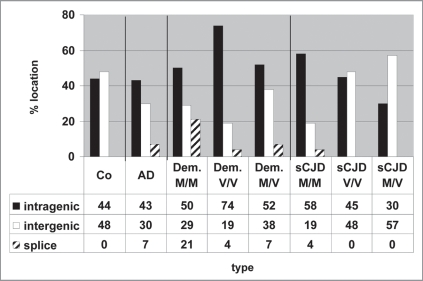
Examination of the chromosomal sites of active Alu elements allowed us to define more precisely the location of the active Alu DNA elements (Suppl. Fig. 4). Residing between genes, these elements may indicate that the respective Alu is transcribed by RNA polymerase III; any sites within genes might point to Alu sequences embedded in RNA polymerase II transcription units. These results are summarized in Figure 5. The patterns are again quite divergent. Some shifts from intragenic to intergenic Alu locations were observed in sCJD M/M cases versus V/V and M/V cases. Also, the Alu transcripts of the dementia V/V group preferentially emerged from intragenic chromosomal regions. Taken together, these results indicate that active Alu DNA elements may serve as markers for differential gene expression, especially when the genes surrounding active Alu DNA elements are known (Kiesel et al., unpublished).
Figure 5.
Intergenic and intragenic locations of active Alu DNA elements. A detailed computer search allowed an exact determination of the genomic location where the transcribed Alu DNA element was located, i.e., within genes or flanking genes: intragenic and splice sites or between genes: intergenic.
Materials and Methods
Human cases and biopsy specimens.
The study was performed in compliance with the Helsinki Declaration. Permission to perform studies with human blood specimens and ethic approval was given by the ethics committee of the University Medical Center in Göttingen to Professor Zerr, head of the TSE Reference Center at the Department of Neurology in Göttingen, Germany. Permit 11/11/93 was given on November 4, 1993, extended on September 18, 1996 and renewed on August 19, 2002, before this study was initiated in the context of CJD surveillance in Göttingen.
We used peripheral buffy coat cells from healthy individuals and patients suffering from sCJD, individuals with non-CJD who were most likely dementia cases and Alzheimer disease (AD) patients who had originally been suspected to have sCJD. Buffy coat was immediately frozen at −80°C when blood sample was taken. About 0.5–1 ml buffy coat was lysed in the tissue lysis buffer of the TriReagent RNA isolation kits for tissue and blood.
RNA isolation and elimination of genomic DNA.
TriReagent was used to extract RNA from the biological samples. The precipitation-based RNA isolation was preferred since it prevented loss of small RNA species.
Yield and purity was determined by optical density and nondegradation of RNA on agarosegels. After DNase treatment (0.25 units/1 µg DNA, 15 min, 4°C) each RNA sample was again subjected to agarosegels to confirm that the RNA was not degraded. In parallel, an actin gene/pseudogene PCR was performed with DNase treated and non-treated RNA samples to exclude detectable contamination with genomic DNA (0.5 µg for β-actin). After these preparatory steps cDNA was made and processed to the Alu-specific PCR.
RT reaction with oligo dT15, Alu primer and PCR for cloning and lightcycler analyses.
Standard RT reactions containing 500 ng RNA and cDNA syntheses were primed with oligo dT 15 as primer, Superscript RT for 50 min at 42°C and 15 min. at 70°C to stop the reaction. Aliqouts were then transferred into the Alu specific PCR. The primers for the Alu-specific amplification were forward: 5 TTC GCG GTG GCT CAC GCC TG 3 and reverse: 5 GAG ACG GAG TCT CGC TCT 3. The amplification product was −277 bp in size since the polyA stretch within Alu elements/transcripts slightly varies. The Alu-specific Hot-Start PCR for cloning the amplicon was performed in a 30 µl volume with 1 µl cDNA, a dUTP containing dNTP mix, 10 picomol primer and 35 cycles. Carry-over was prevented since amplification was performed in the presence of 0.6 units UNG. We carried along three water controls instead of cDNA, and each of the three control PCRs was required to remain negative.
Cloning and sequencing of cDNA made from Alu transcripts.
The amplification products were cloned in an unusual cloning system since newly made dsDNA contained dUTP to prevent contamination with Alu amplification products. Presence of U in newly synthesized dsDNA resembles RNA/ DNA hybrids that are eliminated in bacteria. Therefore, successful cloning of dUTP-containing cDNA required the UNG negative E. coli strain CJ 236 (Takara). Amplification products were cloned into pDRIVE (Qiagen) and introduced by electroporation into CJ 236. Plasmid mini preparations were made to isolate plasmid DNA which contained the Alu cDNA insertions as shown by excision with Eco RI and agarose gel electrophoresis.
Alu-containing pDRIVE plasmids were purified for a second time. We routinely used 600 ng plasmid DNA for sequencing with the enodogenous pDRIVE M13 sites for the forward and reverse primers.
Both strands of the cDNA clones were sequenced since the mutation analyses required both strands to detect mutations or errors introduced by the Taq polymerase. Sequencing was performed by seqlab (Göttingen, Germany). We were able to analyze a total of about 30 cDNA clones of healthy controls, Alzheimer disease cases, dementia and sCJD cases with the prion protein genotype.
Computer-assisted sequence alignment and bioinformatics.
The cloned original Alu RNA/cDNA sequences underwent a first analysis with the BioEdit Editor Version 7.0.5.2 program. In this analysis, both cDNA strands were tested with the NCBI data bank and BLAST 2 to obtain details on sequence similarity and to identify their chromosomal sites. A further step was taken to define the Alu family, the genomic orientation and localization on chromosomes using the UCSC data bank. Sites for deamination were determined with ClustalX Version 2.0; each Alu cDNA sequence was aligned to the chromosomal site with the highest similarity.14 With this information we then used the NCBI/UCSC data bases to identify the exact localization of our Alu cDNA/RNA within the genome either in introns or exons or intergenic chromosomal regions.15,16
Discussion
In the past there have been only few experimental approaches linking Alu DNA elements and their sequences, such as the internal poly A tail, to disease.2,17,18 In the present study we focused on the determination of Alu transcripts which have been functionally discussed as modifiers of translation and, more recently, transcription.4,8 In a reverse experimental approach Bennet et al.9 reported on a data base search for putative active, i.e., transcribed, Alu DNA elements. The authors subsequently confirmed activity by retrotransposition via RNA in HeLa cell culture. At the same time, we had already identified in a “real-time experimental approach” Alu transcripts and assigned them to human chromosomes. To produce reliable results we established robust protocols for Alu transcript identification and characterization. This approach revealed different Alu-specific transcriptional activity in leucocytes from humans including sCJD, dementia and AD cases. Our aim was to investigate small non-coding RNA since such transcripts were hypothesized to play a role in prion disease.19 In Alu DNA element research transcripts of the Alu elements have never before been analyzed in a real-time experimental approach. Our findings are thus promising and novel for Alu transcription in human blood cells.
Working with Alu transcripts is drastically hampered by any trace amounts of genomic DNA that may contaminate RNA preparations. Since small-sized RNA may be lost by silica-based column methods we applied phenol-type precipitation methods to isolate RNA. Tests for DNA contamination were performed with appropriate β-actin primer pairs as single copy gene control, and RNA was separated on agarose gels to visualize the integrity of RNA preparations treated with DNase. We used the dUTP/ UNG method to prevent carry-over; this, however, required plasmid replication/propagation in UNG negative cells that were not prone to degrade cloned Alu cDNA sequences containing U instead of T. After developing these controls, we ventured an interpretation of our sequence data derived from RNA, i.e., the cloned cDNA.
Our data clearly pointed to a dominance of transcripts from “younger” Alu Y elements over those belonging to the S-family.20 Yet transcripts from older Alu family families were also identified, similar to previous reports on Alu transcripts from “old” Alu elements after virus infection.21 The presence of nucleotide G at position 24/25 of the reference Alu DNA element sequence confirmed our interpretation that we were dealing with Alu transcripts from transcriptionally active Alu DNA elements. In addition to this G we detected a putative “GCACUU” target sequence for a micro RNA around position 34 (unpublished, Kiesel et al.).
An obvious drawbrack in this first study is the lack of statistics. Although we had enough cases and controls to generate Alu RNA/cDNA clones, their number did not allow to statistical confidence. We therefore do not yet suggest active Alu DNA elements, i.e., Alu DNA element transcription patterns, as a marker for neurodegenerative diseases. Larger numbers are necessary for statistical evaluation. Single observations might nevertheless be important for developing of future experimental approaches.
Cloning of Alu transcript amplicons products and “resolving” the amplicon population into single clones revealed a preferred transcription of some Alu DNA elements. Furthermore, the disproportionate transcription of Alu DNA elements present in the human genome could easily be seen. Similar findings were reported for a permanently growing cell line but no one has described this for human Alu transcripts so far.22
It is important to note that some RNA/cDNA clones were observed to align perfectly to several Alu DNA elements on several chromosomes, and are thus most likely Alu DNA element paralogues. In fact, these paralogues may have resulted from recent transposition events. This finding and the fact that preferably Alu Y transcripts have been detected may be indicative for recent transposition of these distinct Alu DNA elements. There was most likely not enough time in evolution to introduce mutations by deamination to counteract retrotransposition. Thus, our observations imply that paralogues of active Alu DNA elements exist and were detectable in our real-time approach. Our results demonstrate how real-time Alu DNA element transcription and retrotransposition occur.
The chromosomal patterns of active Alu DNA elements were divergent when we compared healthy humans with those presenting with neurodegenerative diseases such as sCJD, Alzheimer disease or dementia. We were also aware that we were observing Alu transcription in the periphery but not in the brain where these diseases exert their pathogenic potential.
Unexpectedly, control and sCJD as well as dementia and AD source RNA preparations resulted in Alu RNA/cDNA that could be located to their respective Alu DNA elements in different chromosomes and genomic locations. Indeed, we were able to clearly document that different Alu DNA elements are transcribed in living organisms. Our data were compared with previous results from experiments with permanently growing cells under experimental conditions such as viral infection. 23,24 Divergent transcription patterns may point to different Alu-related transcriptomes in individuals. It may be that additional epigenetic mechanisms are responsible for allowing either RNA polymerase III Alu DNA element transcription or RNA polymerase II transcription of certain genes with embedded Alu DNA element sequences.
Stress may also result in a modified transcriptional program even in peripheral leucocytes from patients with neurodegenerative disease. Thus, Alu transcription profiles may serve as an indicator for differential gene expression similar to what is performed on microarrays with high resolution of distinct sets of genes. Those Alu sequences accessible to transcription may then emerge as a surrogate of the cellular transcriptome. In chromosomes in which more than three Alu RNA/cDNA clones could be positioned our impression was that a genome-wide differential Alu DNA element transcription activity is initiated, e.g., on chromosomes 1, 2 and 3 from controls versus 2 and 17 in sCJD cases, or 2 and 11 in AD cases.
In conclusion, our results represent the first real-time study on Alu transcripts transcribed from “active” Alu DNA elements in peripheral blood cells in healthy humans and humans suffering from neurodegenerative diseases. Future research is necessary to confirm the role of Alu trancripts as modifier of gene expression. Although the results of this study may not serve as a basis for developing a marker for prion diseases, they do broaden our limited knowledge of Alu DNA element transcription in human blood cells.
Acknowledgements
Financial support was given by the DFG (ZI 568/3-2 and KA 864/2-1) and by the German Primate Center (DPZ). This study was funded by Robert Koch-Institut through funds of the Federal Ministry of Health (grant no 1369-341 to I.Z.).
Abbreviations
- sCJD
sporadic Creutzfeldt-Jakob disease
- AD
Alzheimer disease
- UNG
uracyl-N-glycosylase
- UCSC
University of California Santa Cruz
- NCBI
National Center for Biotechnology Information
- BLAST
basic local alignment search tool
- CpG
cytosine-guanine phosphodiester bond
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/11965
Supplementary Material
References
- 1.Goodier JL, Kazazian HH. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Deininger PL, Batzer MA. Alu repeats and human diesease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 3.Häsler J, Strub K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res 2. 2006;34:2374–2385. doi: 10.1093/nar/gkl246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubin CM, Kimura RH, Schmid CW. Selective stimulation of translational expression by Alu RNA. Nucleic Acids Res. 2002;30:3253–3261. doi: 10.1093/nar/gkf419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondrashov AV, Kiefmann M, Ebnet K, Khanam T, Muddashetty RS, Brosius J. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly (A)-binding protein (PABP) J Mol Biol. 2005;353:88–103. doi: 10.1016/j.jmb.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 6.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 7.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 8.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promotor DNA within assembled complexes. Proc Natl Acad Sci USA. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, et al. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dewannieux M, Esnault C, Heidmann T. LINEmediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 11.Dewannieux M, Heidmann T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol Biol. 2005;349:241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- 12.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable micro RNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Mattick JS, Mehler MF. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 2008;31:227–233. doi: 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, et al. The UCSC genome browser database: update 2009. Nucleic Acids Res. 2009;37:755–761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2009;37:5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy-Engel AM, Salem AH, Oyeniran OO, Deininger L, Hedges DJ, Kilroy GE, et al. Active Alu element “A-tails”: size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: what causes Alu elements to die? Genome Res. 2009;19:545–555. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson TJ. RuNAway Diesease: A two cycle model for transmissible spongiform encephalopathies (TSEs) wherein SINE proliferation drives PrP overproduction. Genome Biol. 2001;2:6. doi: 10.1186/gb-2001-2-7-preprint0006. [DOI] [PubMed] [Google Scholar]
- 20.Cordaux R, Lee J, Dinoso L, Batzer MA. Recently integrated Alu retrotransposons are essentially neutral residents of the human genome. Gene. 2006;373:138–144. doi: 10.1016/j.gene.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Panning B, Smiley JR. Activation of expression of multiple subfamilies of human Alu elements by adenovirus type 5 and herpes simplex virus type 1. J Mol Biol. 1995;248:513–524. doi: 10.1006/jmbi.1995.0239. [DOI] [PubMed] [Google Scholar]
- 22.Shaik TH, Roy AM, Kim J, Batzer MA, Deininger PL. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J Mol Biol. 1997;271:222–234. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- 23.Williams WP, Tamburic L, Astell CR. Increased levels of B1 and B2 SINE transcripts in mouse fibroblast cells due to minute virus of mice infection. Virology. 2004;327:233–241. doi: 10.1016/j.virol.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 24.Ueda Y, Chaudhuri G. Differential expression of B1-containing transcripts in L Leishmania-exposed macrophages. J Biol Chem. 2000;275:19428–19432. doi: 10.1074/jbc.M001336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.