Abstract
Analyses from diverse eukaryotes reveal that genomes are dynamic, sometimes dramatically so. In numerous lineages across the eukaryotic tree of life, DNA content varies within individuals throughout life cycles and among individuals within species. Novel genome features are discovered and our understanding of the extent of genome dynamism continues to grow as more genomes are sequenced. Though most completed eukaryotic genomes are from animals, fungi, and plants, these lineages represent only three of the 60–200 lineages of eukaryotes. Here, we discuss the diverse genomic strategies in exemplar eukaryotic lineages, including several microbial eukaryotes, to reveal dramatic variation that challenges established views of genome evolution. For example, in the life cycle of some members of the ‘radiolaria’ ploidy increases from haploid (N) to approximately 1000N, while intrapopulation variability of the enteric parasite Entamoeba ranges from 4N to 40N. Variation has also been found within our own species, with substantial differences in both gene content and chromosome lengths between individuals. Data on the dynamic nature of genomes shift the perception of the genome from being fixed and characteristic of a species (typological) to plastic due to variation within and between species.
Keywords: Ploidy, Microbial Eukaryotes, Genome Evolution, Copy Number Variation, Nuclear Life Cycle
Introduction
Genomes are traditionally perceived as having fixed karyotypes within eukaryotic species (e.g. [Lewin 2000; Hartwell et al. 2004]). Under such a model, closely related individuals (within populations or species) share nearly identical genomes and genomic integrity is maintained through the cell cycle. This static notion of the genome is challenged by studies in a variety of lineages that demonstrate considerable variation in genomic DNA content throughout organismal life cycles and among members of a single species. As discussed below, data from diverse eukaryotic lineages reveal extensive intra- and interspecific variation in genome content. These data, along with recognition of the widespread influence of epigenetics on the genome (Cerutti and Casas-Mollano 2006; Katz 2006; Richards 2006; Bird 2007), illuminate an increasingly dynamic picture of the eukaryotic genome.
Examining the phylogenetic distribution of genome dynamics enhances interpretation of the evolution of genome features. The phylogenetic framework of eukaryotes has shifted from the five (Whittaker 1969; Margulis and Schwartz 1988) or six (Cavalier-Smith 2002) kingdom systems emphasizing plants, animals, and fungi towards systems recognizing that these macroscopic groups are only three of an estimated 60–200 lineages (Patterson 1999), the rest are diverse microbial lineages. The current view of eukaryotic classification divides eukaryotes into six “supergroups” that encompass both macrobial and microbial members (Baldauf et al. 2000; Adl et al. 2005; Keeling et al. 2005), although this classification is likely premature as some groups are poorly supported (Parfrey et al. 2006). The diverse microbial lineages employ many genomic strategies and are known to provide extreme examples of some, such as genome processing in some ciliates that generates up to 25,000,000 somatic chromosomes (Raikov 1982; McGrath and Katz 2004; Zufall, Robinson, and Katz 2005). Thus, the consideration of microbial eukaryotes expands understanding of both the distribution and types of genome dynamics.
To elucidate the range of variation in eukaryotic genomes we focus on two aspects of intraspecific genome dynamics: (1) variation in nuclear cycles within lineages, and (2) variation in genome content among individuals within species. Selected examples are presented to highlight the broad phylogenetic distribution of dynamic features of eukaryotic genomes (bold lineages fig. 1; images of representative taxa fig. 2). Elsewhere there are extensive discussions of related topics such as the evolution of sex and meiosis (Kondrashov 1994; Mable and Otto 1998; Archetti 2004; Nuismer and Otto 2004) and changes in ploidy associated with speciation (genome duplication, hybridization, etc; e.g. [Otto and Whitton 2000; Bennett 2004]).
Figure 1. Distribution of genomic features.
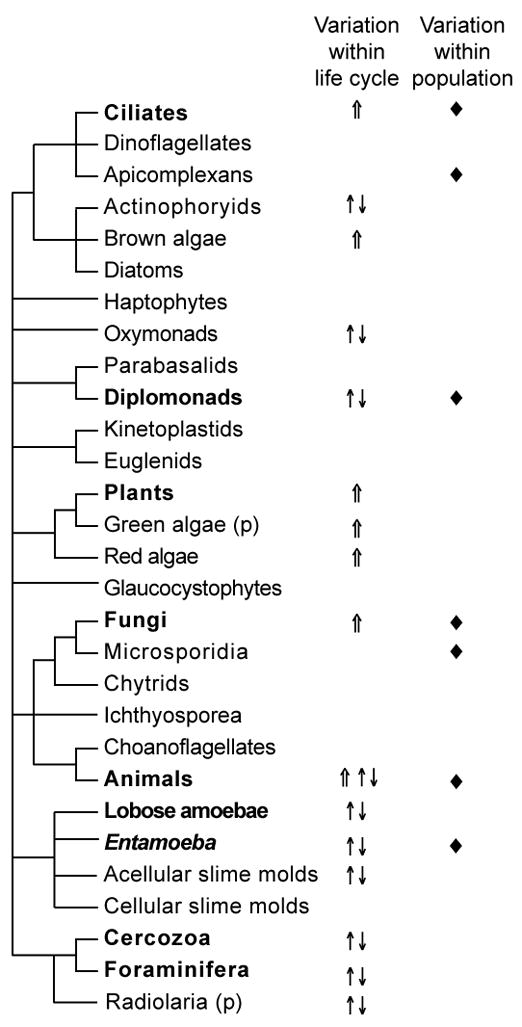
Dynamic genomes are widespread across the eukaryotic tree of life. Occurrence of three metrics of genome dynamism are plotted onto our cartoon of the eukaryotic tree of life, the topology of which is derived from our interpretation of multigene genealogies (Parfrey et al. 2006; Rodriguez-Ezpeleta et al. 2007; Yoon et al. Submitted). (p) indicates paraphyly in radiolaria and green algae. Symbols indicate that the feature is reported in at least one taxon within the lineage. ⇑: Somatic polyploidy, ↑↓: Cyclic polyploidy, ◆: Intraspecific genome variation.
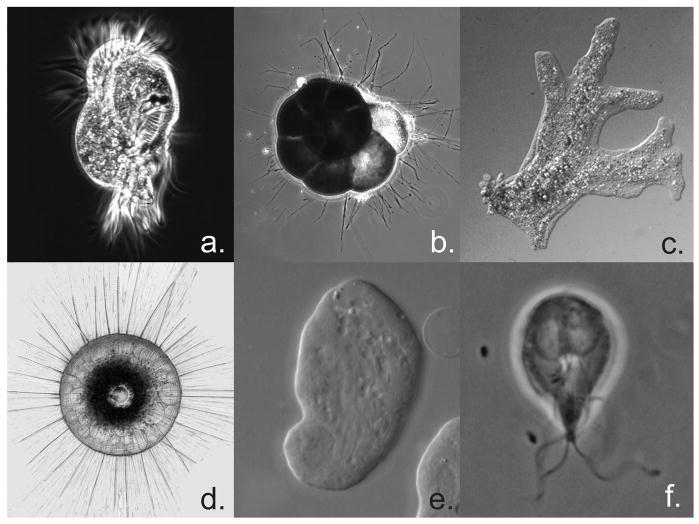
Figure 2. Exemplar microbial eukaryotes.
Images of microbial organisms discussed reveal morphological diversity in addition to genomic diversity described in the text. Approximate size is give for reference. (a) Uronychia (ciliate) – 150μm, (b) Ammonia (Foraminifera) – 300 μm, (c) Amoeba proteus – 300 μm, (d) Aulacantha (Phaeodarea) – 200 μm, (e) Entamoeba – 25 μm, (f) Giardia – 12 μm. All except d are images of live organisms, d is a drawing from Haeckel (1862) from the library of Kurt Stueber (http://caliban.mpiz-koeln.mpg.de/~stueber/haeckel/radiolarien). All images used with permission from micro*scope (http://starcentral.mbl.edu/microscope/portal.php).
We first consider changes in ploidy levels and genome content during the life cycles of exemplar lineages. To facilitate discussion, we present a generalized nuclear cycle for eukaryotes that depicts the progression through meiosis and karyogamy with intervening rounds of mitosis, such as occurs in the alternation of generations in plants (fig. 3a). This generalized nuclear cycle provides a common framework and terminology that enable comparison among diverse eukaryotic lineages. Meiosis and karyogamy, which respectively halve and double genome ploidy (coded yellow and red in fig. 3), are the basis of the sexual haploid-diploid cycle that underlies the typified nuclear cycles of plants, animals, and fungi (fig. 3a–c). Variation in nuclear cycles can be imparted through elimination of some stages (e.g. asexual eukaryotes reproduce via mitosis alone, and in many vertebrate lineages haploid nuclei do not undergo mitosis). Our focus is on the diversity of nuclear cycles that deviate from textbook versions of haploid-diploid cycles (fig. 3d–i).
Figure 3. Diversity of nuclear cycles.
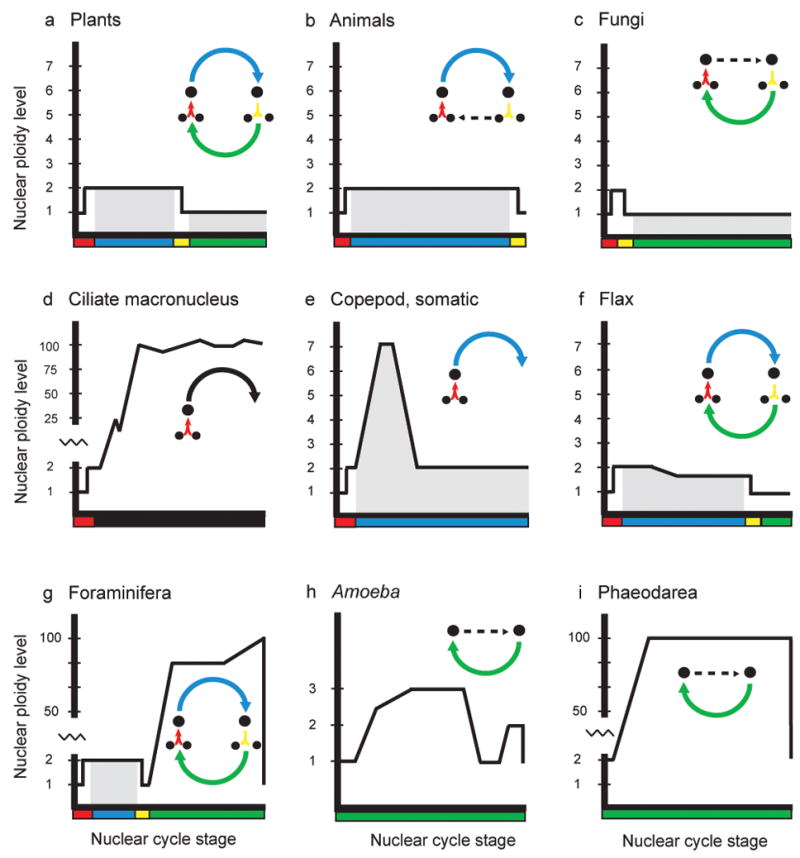
Depiction of changes in DNA content through the nuclear cycles of nine lineages of eukaryotes. Horizontal axis and colors corresponds to the nuclear cycle stage as shown in the inset diagram, while the vertical axis measure approximate DNA content within the nucleus. Gray shading represents periods of multicellularity or multinuclearity. Inset diagram in panel a is the generalized nuclear cycle as exemplified by plants. Arrows represent progression of genome through karyogamy (red), mitosis as a diploid (blue), meiosis (yellow), and mitosis as a haploid (green). Amitosis in ciliates is black. The nuclear cycle of organisms may include some or all components of the generalized nuclear cycle. A dashed arrow indicates the absence of intervening steps. Panels d and e depict the fate of the somatic genomes, therefore they are dead ends.
A second aspect of dynamic genomes explored here is the heterogeneity of DNA content among conspecific individuals (◆ fig. 1; fig. 4). In contrast to the perception that single nucleotide polymorphisms (SNPs) are responsible for intraspecific differentiation, emerging data demonstrate a greater range of variation. This intraspecific genomic variation ranges from insertions and deletions of short stretches of DNA through population differences in karyotype and ploidy. Examples from animals, fungi and microbes are used to illustrate intraspecific genomic variability.
Figure 4. Range of intraspecific variation.
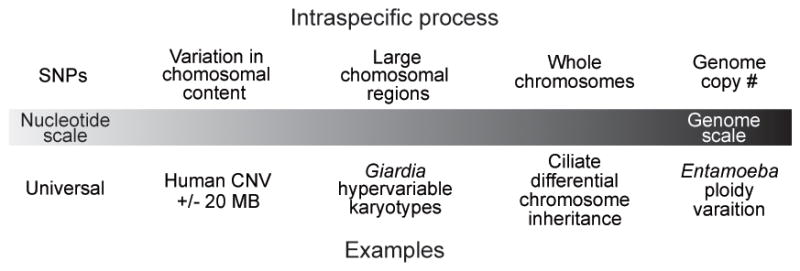
Individuals within a species (and population) do not have identical genomes. Intraspecific genomes can be different from the level of a few nucleotides, to chromosomes, to ploidy of the whole genome. We plot examples of this variation discussed in the text along a gradient of variation. The number of nucleotides involved increases from left to right.
Underlying the diversity of genomic processes are similarities that suggest an ancient origin of the basic features of the genome propagation including meiosis and mitosis. We argue that eukaryotes share mechanisms, possibly epigenetic, to maintain the integrity of genetic material for transmission to future generations while also allowing extensive variation within life cycles and among conspecific individuals. From the observed diversity several trends in genome dynamics emerge such as: 1) elevated ploidy levels in large cells coupled with diverse mechanisms for reducing ploidy (e.g. Foraminifera, Phaeodarea, and the bacterium Epulopiscium fishelsoni); 2) dynamism in the somatic genome in lineages with sequestered germlines (e.g. ciliates and animals); and 3) ploidy cycles in asexual lineages (e.g. Giardia, Entamoeba, and Amoeba). We further suggest that intraspecific variation in genome content is a widespread source of phenotypic variation.
1) Varying DNA content during nuclear cycles
DNA content elevated over the diploid level is the most common manifestation of dynamic eukaryotic genomes (fig. 1), and even occurs in some bacteria and archaea. Two mechanisms by which organisms increase their DNA content are endoreplication resulting in polyploidy and genome amplification (Box 1). There are two fates of polyploid cells: (1) terminal polyploidy and (2) cyclic polyploidy that is later reduced by a non-meiotic mechanism (fig. 1 ⇑ and ↑↓, respectively). Terminal polyploidy occurs in differentiated somatic tissues in many multicellular taxa, including red algae, green algae, and animals, and in the somatic nuclei of ciliates (fig. 3d, e). In lineages characterized by cyclic polyploidy, increasing genome content is a phase in the life cycle and ploidy levels are reduced prior to either sexual or asexual reproduction (fig. 3g–i). These groups have diverse strategies for reducing ploidy level, such as genome segregation in which one polyploid nucleus yields thousands of haploid daughter nuclei (Box 1; [Raikov 1982]). Genomic DNA levels can also be reduced during life cycles of some lineages when portions of the genome are eliminated in response to stress or during development (fig. 3d–f). Below we introduce examples of these types of dynamic genomes, though in many cases the molecular details of genome processes have yet to be elucidated.
Box 1.
Karyotype
The number and length of chromosomes in the haploid chromosome complement.
Polyploidy
Nucleus containing more than two copies of the genome; widespread in eukaryotes.
Endoreplication (syn. endoreduplication, endomitosis)
Increase in nuclear ploidy levels by replication of DNA in the absence of mitosis; widespread in eukaryotes.
Genome amplification
Selective replication of portions of genomic DNA during development; extensive amplification in ciliate macronucleus and rDNA amplification is widespread.
Chromatin diminution
Selective elimination of chromosomes or portions of chromosomes during development; found in animals such as copepods, hagfish, dicyemids, and nematodes.
Genome segregation
Separation of a polyploid genome into haploid genome components prior to reproduction in polyploid protists such as Phaeodarea and other ‘radiolaria’.
Depolyploidization
Enigmatic reduction in DNA levels in the absence of nuclear division or mitosis reported in Amoeba proteus.
Reduction
Ploidy is reduced by nuclear division not preceded by replication, occurs in Entamoeba, Giardia, and human fibroblasts.
Ciliates are microbial eukaryotes (fig. 2a) that undergo extensive genomic processing through genome amplification and chromosome fragmentation during the development of the somatic macronucleus (Box 1; fig. 3d; [Prescott 1994; Katz 2001]). During conjugation two cells exchange meiotically produced micronuclear-derived gametic nuclei that fuse to become the zygotic nucleus. Both the macronucleus and the germline micronucleus, which does not undergo genome processing, differentiate from the zygotic nucleus following conjugation. The nuclear cycle of the germline micronucleus is similar to animals in that it is sequestered from the somatic genome, though in ciliates this all occurs within a single cell. Macronuclear genomes are highly polyploid, with a range of 60 copies of each chromosome in Tetrahymena to as many as 1000 copies of a chromosome in Stylonychia. The parental macronucleus degrades during conjugation, but influences the developing macronucleus through epigenetic processes (Mochizuki and Gorovsky 2005). The somatic macronculear genomes of the model ciliates Tetrahymena (Eisen et al. 2006) and Paramecium (Aury et al. 2006) have recently been completed, and planned micronuclear genome sequencing will elucidate further genome processing in this lineage.
Selective elimination of germline-limited DNA during somatic development also occurs in widely distributed lineages of animals, including hagfish (Vertebrata), dicyemid worms (Mesozoa), ascarids (Nematoda), and copepods (Crustacea) (Kloc and Zagrodzinska 2001; Redi et al. 2001; Zufall, Robinson, and Katz 2005; Awata, Noto, and Endoh 2006). In animals this process is referred to as chromatin diminution and results in the loss of 15 – 95% of germline DNA (Kloc and Zagrodzinska 2001). Following fertilization in copepods, the genome is endoreplicated five to ten-fold. Roughly half of the germline genome, predominately highly repetitive heterochromatin, is then eliminated during diminution (fig. 3e; [Beerman 1977]). The resulting diminuted somatic nuclei (diploid) contain the same amount of DNA as the haploid sperm cells (Wyngaard et al. 2001; Drouin 2006). The broad phylogenetic distribution of somatic chromosome processing in animals indicates that extensive modification of the somatic genome may be ancient in lineages with sequestered germline genomes. We anticipate that numerous additional examples will be discovered.
In the plant flax (Linum usitatissimum), reduction of DNA content occurs as a response to stress in the lifetime of the individual (Fig. 3i; [Cullis 2005]). Several studies find that the amount of DNA in the genome can be reduced by 15% when the plant is grown under stress (fig. 3f), and this reduction appears to effect high, middle, and low repetitive classes of DNA equally (Evans, Durrant, and Rees 1966; Cullis 1973; Cullis 1980). Thus, it is possible that coding regions are lost in addition to highly repetitive, non-coding DNA. The genomic response to stress is non-random, as the same stress induced genomic changes occur repeatedly (Cullis 2005). Intriguingly, an increase in homologous recombination has also been measured in Arabidopsis when this model plant is exposed to stress by ultraviolet light or pathogens (Molinier et al. 2006). In the case of flax, Cullis (2005) argues that genomic changes are reversible as after six generations in native conditions the full genome complement returns in new tissues. These observations suggest that this response may be epigenetic in nature.
Foraminifera, a diverse group of amoebae (fig. 2b), have dynamic genomes that provide examples of genome segregation and possibly genomic processing. Nuclear cycles have been studied in fewer than 1% of extant Foraminifera species (Goldstein 1999), but are generally characterized by an alternation between multinucleated diploid agamonts and uninucleate haploid gamonts (fig. 3g). The large agamonts reproduce by meiosis and multiple fission, yielding numerous haploid gamonts. The single gamontic nucleus then increases in size and DNA content as the organism grows, reaching 400 μm in diameter in some species (Goldstein 1997; Bowser, Habura, and Pawlowski 2006). It is not known whether DNA content increases through polyploidization, by differentially amplifying some portion of the genome, or through a combination of the two. Just prior to gametogenesis nuclear products, predominately nucleoli plus some DNA, are expelled from the gamontic nucleus and degraded in a process referred to as ‘Zerfall’ (Føyn 1936). This ‘nuclear cleansing’ during ‘Zerfall’ may facilitate a return to the haploid genome before reproduction (Bowser, Habura, and Pawlowski 2006). The retained DNA proliferates through many rounds of mitosis (Føyn 1936; Arnold 1955) to generate hundreds to thousands of small (~ 5 μm) haploid gametes (Bé and Anderson 1976; Goldstein 1997).
Amoeba proteus is reported to reduce the DNA levels in its nucleus by depolyploidization in the absence of mitosis (fig. 2c, 3h). A. proteus, an asexual lobose amoebae, contains over 500 chromosomes and is believed to be polyploid (Raikov 1982). Though there are no direct measurements, several indirect methods have been applied to show ploidy variations (Afonkin 1986). Cytofluorimetric data shows that the DNA content of A. proteus increases and decreases by up to 2.9 fold during interphase of the cell cycle (Makhlin, Kudryavtseva, and Kudryavtsev 1979). DNA synthesis is reported to occur in bursts, with one major peak right after mitosis and another smaller peak immediately before subsequent mitosis and cell division (Ord 1968). Taken together these studies suggest that A. proteus becomes polyploid during interphase and that prior to reproduction, this surplus of DNA is eliminated by depolyploidization to recover the haploid genome. Under this hypothesis, the genome is then replicated and passed on to daughter A. proteus cells by mitosis (fig. 3h). Intriguingly, A. proteus is a congener of Amoeba dubia, the eukaryote with the largest reported genome at 670,000 MB (Friz 1968), and may share mechanisms for managing huge amounts of DNA during its life cycle with A. proteus.
Phaeodarea, a member of the polyphyletic ‘radiolaria’ that is currently placed within the ‘Cercozoa’, reduce ploidy levels in their large polyploid nucleus by genome segregation (fig. 2d, 3i; [Grell 1953; Raikov 1982]). While the complete nuclear cycle of this lineage has not been fully elaborated, in part as no member of this clade has yet to be maintained in culture, Phaeodarea members are reported to be asexual and have a single nucleus that increases in ploidy by endomitosis. Ploidy is reduced prior to reproduction as the chromatin in this large (~100 μm in diameter) nucleus condenses into thousands of polytene “chromosomes” that segregate along microtubules, each becoming a nucleus (fig. 3i; [Grell and Ruthmann 1964]). These polytene “chromosomes” later break down into 10–12 smaller chromosomes, presumably the haploid genome complement. The subsequent nuclei undergo rounds of mitosis yielding thousands of daughter nuclei that are individually packaged into biflagellate spores along with other organelles. Production of spores consumes the entire cytoplasm of the parent, and allows alternation between a large vegetative cell and small reproductive cells (Raikov, 1982).
Although not the focus of this manuscript, variation in ploidy levels is also reported from bacteria and archaea. DNA content in Escherichia coli varies from two, four, or eight genome equivalents in stationary phase up to 11 genome equivalents per cell in early exponential phase (Akerlund, Nordstrom, and Bernander 1995). Similar variability is reported for Methanococcus jannaschii, Micrococcus radiodurans, Synechococcus PCC6301, Desulfovibrio gigas, Borrelia hermsii, Azotobacter vinlandii ([Bendich and Drlica 2000] and references therein). The giant bacterium Epulopiscium fishelsoni (up to 600 μm in length) has a 3,000-fold range in ploidy variation that corresponds to a similar range in cell size (Bresler and Fishelson 2003), suggesting that polyploidy is associated with large cell sizes in this domain of life as well. Ploidy levels are reduced in during reproduction E. fishelsoni when numerous daughter cells are produced simultaneously in the mother cell (Angert 2005).
2) Intraspecific variation in DNA content
Genomes also vary dramatically among individuals within species of diverse eukaryotic lineages (◆ fig. 1). The portion of the genome that varies ranges from polyploidization of the entire genome to insertions and deletions of megabase stretches of genomic DNA (fig. 4). Such variation contrasts markedly with the SNP variants that are the focus of many current studies of genomic variation within species.
Substantial levels of among individual variation in DNA content have recently been found in humans, where the phenomenon is called copy number variation (CNV; [Freeman et al. 2006; Redon et al. 2006]). Redon et al. (2006) found insertions and deletions of kilobase to megabase segments of DNA that lead to polymorphisms in the presence/absence of chromosome regions and genes contained within them. The scale of this variation is enormous; a survey of 270 individuals found that 360 MB (13% of the genome) varied (Redon et al. 2006), presenting a marked contrast to the genomic conservation symbolized by the 99% similarity in orthologous sequences between humans and chimps (Mikkelsen et al. 2005). Geneticists are struggling to conceptualize the species genome and redefine ‘normal’ in this context as they search for the disease implications of this variability (Kehrer-Sawatzki 2007).
The ciliate macronucleus displays intraspecific variability on the level of whole chromosomes. The macronucleus is inherited by amitosis during asexual reproduction, an imprecise mechanism resembling binary fission or budding in which chromosomes are replicated and distributed to daughter nuclei without a mitotic spindle. Amitosis can lead to differential inheritance of alleles or paralogs (Robinson and Katz 2007) and contribute to overall elevated rates of protein evolution seen in ciliates (Katz et al. 2004; Zufall and Katz 2007). In both cases, epigenetic mechanisms likely play a role in regulating genome dynamics. Hence, ciliates within a population may have identical, or very similar, germline nuclei while their somatic nuclei can vary in the presence/absence of chromosomes.
Populations of Entamoeba, the causative agent of amoebic dysentery in humans (fig. 2e; [Stauffer and Ravdin 2003]), demonstrate heterogeneity in nuclear ploidy due to varying levels of endomitosis. Entamoeba alternates between infective resting cysts with four haploid nuclei and metabolically active trophozoites with one or more nuclei. DNA levels within a population of trophozoites exhibit continuous variation from 4N to 40N, and this variation is present both within multinucleate individuals and among the nuclei of separate individuals (Lohia 2003). Populations can be synchronized to 4N by starvation, but achieve the same 10-fold range of variation within two hours of addition of serum (Lohia 2003).
The diplomonad Giardia, a causative agent of diarrhea in humans, is an anaerobic flagellate with a hypervariable karyotype whereby the number and lengths of chromosomes vary among isolates (fig. 2f; [Hou et al. 1995; Adam 2000]). Though Giardia is a putative asexual lineage it has a ploidy cycle due to endoreplication and reduction (Bernander, Palm, and Svard 2001). Analysis of Giardia chromosomes by pulse field gel electrophoresis reveals several size variants for each chromosome and there is no fixed karyotype for this species (Le Blancq and Adam 1998). While the core portions of the chromosomes appear to be stable, there is considerable variation in the subtelomeric region (Le Blancq and Adam 1998; Adam 2000). Heterogeneity in karyotypes contrasts starkly with the almost complete lack of nucleotide heterogeneity in protein-coding genes between strains (Morrison et al. 2007; Teodorovic, Bravermanj, and Elmendorf 2007; Lasek-Nesselquist personal communication). As Giardia is asexual, one would expect large amounts of allelic variation to accumulate.
Intraspecific DNA sequence variation has also been found in Arbuscular Mycorrhizal Fungi (AMF), which supply essential nutrients to plant roots (Smith and Read 1997). Vegetative AMF cells contain numerous nuclei, as do fungi in general; however AMF are unusual in that hundreds to thousands of nuclei appear to be transferred to each spore (Pawlowska 2005). Within individual and within spore genetic variation is documented for rDNA and protein coding genes in several species of AMF (reviewed in [Pawlowska 2005]). These results contradict the expected clonal population structure and haploid genome expected for fungi considered ancient asexuals (Judson and Normark 1996). It remains to be seen whether this variation is harbored in each nuclei as either duplicated genes or polyploid genomes (Pawlowska and Taylor 2004) or in genetically distinct nuclei (each presumably haploid) that are passed to each spore (Kuhn, Hijri, and Sanders 2001; Hijri and Sanders 2005).
Synthesis
We demonstrate that genomes are dynamic within and between eukaryotic lineages. This dynamism elaborates the generalized ‘textbook’ view of nuclear cycles (fig. 3a–c) through addition of ploidy cycles, modification of genome content during development or in response to stress, and/or generation of individual variation in karyotypes (fig. 3d–i; fig. 4). For example, cyclic changes in ploidy occur during the life cycle in many ‘large’ organisms, such as Phaeodarea, Foraminifera, and large prokaryotes. Ploidy cycles may be advantageous as they allow both reproduction via haploid gametes or spores, which is suggested to reduce the mutational load (Kondrashov 1997), and the high levels of DNA that are correlated with large vegetative cells (Mortimer 1958; Kondorosi et al. 2000). These nuclear features are broadly distributed (fig. 1) and likely involve shared mechanisms. The observed variation in genome content is underlain by conservation in at least some of the molecular machinery regulating meiosis and mitosis—two basic components of the nuclear cycle—across the diversity of eukaryotes (Nurse 1990; Ramesh, Malik, and Logsdon 2005). Hence, the broad phylogenetic distribution of ploidy cycles suggest that mode of genome propagation evolves quickly, but within the constraints of the conserved regulatory framework that was likely present in the last common ancestor of eukaryotes. The limited data on ploidy levels in prokaryotes hint that the molecular machinery may be even more ancient.
Data on the population level variation in DNA content are changing further perceptions on the nature of the eukaryotic genome. Recognized cases of intraspecific genomic heterogeneity are already widespread in eukaryotes (fig. 4), with examples coming from genome array studies on humans and limited data on microbial eukaryotes such as Giardia and Entamoeba. Intraspecific genome variability is likely a broad phenomenon that awaits detection in many more groups, and may be a common source of phenotypic variation.
Elucidating the scope of genome dynamics is essential for understanding the relationship between genomes and phenotypes. Recent studies suggest that genome features can affect evolution by altering population genetic parameters such as effective population size (Lynch 2007) and rates of molecular evolution (Zufall et al. 2006). Understanding the wide range of ploidy levels found in diverse eukaryotes may also shed light on the intolerance of vertebrate cells to such fluctuations, where departure from diploidy often leads to cancer and aneuploid defects such as trisomy 21 (Ganem, Storchova, and Pellman 2007).
Though much remains to be discovered about the mechanisms behind genome dynamics during life cycles and within species, candidate mechanisms are most likely epigenetic. Epigenetic phenomena such as methylation, acetylation and genome scanning through RNAi (e.g [Mochizuki and Gorovsky 2005; Cerutti and Casas-Mollano 2006; Goldberg, Allis, and Bernstein 2007]) may enable cells to differentiate between germline and somatic genomes, even in the context of a single nucleus. Under such scenarios, nuclei mark ‘germline’ genetic information for transmission to subsequent generation. Such a distinction between germline and soma may be key in enabling variation in genomes within life cycles and among individuals within populations while also maintaining integrity between generations.
Acknowledgments
Thanks to Ben Normark and David Lahti for providing helpful comments, and to David Patterson for permission to use micro*scope images. The work was supported by grant NIH (1R15GM081865-01) and NSF ATOL (DEB 043115) to LAK.
References
- Adam RD. The Giardia lamblia genome. International Journal for Parasitology. 2000;30:475–484. doi: 10.1016/s0020-7519(99)00191-5. [DOI] [PubMed] [Google Scholar]
- Adl SM, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, Barta JR, Browser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor M. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. Journal of Eukaryotic Microbiology. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Afonkin SJ. Spontaneous depolyploidization of cells in Amoeba clones with increased nuclear-DNA content. Archiv Für Protistenkunde. 1986;131:101–112. [Google Scholar]
- Akerlund T, Nordstrom K, Bernander R. Analysis of cell-size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. Journal of Bacteriology. 1995;177:6791–6797. doi: 10.1128/jb.177.23.6791-6797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angert ER. Alternatives to binary fission in bacteria. Nature Reviews Microbiology. 2005;3:214–224. doi: 10.1038/nrmicro1096. [DOI] [PubMed] [Google Scholar]
- Archetti M. Loss of complementation and the logic of two-step meiosis. Journal of Evolutionary Biology. 2004;17:1098–1105. doi: 10.1111/j.1420-9101.2004.00726.x. [DOI] [PubMed] [Google Scholar]
- Arnold ZM. Life history and cytology of the foraminiferan Allogromia laticollaris. University of California Publications in Zoology. 1955;61:167–252. [Google Scholar]
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Camara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le Mouel A, Lepere G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Betermier M, Weissenbach J, Scarpelli C, Schachter V, Sperling L, Meyer E, Cohen J, Wincker P. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Awata H, Noto T, Endoh H. Peculiar behavior of distinct chromosomal DNA elements during and after development in dicyemid mesozoan Dicyema japonicum. Chromosome Research. 2006;14:817–830. doi: 10.1007/s10577-006-1084-z. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Bé AWH, Anderson OR. Gametogenesis in planktonic foraminifera. Science. 1976;192:890–892. doi: 10.1126/science.946914. [DOI] [PubMed] [Google Scholar]
- Beerman S. The diminution of heterochromatic chromosomal segments in Cyclops (Crustacea, Copepoda) Chromosoma. 1977;60:297–344. doi: 10.1007/BF00292858. [DOI] [PubMed] [Google Scholar]
- Bendich AJ, Drlica K. Prokaryotic and eukaryotic chromosomes: what’s the difference? Bioessays. 2000;22:481–486. doi: 10.1002/(SICI)1521-1878(200005)22:5<481::AID-BIES10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Perspectives on polyploidy in plants - ancient and neo. Biological Journal of the Linnean Society. 2004;82:411–423. [Google Scholar]
- Bernander R, Palm JED, Svard SG. Genome ploidy in different stages of the Giardia lamblia life cycle. Cellular Microbiology. 2001;3:55–62. doi: 10.1046/j.1462-5822.2001.00094.x. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bowser SS, Habura A, Pawlowski J. Molecular evolution of Foraminifera. In: Katz LA, Bhattacharya D, editors. Genome Evolution in Eukaryotic Microbes. Oxford University Press; Oxford: 2006. [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of protozoa. International Journal of Systematic and Evolutionary Microbiology. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Current Genetics. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis CA. Mechanisms and control of rapid genomic changes in flax. Annals of Botany. 2005;95:201–206. doi: 10.1093/aob/mci013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis CA. DNA differences between flax genotypes. Nature. 1973;243:515–516. doi: 10.1038/243515a0. [DOI] [PubMed] [Google Scholar]
- Cullis CA. DNA sequence organization in the flax genome. Biochemica et Biophysica Acta. 1980;652:1–15. doi: 10.1016/0005-2787(81)90203-3. [DOI] [PubMed] [Google Scholar]
- Drouin G. Chromatin diminution in the copepod Mesocyclops edax: diminution of tandemly repeated DNA families from somatic cells. Genome. 2006;49:657–665. doi: 10.1139/g06-022. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu DY, Thiagarajan M, Wortman JR, Badger JH, Ren QH, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang YF, Cai H, Collins K, Stewart A, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA, Krieger CJ, del Toro C, Ryder HF, Williamson SC, Barbeau RA, Hamilton EP, Orias E. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biology. 2006;4:1620–1642. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans GM, Durrant A, Rees H. Associated nuclear changes in the induction of flax genotrophs. Nature. 1966;212:697–699. [Google Scholar]
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, Carter NP, Scherer SW, Lee C. Copy number variation: New insights in genome diversity. Genome Research. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- Friz CT. The biochemical composition of the free-living Amoebae Chaos chaos, Amoeba dubia and Amoeba proteus. Comp Biochem Physiol. 1968;26:81–90. doi: 10.1016/0010-406x(68)90314-9. [DOI] [PubMed] [Google Scholar]
- Føyn B. Über die Kernverhaltnisse der Foraminifere Myxotheca arenilega Schaudinn. Archiv Für Protistenkunde. 1936;87:272–295. [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Current Opinion in Genetics & Development. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Goldstein ST. Gametogenesis and the antiquity of reproductive pattern in the Foraminiferida. Journal of Foraminiferal Research. 1997;27:319–328. [Google Scholar]
- Goldstein ST. In: Foraminifera: A biological overview. Sen Gupta BK, editor. Modern Foraminifera; Kluwer: 1999. pp. 37–56. [Google Scholar]
- Grell KG. Die Chromosomen von Aulacantha scolymantha Haeckel. Archiv für Protistenkunde. 1953;99:1–54. [Google Scholar]
- Grell KG, Ruthmann A. Über die Karyologie des Radiolars Aulacantha scolymantha und die Feinstruktur seiner Chromosomen. Chromosoma. 1964;15:158–211. doi: 10.1007/BF00285729. [DOI] [PubMed] [Google Scholar]
- Haeckel E. Die Radiolarien (Rhizopoda Radiaria): Eine Monographie. G. Reimer; Berlin: 1862. [Google Scholar]
- Hartwell LH, Hood L, Goldberg ML, Reynolds AE, Silver LM, Veres RC. Genetics: From Genes to Genomes. McGraw Hill; New York: 2004. Genetics: The Study of Biological Information; pp. 1–12. [Google Scholar]
- Hijri M, I, Sanders R. Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nature. 2005;433:160–163. doi: 10.1038/nature03069. [DOI] [PubMed] [Google Scholar]
- Hou GY, Leblancq SM, Yaping E, Zhu HX, Lee MGS. Structure of a frequently rearranged ribosomal RNA encoding chromosome in Giardia lamblia. Nucleic Acids Research. 1995;23:3310–3317. doi: 10.1093/nar/23.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson OP, Normark BB. Ancient asexual scandals. Trends in Ecology & Evolution. 1996;11:A41–A46. doi: 10.1016/0169-5347(96)81040-8. [DOI] [PubMed] [Google Scholar]
- Katz LA. Genomes: Epigenomics and the future of genome sciences. Current Biology. 2006;16:R996–R997. doi: 10.1016/j.cub.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Katz LA. Evolution of nuclear dualism in ciliates: a reanalysis in light of recent molecular data. Int J Syst Evol Microbiol. 2001;51:1587–1592. doi: 10.1099/00207713-51-4-1587. [DOI] [PubMed] [Google Scholar]
- Katz LA, Lasek-Nesselquist E, Bornstein J, Muse SV. Dramatic diversity of ciliate histone H4 genes revealed by comparisons of patterns of substitutions and paralog divergences among eukaryotes. Molecular Biology and Evolution. 2004;21:555–562. doi: 10.1093/molbev/msh048. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends in Ecology & Evolution. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H. What a difference copy number variation makes. Bioessays. 2007;29:311–313. doi: 10.1002/bies.20554. [DOI] [PubMed] [Google Scholar]
- Kloc M, Zagrodzinska B. Chromatin elimination - an oddity or a common mechanism in differentiation and development? Differentiation. 2001;68:84–91. doi: 10.1046/j.1432-0436.2001.680202.x. [DOI] [PubMed] [Google Scholar]
- Kondorosi E, Roudier F, Gendreau E. Plant cell-size control: growing by ploidy? Current Opinion in Plant Biology. 2000;3:488–492. doi: 10.1016/s1369-5266(00)00118-7. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Evolutionary genetics of life cycles. Annual Review of Ecology and Systematics. 1997;28:391–435. [Google Scholar]
- Kondrashov AS. The asexual ploidy cycle and the origin of sex. Nature. 1994;370:213–216. doi: 10.1038/370213a0. [DOI] [PubMed] [Google Scholar]
- Kuhn G, Hijri M, Sanders IR. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature. 2001;414:745–748. doi: 10.1038/414745a. [DOI] [PubMed] [Google Scholar]
- Le Blancq SM, Adam RD. Structural basis of karyotype heterogeneity in Giardia lamblia. Molecular and Biochemical Parasitology. 1998;97:199–208. doi: 10.1016/s0166-6851(98)00150-9. [DOI] [PubMed] [Google Scholar]
- Lewin B. Genes VII. Oxford University Press; 2000. [Google Scholar]
- Lohia A. The cell cycle of Entamoeba histolytica. Molecular and Cellular Biochemistry. 2003;253:217–222. doi: 10.1023/a:1026055631421. [DOI] [PubMed] [Google Scholar]
- Lynch M. Origins of Genome Architecture. Sinauer Associates; Sunderland, MA: 2007. [Google Scholar]
- Mable BK, Otto SP. The evolution of life cycles with haploid and diploid phases. Bioessays. 1998;20:453–462. [Google Scholar]
- Makhlin E, Kudryavtseva M, Kudryavtsev B. Peculiarities of changes in DNA content of Amoeba proteus nuclei during interphase - cytofluorometric study. Experimental Cell Research. 1979;118:143–150. doi: 10.1016/0014-4827(79)90592-5. [DOI] [PubMed] [Google Scholar]
- Margulis L, Schwartz K. Five Kingdoms: An Illustrated Guide to the Phyla of Life on Earth. 2. W. H. Freeman and Company; New York: 1988. [Google Scholar]
- McGrath CL, Katz LA. Genome diversity in microbial eukaryotes. Trends Ecol Evol. 2004;19:32–38. doi: 10.1016/j.tree.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hillier LW, Eichler EE, Zody MC, Jaffe DB, Yang SP, Enard W, Hellmann I, Lindblad-Toh K, Altheide TK, Archidiacono N, Bork P, Butler J, Chang JL, Cheng Z, Chinwalla AT, deJong P, Delehaunty KD, Fronick CC, Fulton LL, Gilad Y, Glusman G, Gnerre S, Graves TA, Hayakawa T, Hayden KE, Huang XQ, Ji HK, Kent WJ, King MC, Kulbokas EJ, Lee MK, Liu G, Lopez-Otin C, Makova KD, Man O, Mardis ER, Mauceli E, Miner TL, Nash WE, Nelson JO, Paabo S, Patterson NJ, Pohl CS, Pollard KS, Prufer K, Puente XS, Reich D, Rocchi M, Rosenbloom K, Ruvolo M, Richter DJ, Schaffner SF, Smit AFA, Smith SM, Suyama M, Taylor J, Torrents D, Tuzun E, Varki A, Velasco G, Ventura M, Wallis JW, Wendl MC, Wilson RK, Lander ES, Waterston RH. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Studies on an RNAi-like mechanism of germline DNA elimination in Tetrahymena thermophila. Developmental Biology. 2005;283:576–576. [Google Scholar]
- Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A, Nixon JEJ, Palm D, Passamaneck NE, Prabhu A, Reich CI, Reiner DS, Samuelson J, Svard SG, Sogin ML. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Mortimer RK. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiation Research. 1958;9:312–326. [PubMed] [Google Scholar]
- Nuismer SL, Otto SP. Host-parasite interactions and the evolution of ploidy. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11036–11039. doi: 10.1073/pnas.0403151101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Ord MJ. Synthesis of DNA through cell cycle of Amoeba proteus. Journal of Cell Science. 1968;3:483–491. doi: 10.1242/jcs.3.4.483. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Barbero E, Lasser E, Dunthorn M, Bhattacharya D, Patterson DJ, Katz LA. Evaluating support for the current classification of eukaryotic diversity. PLoS Genetics. 2006;2:2062–2073. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson DJ. The diversity of eukaryotes. American Naturalist. 1999;154:S96–S124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- Pawlowska TE. Genetic processes in arbuscular mycorrhizal fungi. FEMS Microbiology Letters. 2005;251:185–192. doi: 10.1016/j.femsle.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Pawlowska TE, Taylor JW. Organization of genetic variation in individuals of arbuscular mycorrhizal fungi. Nature. 2004;427:733–737. doi: 10.1038/nature02290. [DOI] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikov IB. The Protozoan Nucleus: Morphology and Evolution. Springer-Verlag; Wien: 1982. [Google Scholar]
- Ramesh MA, Malik SB, Logsdon JM. A phylogenomic inventory of meiotic genes: Evidence for sex in Giardia and an early eukaryotic origin of meiosis. Current Biology. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Redi CA, Garagna S, Zacharias H, Zuccotti M, Capanna E. The other chromatin. Chromosoma. 2001;110:136–147. doi: 10.1007/s004120000114. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen WW, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang FT, Zhang JJ, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. Inherited epigenetic variation - revisiting soft inheritance. Nature Reviews Genetics. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Robinson T, Katz LA. Non-mendelian inheritance of paralogs of two cytoskeletal genes in the ciliate Chilodonella uncinata. Molecular Biology and Evolution. 2007;24:2495–2503. doi: 10.1093/molbev/msm203. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Burger G, Roger AJ, Gray MW, Philippe H, Lang BF. Toward resolving the eukaryotic tree: The phylogenetic positions of jakobids and cercozoans. Current Biology. 2007;17:1420–1425. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. Mycorrhizal Symbiosis. Academic; San Diego: 1997. [Google Scholar]
- Stauffer W, Ravdin JI. Entamoeba histolytica: an update. Current Opinion in Infectious Diseases. 2003;16:479–485. doi: 10.1097/00001432-200310000-00016. [DOI] [PubMed] [Google Scholar]
- Teodorovic S, Bravermanj JM, Elmendorf HG. Unusually low levels of genetic variation among Giardia lamblia isolates. Eukaryotic Cell. 2007;6:1421–1430. doi: 10.1128/EC.00138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker RH. New concepts of kingdoms of organisms. Science. 1969;163:150–160. doi: 10.1126/science.163.3863.150. [DOI] [PubMed] [Google Scholar]
- Wyngaard GA, Domangue R, Gerken S, Rasch EM. A mechanistic role for chromatin diminution in the evolution of genome size and life history characteristics in zooplankton. American Zoologist. 2001;41:1632–1632. [Google Scholar]
- Yoon HS, Grant J, Tekle YI, Wu M, Chaon BC, Cole JC, Logsdon JM, Patterson DJ, Bhattacharya D, Katz LA. Broadly sampled multigene trees of eukaryotes. Bmc Evolutionary Biology. 2008;8 doi: 10.1186/1471-2148-1188-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufall RA, Katz LA. Micronuclear and macronuclear forms of beta-tubulin genes in the ciliate Chilodonella uncinata reveal insights into genome processing and protein evolution. Journal of Eukaryotic Microbiology. 2007;54:275–282. doi: 10.1111/j.1550-7408.2007.00267.x. [DOI] [PubMed] [Google Scholar]
- Zufall RA, McGrath CL, Muse SV, Katz LA. Genome architecture drives protein evolution in ciliates. Molecular Biology and Evolution. 2006;23:1681–1687. doi: 10.1093/molbev/msl032. [DOI] [PubMed] [Google Scholar]
- Zufall RA, Robinson T, Katz LA. Evolution of developmentally regulated genome rearrangements in eukaryotes. Journal of Experimental Zoology Part B-Molecular and Developmental Evolution. 2005;304B:448–455. doi: 10.1002/jez.b.21056. [DOI] [PubMed] [Google Scholar]