Abstract
Building reliable structural models of G protein-coupled receptors (GPCRs) is a difficult task due to the paucity of suitable templates, low sequence identity, and the wide variety of ligand specificities within the superfamily. Template-based modeling is known to be the most successful method for protein structure prediction. However, refinement of homology models within 1–3 Å Cα RMSD of the native structure remains a major challenge. Here we address this problem by developing a novel protocol (foldGPCR) for modeling the transmembrane (TM) region of GPCRs in complex with a ligand, aimed to accurately model the structural divergence between the template and target in the TM helices. The protocol is based on predicted conserved inter-residue contacts between the template and target, and exploits an all-atom implicit membrane force field. The placement of the ligand in the binding pocket is guided by biochemical data. The foldGPCR protocol is implemented by a stepwise hierarchical approach, in which the TM helical bundle and the ligand are assembled by simulated annealing trials in the first step, and the receptor-ligand complex is refined with replica exchange sampling in the second step. The protocol is applied to model the human β2-adrenergic receptor (β2AR) bound to carazolol, using contacts derived from the template structure of bovine rhodopsin. Comparison to the X-ray crystal structure of the β2AR shows that our protocol is particularly successful in accurately capturing helix backbone irregularities and helix-helix packing interactions that distinguish rhodopsin from β2AR.
Keywords: class A GPCR, structure prediction, simulated annealing, ligand binding, implicit solvent, membrane protein
INTRODUCTION
G protein-coupled receptors (GPCRs) comprise a large family of integral membrane proteins that mediate signal transduction across the cell membrane.1 GPCRs are activated by a wide variety of extracellular stimuli and interact primarily with G proteins to trigger a cascade of responses inside the cell. The human genome encodes about 800 GPCRs and they can be phylogenetically classified into five main classes, with the rhodopsin-like class A being the largest with about 670 members.2,3 Members of this family are important pharmaceutical targets because of their association with numerous diseases,4 thus structural models of GPCRs have useful applications in rational drug discovery.
It is believed that GPCRs share a common architecture of seven transmembrane (TM) helices packed into a 7-TM helical bundle, with three intracellular and three extracellular loops.5 Sequence analysis shows that class A receptors can be characterized by a set of highly conserved residues in each TM helix.6 Ligands for class A GPCRs are chemically diverse, including photons, ions, biogenic amines, nucleosides and nucleotides, peptides and protein hormones, lipids and eicosanoids, and they bind mainly within a pocket confined to the TM region and extracellular loops.7 Understanding the structural basis for how such a large diversity of ligands is accommodated within a common TM helical bundle architecture has been limited by a lack of structural information. Experimental structure determination of membrane proteins is technically challenging, and high-resolution structures are available for only few members from the eukaryotic class A GPCRs.8–19 Structural models predicted in silico can provide insights to better understand ligand-binding specificity and the activation mechanism of GPCRs.
Protein structure prediction methods, ranging from homology modeling to de novo methods, have been applied to the modeling of GPCRs. Before the first X-ray crystal structure for a 7-TM receptor was solved, models for bacteriorhodopsin and rhodopsin were built using a low-resolution electron density map and restraints derived from sequence analysis or biophysical experiments.20–22 Mosberg et al. built models of 26 GPCRs by distance geometry calculations based on interhelical hydrogen bonds.23,24 After the rhodopsin structure was solved in 2000, homology modeling and fragment-based threading methods were applied to build models for GPCRs using the rhodopsin structure as the template.25,26 Ab initio approaches have also been applied to model class A receptors.27,28 Within this set of techniques, template-based modeling methods are arguably the most reliable approach for predicting protein structure from amino acid sequence.29 However, the suitability of the rhodopsin template to accurately predict the structure of other GPCRs is questionable,30 as the average sequence identity to bovine rhodopsin of human GPCRs is relatively low at ~20%,26 and the GPCR superfamily is highly diverse in its ligand-binding properties. Reliable homology models are less confidently obtained as the sequence and structure similarity between the template and target decrease.29,31 While the overall helical bundle architecture is thought to be similar for class A GPCRs, detailed structural features are expected to differ for receptors binding to different ligands.
Indeed, the crystal structures of bovine rhodopsin and human β2-adrenergic receptor (β2AR) bound to an antagonist show that the two structures are similar, with a 2.7 Å Cα RMSD in the TM region, despite a relatively low sequence identity of 23%.12 However, the structures also show subtle differences in the position, rotational orientation, and kink angles of helices I, III, IV, V and VI. Other class A GPCRs will likely possess such subtle differences.
Effective use of multiple templates and successful application of fragment-based modeling or model refinement methods have greatly improved the accuracy of homology models. Nonetheless, obtaining models closer to the target than the template remains a major challenge in comparative modeling methods.29,32 Alternatively, recent progress in modeling of membrane proteins has demonstrated that an implicit membrane generalized Born (GB) force field, combined with advanced sampling techniques, performs well in the study of folding and assembly of membrane proteins, as well as in de novo structure prediction and refinement of homology models.33–36 Implicit solvent force fields in general have also been shown to be effective in refining NMR structures with a limited number of experimental observables or near-native decoys and predicted structures sufficiently close to the native basin.37–39
This paper describes a novel modeling approach (foldGPCR) for predicting the structure of a class A GPCR 7-TM helical bundle in complex with its ligand. The protocol aims to accurately model the structural divergence between the template and target in the TM helices. Building on the idea of using secondary structure and tertiary restraints to predict 3D structures, the protocol uses distance restraints derived from a template structure to impose the overall helical bundle topology and conserved inter-residue contacts. Additional distance restraints suggested mostly from experimental data are imposed on receptor-ligand interactions to model the ligand binding geometry. These restraints guide the conformational sampling process in simulated annealing trials during the assembly of the helical bundle and the ligand. The receptor-ligand complex model is then refined in an implicit membrane GB force field with replica exchange sampling.
The intracellular and extracellular loops in GPCRs are known to have important functional roles in ligand-binding and G-protein interaction,40 but they are challenging to model accurately and are not modeled in the current foldGPCR protocol. The crystal structures of rhodopsin and β2AR show that the structure of the loop regions can be vastly different, with the extracellular loop 2 (ECL2) forming a β sheet lid in rhodopsin, and a helix in β2AR.8,12 The loop regions are more variable in sequence and length,6 and also more dynamic and conformationally flexible than the TM regions,40 making them difficult to predict by both homology modeling and de novo modeling approaches.41,42 The recent community-wide assessment of GPCR structure modeling also showed that the loop regions are significantly more difficult than the TM regions to model accurately.43
The foldGPCR protocol is applied to model the human β2AR bound to carazolol using contact restraints derived from the bovine rhodopsin template. As a control, the protocol is also applied to model the bovine rhodopsin using the same set of contact restraints. We will report on the accuracy of the models obtained for β2AR (see supplementary material for the results on the rhodopsin models), then discuss some of the key properties of the protocol and its strengths compared to other GPCR modeling approaches.
METHODS
FoldGPCR: a protocol for GPCR transmembrane structure prediction
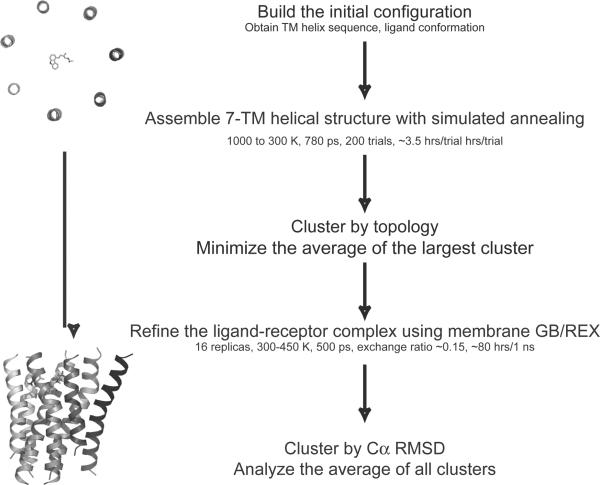
The foldGPCR method uses all-atom restrained molecular dynamics, and is implemented in a stepwise hierarchical manner, in which the helical bundle and the ligand are assembled by simulated annealing trials using tertiary restraints in the first step, and the receptor-ligand complex is refined using an implicit membrane GB force field and replica exchange sampling in the second step (Fig 1). The protocol aims to accurately model the structural divergence between the template and target in the TM helices, by using tertiary restraints derived from the template structure, while relying on an improved physics-based force field to yield conformations capturing structural features unique to the target. The molecular dynamics simulations are performed using the CHARMM program.44,45 We will first describe the two steps of the foldGPCR method, and then give details of the distance restraints used throughout the protocol.
Figure 1.
The foldGPCR protocol is implemented by a two-step hierarchical approach, in which the 7-TM helical bundle and the ligand are assembled in the first step, and the receptor-ligand complex is refined in the second step.
Step 1: Assembly of the 7-TM helical bundle
Initial configuration
The seven TM helices are modeled independently as disjoint helical segments. A canonical α-helix (ψ=−57, Φ=−47; side chains are in extended conformations) is built for each helix, then it is oriented perpendicularly to the membrane plane (xy-plane), centered at the mid-plane, z=0, and positioned around the perimeter of a circle sequentially, with each helix axis displaced 25 Å from the center of the circle (Fig. 2A). The N-terminus to C-terminus directionality of TM helices I, III, V, and VII are flipped relative to TM helices II, IV, and VI to reflect the membrane traversal directions of the helices in the native helical bundle. The putative seven TM segments are obtained from a previously published multiple sequence alignment of class A GPCRs.46 The alignment was checked that the following conserved motifs are aligned across the family: N1.50 in TM helix I, L2.46, A2.47, D2.50 in TM helix II, D/E3.49, R3.50, Y3.51 in TM helix III, W4.50 in TM helix IV, F5.47, P5.50, Y5.58 in TM helix V, F6.44, W6.48, P6.50 in TM helix VI, N7.49, P7.50, Y7.53 in TM helix VII; the Ballesteros-Weinstein numbering scheme is used to refer to residues.47 Helix lengths and boundaries are assumed to be the same as rhodopsin. However, this assumption is not an essential requirement for the protocol, and can be substituted by predictions of the helical segments using methods based on sequence information or potential energy profiles.35
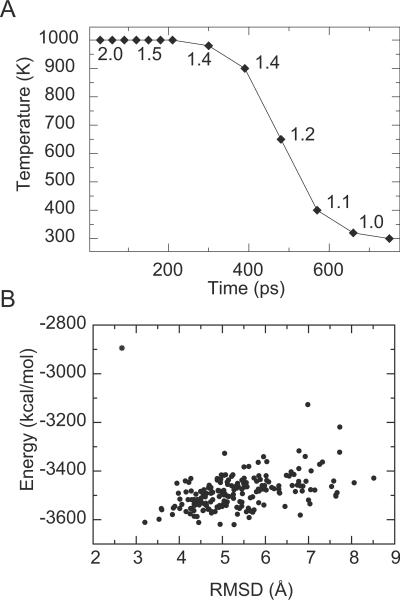
Figure 2.
FoldGPCR Step 1. Assembly of the helical bundle by simulated annealing trials. (A) The cooling schedule and the coupled distance scaling factor (2.0 to 1.0). (B) Energy versus Cα RMSD to the native crystal structure is plotted for the ensemble of annealed structures (dots) and the average structure (star) from a single modeling trial for β2AR.
The ligand is positioned so that its center of mass is approximately at the center of the circle and translated vertically to the average z-coordinate of the residues used in receptor-ligand distance restraints. The conformation of the ligand is adopted from the crystal structures (PDB ID code 1U19 for 11-cis-retinal bound to rhodopsin and 2RH1 for carazolol bound to β2AR). For other class A receptors with no known ligand structure, the initial ligand conformation can be built de novo prior to the annealing step by an additional step such as docking or other conformational sampling techniques. LPDB CHARMM parameters are used,48,49 and the atom types of the ligands are assigned with the MATCH toolset (Price and Brooks III, MATCH: primitive chemical pattern-matching engine provides robust atom-typing toolset for molecular mechanics force fields, manuscript in preparation). This initial 7-TM-ligand configuration is minimized with 200 steps of mixed steepest descent and adopted basis Newton-Raphson (ABNR) minimization using positional harmonic restraints on all heavy atoms.
Simulated annealing protocol
The TM helical bundle is assembled by simulated annealing trials, in vacuum using a distance-dependent dielectric function. The nonbonded interactions are smoothly switched off from 6.5 Å to 8 Å. The conformational sampling in this step is focused on local interactions within the helix and between adjacent helices, hence short distance cutoffs are used to expedite the calculations. During each trial, the temperature is decreased exponentially from 1000K to 300K over 780ps (Fig. 2A). The cooling schedule is coupled to a gradually decreasing distance scaling factor such that as the temperature decreases from 1000K to 300K, the distances used in the restraint potentials are scaled by a factor of 2.0 to 1.0. Sampling time is maximized at distances where the side chains of the helices are just coming into contact but not yet tightly packed (scaling factor of ~1.4), to allow structural rearrangements necessary for sampling alternative interhelical side chain packing interactions.
At the beginning of every annealing trial, each helix is rotated about its axis and the ligand about the x-axis by random angles. Each trial results in a single annealed structure. For each modeling attempt, an ensemble of 200 annealed structures is generated. The tertiary restraints are satisfied by nearly all of the annealed structures. The averaged structure is then minimized and refined in the subsequent step.
The number of annealing trials per modeling attempt was arbitrarily chosen so that the average structures from any two distinct attempts would be within 1.0 Å Cα RMSD from each other. The total length of the annealing time was optimized with respect to the RMSD distribution of the ensemble. The average structure is used as the initial structure for refinement because it tends to be closer to the native than any one structure in the ensemble (Fig. 2B). The effect of averaging in increasing the level of similarity to a given reference structure has previously been demonstrated by Zagrovic and Pande.50 The averaging process likely leads to a more native-like structure by cancellation of random errors sampled in the ensemble. Individual structural features usually occur with a unimodal distribution in sampled ensemble, hence the average structure represents the most populated conformation in the ensemble. Distortions resulting from the averaging process are largely corrected in the refinement step.
Step 2: All-atom refinement in implicit membrane
Refinement protocol
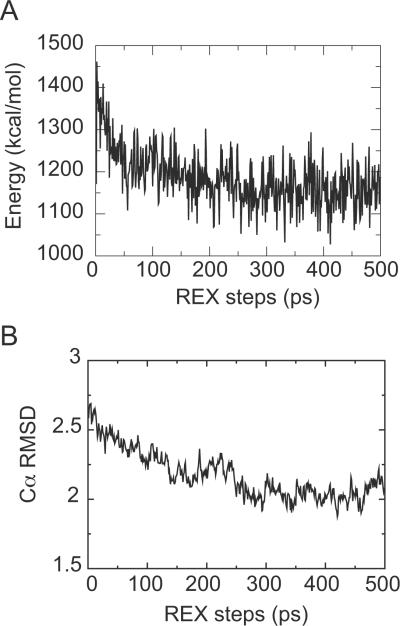
The use of a GB implicit membrane model combined with replica exchange (REX) sampling has been shown to be effective in obtaining native-like conformations in assembly and refinement of TM proteins.33–36 The replica exchange method is known to enhance conformational sampling by simulating multiple replicas of the system at different temperatures independently and simultaneously, and exchanging pairs of replicas at neighboring temperatures at preset intervals with a specified transition probability, allowing random walks in temperature space to help replicas escape from local energy minima.51,52 GB/REX refinement is applied in the second step of foldGPCR to refine the average structure from the simulated annealing trials. An all-hydrogen CHARMM parameter set (PARAM22) with the CMAP backbone torsional correction term is used.53,54 The GB implicit membrane model implemented in the GB model with a simple SWitching function (GBSW) module of CHARMM represents the membrane environment as a low-dielectric slab.55 The membrane thickness (Tmemb) is set to 35 Å, the membrane switching length (msw) is set to 2.5 Å, the surface tension coefficient (γ) is set to 0.04 kcal/(mol·Å2), and other parameters are set to default values. The values for Tmemb and msw were chosen to match the hydrophobic thickness of 31 Å estimated for rhodopsin in the OPM database.56 The nonbonded interactions are switched off at 20 Å. The replica exchange simulations are carried out using the MMTSB toolset (www.mmtsb.org).57 We use 16 replicas exponentially spaced between 300 and 450 K. Exchanges between adjacent replicas are attempted once every picosecond, with a total length of 500 ps. The exchange acceptance ratio ranged between 0.15 and 0.2. The energy profile from the lowest temperature ensemble shows that it converges within 500 ps (Fig. 3A). The last 20% of the structures (100 models) from the lowest temperature ensemble are minimized and clustered based on Cα RMSD to extract a few representative structures. Clustering was carried out with the MMTSB toolset using a Cα RMSD-based hierarchical and divisive method.57 The average structures from all clusters as well as the average structure of all 100 models are presented as the final predictions. The refinement step generally improves the packing interaction of helices, thereby increases the fraction of native contacts by ~10% and lowers the overall RMSD with respect to the native structure (Fig. 3B).
Figure 3.
FoldGPCR Step 2. Membrane GB/REX refinement of the receptor-ligand complex. Energy profile of the lowest temperature ensemble (A) and Cα RMSD profile for a replica that contributes to the last 100ps of the lowest temperature ensemble (B) during a 500 ps refinement run from a modeling trial for β2AR.
Distance restraints
Distance restraints are applied using a flat-bottom harmonic potential function with a soft asymptote implemented in the NOE module of CHARMM. The flat-bottom widths and the force constants for the harmonic restraints were chosen so that the restraints would be weakly imposed to allow sufficient structural flexibility during the sampling procedure. These parameters were chosen empirically, based on our previous experience with protein structure refinement in implicit solvent,33,37 and by trial-and-error. Three sets of distances are derived and the corresponding restraints are applied with slightly different parameter values, as described below. The optimal distances were estimated from known structures.
Topology of helical bundle
A set of 40 interhelical distances parallel to the membrane plane at the top, middle, and bottom of the helical bundle (z=0, ±~12 Å) is used to impose a particular topology. This set of restraints mimics the use of low-resolution cryo-EM structural data to approximate the spatial organization of the helices without defining the position of individual amino acids.21,58 The distance measurements that define the relative positions and tilt angles of the helices are obtained either from the β2AR (PDB ID code 2RH1) or the rhodopsin (PDB ID code 1U19) crystal structure.8,12 Each topological distance restraint is applied between centers of mass of sets of four adjacent Cα atoms from the two helices, to avoid biasing the rotational orientations of the helices. The flat-bottom half width is set to 2.5 Å and the force constant is set to 5 kcal/(mol·Å2).
Conserved inter-residue contacts
Structurally conserved inter-residue contacts are predicted by using sequence conservation as filters to select from contacts present in the template rhodopsin structure (PDB ID code 1U19). The contact prediction method is described elsewhere.59 Briefly, a contact in rhodopsin is predicted to be structurally conserved in another GPCR if the contact-forming residues or the fragments around these residues are conserved in sequence between the two sequences and across class A GPCRs. Out of 183 interhelical contacts in the rhodopsin TM region, ~40–50 are typically predicted to be conserved in another GPCR. The method predicts 45 contacts for the human β2-adrenergic receptor (Supplementary Table I). The restraints are applied between the side chain heavy atoms (Cα atom for Gly) with distance summation . The distance is set to 3.2 ± 2.0 Å (4.2 ± 2.0 Å for pairs containing Gly) and the force constant is set to 5 kcal/(mol·Å2).
Receptor-ligand interactions
Specific receptor-ligand interactions such as hydrogen bonds are restrained to model the binding geometry of the ligand. While the topological and contact restraints are derived directly from the template structure, the ligand restraints are mostly derived from experiments such as mutagenesis studies that suggest particular residues are involved in ligand-binding. The restraints are applied between the side chain heavy atoms and particular functional groups in the ligand. The distance is set to 3.5 ± 0.5 Å, 3.0 ± 0.5 Å, and 5.0 ± 2.0 Å for salt bridge, hydrogen bonding, and hydrophobic interactions, respectively. The force constant is set to 50 kcal/(mol·Å2). For the β2AR-carazolol interaction, residues D1133.32, N3127.39, F2906.52, S2035.42, Y3167.43, V1143.33, and W1093.28 are used; for the rhodopsin-11-cis-retinal interaction, residues K2967.43, E1133.28, W2656.48, F2125.47, and M2075.42 are used. In modeling class A receptors with limited and ambiguous experimental data on receptor-ligand interactions, several different sets of ligand restraints may need to be tested and optimized to obtain an average annealed model that agrees well with the experimental data.
Additional restraints
The dihedral angles of the helices and the ligands are restrained by weak harmonic potentials to minimize severe distortions at high temperature. The backbone torsion angles of the helices are restrained near ideal values but with significant latitude for fluctuation, ψ=−57 ± 30, Φ=−47 ± 30 degrees. The dihedral angles of all rotatable bonds in the ligand are restrained so that the ligand conformation is maintained within ~1.0 Å heavy atom RMSD from the initial structure. In the case that the receptor-bound ligand structure is not known, and the initial ligand conformation is built de novo, the force constants can be adjusted to allow sampling of other ligand conformations.
RESULTS AND DISCUSSION
Accuracy of β2AR models
The foldGPCR protocol is applied to model the human β2AR structure. In this section, we report the accuracy of the β2AR models generated using contact restraints derived from the rhodopin template, topological restraints with distance measurements from the β2AR template, and ligand conformation and restraints obtained from the β2AR crystal structure. In a blind prediction scenario, the protocol would be run without any restraints derived from the high-resolution structure of β2AR, but this realistic case would incorporate a spectrum of errors inherent in the source of each set of restraints. The models described here represent the optimal case in which the protocol is run with restraint sets that are mostly correct. We report the accuracy of the models generated without any restraints from the β2AR template in the context of the discussion on how the three restraint sets affect model accuracy in a later section.
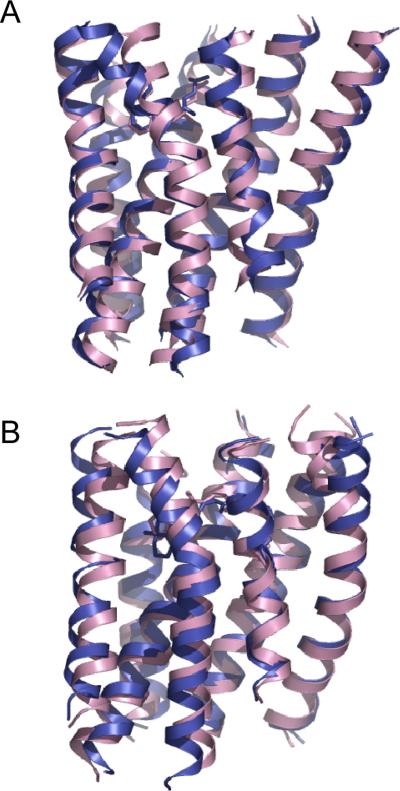
The predicted models are ~2.0 Å Cα RMSD for the TM helices and ~1.0 Å heavy atom RMSD for the ligand carazolol with respect to the β2AR crystal structure (PDB ID code 2RH1). We ran three independent modeling trials, and obtained representative models ranging from 2.0 to 2.9 Å Cα RMSD. The average model of the entire final ensemble of refined models tends to be more accurate and converged across repeated trials than the cluster average models; the ensemble average model is 2.1 ± 0.06 Å Cα RMSD from native in the three trials. These models show that the overall topology of the helical bundle is in good agreement with the crystal structure (Fig. 4A). The ligand is positioned accurately in the binding pocket and the binding geometry agrees well with the crystal structure. The alkylamine and alcohol moieties of carazolol make several hydrogen bonds with the D1133.32 and N3127.39 side chains. N7 of the carbazole heterocycle forms a hydrogen bond with the side chain hydroxyl of S2035.42. The “hydrophobic sandwich” around the C8-to-C13 ring of the carbazole heterocycle is formed by V1143.33 and F2906.52.
Figure 4.
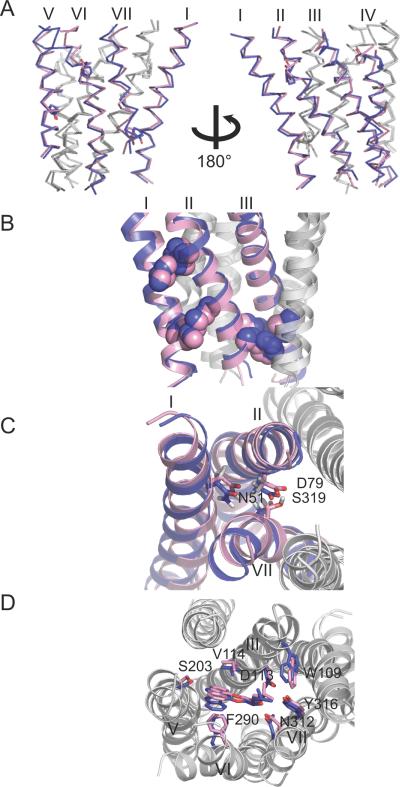
Accuracy of the predicted models for β2AR (A) and rhodopsin (B). The models are superimposed to their respective crystal structures (PDB ID codes 2RH1 for β2AR; 1U19 for rhodopsin). The model shown is the average structure of the entire refined ensemble. Model (magenta); crystal structure (blue).
Helix irregularities such as kinks, twists, bends, and bulges are modeled in near-native conformations. The average backbone RMSD from the crystal structure for each individual helix is < ~2.0 Å for all seven helices. Kinks induced by Pro residues are modeled well in helices II, V, VI, and VII. The models capture the varying degree of kink angles induced by Pro residues. The lack of kink in helix I due to the absence of a Pro residue is also modeled accurately. Other backbone features, such as the bulge in helix V, are successfully modeled. The bulge in helix V is formed by a backbone hydrogen bond between the carbonyl oxygen of I2055.44 and the amide hydrogen of V2105.49, preceding P2115.50.
Helix-helix packing interactions are modeled particularly well for helices I, II, III, and V. The average side chain RMSD from the crystal structure is < ~3.5 Å for helix pairs I–II, II–III, III–V, VII–I and ~3.5–4.5 Å for helix pairs III–IV, V–VI, VI–VII. The close-packing of residues V541.53-I551.54-A762.47, I471.46-V481.47-G832.54, and F712.42-V1263.45-I1273.46, and the interhelical hydrogen bonds formed between highly conserved residues, S742.45-T1233.42-W1584.50 and N511.50-D792.50-S3197.46, show that the side chain packing interactions between helices I, II, and III are captured accurately. Some of the correctly modeled interactions overlap with the conserved contact restraints imposed in the protocol. However, it is notable that near-native interactions are also modeled in the absence of explicit restraints. The current protocol is able to re-model ~60% of the native contacts in the final predicted models using just 45 contacts, or ~10% native contacts. The majority of these native contacts are not immediately adjacent to the restrained contacts; for the β2AR/pred/struct models, on average 11% are from contacts used in the restraints, 16% are within ±1 residue of those restrained contacts, and 17% are within ±2 residues.
Accuracy of rhodopsin models
The performance of the foldGPCR protocol is assessed on modeling the bovine rhodopsin structure using contact restraints analogous to those predicted for β2AR, topological restraints with distance measurements from the rhodopsin template, and ligand conformation and restraints obtained from the rhodopsin crystal structure. Similarly to the β2AR models reported in the previous section, these rhodopsin models represent the optimal case in which the protocol is run with restraint sets that are highly reliable.
The predicted models are ~2.5 Å Cα RMSD for the TM helices and ~2.0 Å heavy atom RMSD for the ligand 11-cis-retinal with respect to the rhodopsin crystal structure. In the three repeated trials, representative cluster average models range from 2.4 to 2.9 Å Cα RMSD, and the ensemble average model is 2.5 ± 0.06 Å Cα RMSD. The overall topology of the helical bundle agrees well with the crystal structure (Fig. 4B). Most of the ligand binding interactions are modeled accurately: the carboxylate oxygen atoms of E1133.28 form salt bridges with the Schiff base nitrogen atom of K2967.43; the indole ring of W2656.48 comes close to the C13-methyl group; the polyene chain and the β-ionone ring interact with side chains from helices III (A1173.32, T1183.33), V (M2075.42, F2125.47) and VI (Y2686.51, A2696.52). The ligand is positioned less deeply in the binding pocket, and residues at the bottom of the pocket, e.g. G1213.36 and F2616.44, are not proximal to the ligand. In general, residues lining the ligand-binding pocket are positioned accurately to within 1.0–1.5 Å heavy atom RMSD with respect to the crystal structures.
Helix backbone irregularities are modeled accurately for the Pro-induced kinks in helices VI and VII, and the bulge in helix V. Helix II is modeled as a straight helix and the kink induced by the Gly-Gly motif is not modeled; the kink angle in helix I is slightly too large, perhaps caused by the inaccuracy in helix II. The average backbone RMSD from the crystal structure for each individual helix is < ~2.0 Å for helices III, IV, V, VI, VII, and 2.3 Å for helices I and II.
Helix-helix packing interactions are modeled well for helices III, IV, V, VI, and VII. The average side chain RMSD from the crystal structure is < ~3.5 Å for helix pairs III–IV, III–V, VI–VII and ~3.5–4.5 Å for helix pairs I–II, II–III, V–VI, VII–I. The inaccuracy in helix packing interactions for helix II, especially on the extracellular side, is likely caused by the inaccuracy in the backbone structure of helix II. The “knobs-into-holes” packing of residues T1604.49-I1233.38-W1263.41-S1273.42-V1303.45 and I2195.54-L1253.40-L1283.43-V1293.44-A1323.47 shows that the interfaces between helices III, IV, and V are captured accurately.
The role of the restraints on model accuracy
The conformational search process in the assembly step of the protocol is guided by distance restraints that impose the helical bundle topology, inter-residue contacts, and receptor-ligand interactions. These restraints effectively reduce the conformational search space so that sufficiently accurate models can be generated with the current sampling protocol, and although more extensive sampling may improve the overall structure in the presence of the restraints, the nature of the underlying force field and representation of the environment will likely limit such models to Cα RMSD values of ~2 Å (based on unrestrained simulations of the native GPCR structures). The three restraint sets are hierarchical in nature and operate at different scales: the topological restraints are coarsest and impose a general topology, analogous to the use of restraints based on lower resolution cryo-EM structural data,21,58 the contact restraints are analogous to the NOE-like restraints used in NMR structure calculations and guide the packing interaction of secondary structural elements; the ligand-side chain interaction restraints are at the finest scale and determine the side chain orientation and rotamer state of ligand-binding residues. Because the three restraint sets contribute to the models at different scales, it is expected that the errors incorporated in them would have distinct effects on the accuracy of the predicted models. To better understand the role of each restraint set to model accuracy, we ran a series of control modeling trials for β2AR using different combinations of restraint sets with varying levels of errors (Table I). In applying this protocol to modeling other members of class A GPCRs, for which the reliability of the restraint sets may be unknown, it is important to assess the performance of the protocol given these errors. The resulting models are described below and organized around discussion of each restraint set.
Table I.
Accuracy of the β2AR models generated with different sets of restraints for each of the three types of restraints: topological, contacts, and ligand.
Topological restraints: β2AR (all distance measurements are from the β2AR crystal structure), Rhod (all distance measurements are from the rhodopsin crystal structure), Rhod* (same as Rhod, except the distance between TM helices I and VII on the extracellular side is from the β2AR structure), - (not used); Contact restraints: pred (predicted conserved contacts), - (not used), correct (true conserved contacts), all (unfiltered set of contacts from the template structure); Ligand restraints: struct (obtained from the β2AR crystal structure), mut (inferred from mutagenesis data), - (ligand is not modeled, ligand restraints are not used). Model accuracy is reported as the Cα RMSD (Å) of the ensemble or cluster average structure from the crystal structure (PDB ID codes 2RH1 for β2AR; 1U19 for rhodopsin) ± Cα RMS fluctuation (Å) around the average structure after each step of the protocol, with the number of models in the ensemble or cluster in brackets.
| Restraint Type | Models | Step 1: Assembly | Step 2: Refinement |
|---|---|---|---|
| Topology | Rhod/pred/struct | 3.4 ± 4.7 (199) | 2.9 ± 1.2 (100) |
| 3.1 ± 0.5 (36) | |||
| 3.2 ± 0.6 (30) | |||
| 3.1 ± 0.4 (24) | |||
| 3.0 ± 0.4 (10) | |||
|
|
|||
| Rhod*/pred/struct | 3.1 ± 4.9 (197) | 1.9 ± 1.1 (100) | |
| 1.9 ± 0.5 (66) | |||
| 2.5 ± 0.5 (27) | |||
| 2.3 ± 0.5 (7) | |||
|
|
|||
| -/pred/struct | 4.3 ± 6.9 (199) | 2.9 ± 1.3 (100) | |
| 3.2 ± 0.5 (32) | |||
| 3.2 ± 0.5 (31) | |||
| 3.0 ± 0.5 (29) | |||
| 2.8 ± 0.3 (8) | |||
|
| |||
| Contacts | β 2 AR/all/struct | 2.2 ± 2.4 (198) | 2.3 ± 0.7 (100) |
| 2.3 ± 0.5 (71) | |||
| 2.2 ± 0.5 (29) | |||
|
|
|||
| β 2 AR/correct/struct | 1.6 ± 3.4 (198) | 1.8 ± 0.9 (100) | |
| 1.9 ± 0.5 (35) | |||
| 2.1 ± 0.5 (34) | |||
| 1.8 ± 0.5 (31) | |||
|
|
|||
| β 2 AR/-/struct | 3.8 ± 6.1 (67) | 3.5 ± 1.0 (100) | |
| 3.5 ± 0.6 (87) | |||
| 3.8 ± 1.4 (13) | |||
|
|
|||
| β2AR/-/- | 4.8 ± 7.0 (87) | 4.1 ± 2.0 (100) | |
| 4.5 ± 0.6 (54) | |||
| 4.7 ± 0.6 (30) | |||
| 4.2 ± 0.4 (16) | |||
|
| |||
| Ligand | β 2 AR/pred/struct | 2.7 ± 4.9 (190) | 2.0 ± 1.0 (100) |
| 2.0 ± 0.6 (63) | |||
| 2.5 ± 0.8 (37) | |||
|
|
|||
| β 2 AR/pred/mut | 2.9 ± 4.9 (197) | 2.2 ± 1.5 (100) | |
| 2.9 ± 0.7 (42) | |||
| 2.3 ± 0.7 (37) | |||
| 2.6 ± 0.5 (21) | |||
|
|
|||
| β2AR/pred/- | 2.9 ± 5.3 (86) | 2.4 ± 1.2 (100) | |
| 2.6 ± 0.5 (42) | |||
| 2.5 ± 0.6 (32) | |||
| 2.6 ± 0.7 (26) | |||
The ligand interaction restraints guide the placement of the ligand within the binding pocket and the conformation of the side chains of the ligand-binding residues. Models generated with and without the ligand are comparable in backbone accuracy, ~2.0–2.5 Å Cα RMSD from native, but the average side chain heavy atom RMSD of residues within 4 Å of the ligand increases from 2.7 Å for models with the ligand to 4.1 Å for models without the ligand. Models generated with a smaller set of ligand interaction restraints (restraint sets β2AR/pred/mut), using those just inferred from mutagenesis data, are also comparable in backbone accuracy to models generated with restraint sets β2AR/pred/struct, but the average heavy atom RMSD of binding site residue side chains increases to 3.6 Å. While models that agree well with experimental data may be generated, these models show that the side chain accuracy of the binding site residues is quite sensitive to the ligand restraints used in the protocol, suggesting that an erroneous set of restraints may lead to models with inaccurate ligand placement. The biochemical data used to infer ligand restraints thus needs to be carefully selected for those showing direct interactions between the receptor residues and the ligand. Experiments such as affinity labeling and photo-crosslinking or functional complementation studies would be more informative than site-directed mutagenesis studies.60
The topological restraints impose the topology of the overall helical bundle without restricting on the rotational orientations of the helices. Models built with topological restraints alone (restraint sets β2AR/−/−) are ~4.0–4.5 Å Cα RMSD from native, and have <20% native contacts. Models generated without the topological restraints (restraint sets -/pred/struct) are about ~3.0 Å Cα RMSD from native. In these models, the tilt angles of helices tend to be less accurate. Models generated with topological distance measurements from the rhodopsin template (restraint sets Rhod/pred/struct) are ~3.0 Å Cα RMSD from native, and are worse than models generated with restraint sets β2AR/pred/struct. These models are ~2.5 Å Cα RMSD from the rhodopsin structure, and hence are closer to the rhodopsin template than to β2AR. Although the topology of the rhodopsin and the β2AR structures are similar, and the topological distance measurements from the two structures are mostly within ~2.5 Å of one another (Supplementary Table III), there is one distance between TM helices I and VII on the extracellular side that differs as much as ~5 Å between the two structures due to the lack of Pro-induced kink in helix I of β2AR. Substituting this one distance measurement for the native distance (restraint sets Rhod*/pred/struct) results in models that are ~2.0 Å Cα RMSD from native, and comparable in accuracy to the β2AR/pred/struct models. These models show that the topological distance measurements need to be accurate within 5 Å, and any shifts in helix position of >5 Å would be difficult to model accurately. Receptors lacking a Pro residue in helix I are likely to be better modeled using the distance measurements from the β2AR template than the rhodopsin template.
The contact restraints guide the helix-helix packing interactions. Models generated without contact restraints (restraint sets β2AR/-/struct) are ~3.5 Å Cα RMSD from native. In these models, helix packing interactions are not modeled very accurately and the fraction of native contacts is ~35%, significantly lower than ~60% native contacts achieved in the β2AR/pred/struct models. The contact restraints are derived from a subset of contacts in the rhodopsin template that are predicted to be structurally conserved. Using the entire set of contacts from rhodopsin (restraint sets β2AR/all/struct) results in models that are overall slightly less accurate than the β2AR/pred/struct models, especially in the tilt angle of helix I, because contacts with respect to the extracellular side of helix I are not structurally conserved between β2AR and rhodopsin. The distribution and accuracy of the predicted conserved contacts may affect how well different regions of the helical bundle are modeled. Although the accuracy of predicted contacts is expected to be high in general,59 two of the 45 predicted contacts for β2AR are clear false positives and both involve residues in helix IV. Some of the contacts between helix IV and other helices in rhodopsin are not formed in β2AR, because helix IV in β2AR is shifted farther away from the binding site compared to rhodopsin. These false positive contact restraints may have led to the less accurate side chain packing interactions between helices III and IV in the β2AR/pred/struct models. Using the true conserved contacts (restraint sets β2AR/correct/struct) results in models that are slightly more accurate than the β2AR/pred/struct models (Table III). The majority of the predicted conserved contacts are on the cytoplasmic side of the helical bundle owing to the higher level of sequence conservation towards this half of the helical bundle.6 The contacts formed in this region may undergo significant rearrangements upon receptor activation,61 hence the template structure from which the contact restraints are derived likely determines the conformational state of model.
Table III.
Independent contributions to model accuracy of the use of conserved contact restraints in the assembly step, and the GB implicit membrane model in the refinement step.
Model accuracy for the β2AR models is reported as the Cα RMSD (Å) for all seven TM helices as well as partially (all seven TM helices / helices II, III, V / all helices except helix VI). The β2AR homology model based on the rhodopsin template was generated by Modeller 31.
| Step 1: Rough model generation | Step 2: Refinement |
|---|---|
| FoldGPCR assembly | GB implicit membrane |
| β 2 AR/pred/struct | 2.0 ± 1.0 (100) / 1.2 / 1.8 |
| 2.7 ± 4.9 (190) / 2.4 / 2.6 | 2.0 ± 0.6 (63) / 1.3 / 1.7 |
| 2.5 ± 0.8 (37) / 1.4 / 2.2 | |
| β 2 AR/correct/struct | GB implicit membrane |
| 1.6 ± 3.4 (198)/ 1.3/1.4 | 1.8 ± 0.9 (100) / 1.4 / 1.4 |
| 1.9 ± 0.5 (35) / 1.6 / 1.7 | |
| 2.1 ± 0.5 (34) / 1.6 / 1.7 | |
| 1.8 ± 0.5 (31) / 1.3 / 1.4 | |
| β 2 AR/all/struct | GB implicit membrane |
| 2.2 ± 2.4 (198) / 1.6 / 2.1 | 2.3 ± 0.7 (100) / 1.5 / 2.1 |
| 2.3 ± 0.5 (71) / 1.5 / 2.2 | |
| 2.2 ± 0.5 (29) / 1.5 / 2.1 | |
| β 2 AR/pred/struct | vacuum |
| 2.7 ± 4.9 (190) / 2.4 / 2.6 | 3.0 ± 1.7 (100) / 2.2 / 2.8 |
| 3.3 ± 0.8 (50) / 2.8 / 3.1 | |
| 3.0 ± 0.7 (37) / 2.0 / 2.8 | |
| 3.6 ± 0.6 (13) / 2.9 / 3.6 | |
|
| |
| Rhodopsin-based homology model | GB implicit membrane |
| 2.1 / 2.2 / 2.2 | 2.0 ± 1.5 (100) / 2.0 / 2.0 |
| 2.7 ± 0.4 (34) / 2.9 / 2.7 | |
| 2.3 ± 0.4 (23) / 2.1 / 2.2 | |
| 2.8 ± 0.4 (22) / 2.5 / 2.8 | |
| 2.1 ± 0.4 (21) / 1.2 / 2.1 | |
Highlights of the foldGPCR protocol in comparison to other approaches
We find that helices I, II, III, and V in our β2AR models are modeled particularly well, as indicated by an average Cα RMSD of 1.4 Å. The lack of kink in helix I, and the bend angles in helices II, III, V are captured accurately. The packing interactions for helices II, III, V are modeled much closer to the β2AR structure than to the rhodopsin template, with an average Cα RMSD of 1.4 Å to β2AR and 1.8 Å to rhodopsin (Fig. 5), and improved from the rhodopsin template which differs by 2.1 Å Cα RMSD to the β2AR structure in helices II, III, V.
Figure 5.
Modeling the structural divergence between rhodopsin and β2AR for helices II, III, and V. The β2AR model (magenta) generated by the restraint sets β2AR/pred/struct (see Table I for annotation) is superimposed to the rhodopsin (green) and the β2AR (blue) crystal structures using backbone atoms of helices II, III, and V.
In comparison to other structure prediction methods that have previously been applied to model β2AR,23,26,62 our current protocol appears to more accurately model the backbone divergence between the target and the best available template (Table II). TASSER models (C-score: 1.4, template: 1F88_A) are ~1.0 Å Cα RMSD to rhodopsin, and do not recapitulate the structural features unique to β2AR: the bend angles in helices I, III, V, and the packing interactions for helices II, III, V are not improved much from the rhodopsin template. Although the TASSER method is reported to often refine structures closer to native,63 the improvement relative to the template for the β2AR models seems rather small. The de novo models generated by the MembStruk method and the Mosberg distance geometry approach are in general further away from both β2AR and rhodopsin. The MembStruk model captures the lack of kink in helix I, but the backbone of helix III is distorted and the rotational orientation and the vertical alignment of helix V are inaccurate. The Mosberg model shows some trends towards the native β2AR structure, in that the bend angle in helix III is native-like, but helix V is positioned too close to the center of the receptor.
Table II.
Comparison of model accuracy for the β2AR models generated by foldGPCR using the restraint sets β2AR/pred/struct, and other methods 23,26,62.
Model accuracy is reported as the Cα RMSDs (Å) for all seven TM helices and for TM helices II, III, V from the β2AR crystal structure (PDB ID code 2RH1). The ensemble average models from the three trials are shown for the foldGPCR models. For the TASSER models, those with RMSD to native of below 6.5 Å are shown.
| Methods | Cα RMSD all helices / helices II, III, V |
|---|---|
| FoldGPCR β2AR/pred/struct |
2.0 / 1.2 |
| 2.1 / 1.4 | |
| 2.1 / 1.7 | |
|
| |
| TASSER | 2.1 / 1.9 |
| 2.1 / 1.7 | |
| 2.1 / 1.9 | |
|
| |
| Mosberg de novo | 3.4 / 2.5 |
|
| |
| MembStruk | 4.0 / 4.4 |
The foldGPCR protocol is designed to optimize interactions within and between the helices using tertiary restraints combined with an improved physics-based force field, and an efficient hierarchical sampling strategy. To investigate the independent contributions to model accuracy of the use of conserved contact restraints in the assembly step and the GB implicit membrane model in the refinement step, we ran modeling trials with i) a homology model generated by Modeller31 instead of the foldGPCR assembled model, and ii) refinement in vacuum instead of the GB implicit membrane model (Table III). The homology model refined with GB/REX results in models of ~2.0 Å Cα RMSD and ~60% native contacts, and are comparable in overall accuracy to the foldGPCR β2AR/pred/struct models, although helices II, III, and V are more accurate in the foldGPCR models. Models refined in vacuum are ~3.0 Å Cα RMSD, and are clearly worse than models refined with the GB implicit membrane model. These trials demonstrate that the assembly step using the conserved contact restraints and the refinement step using the GB implicit membrane model both contribute to model accuracy.
The protocol can be used to predict divergent structural features for GPCRs of unknown structure. We applied the protocol to model the human vasopressin V1a receptor (Swiss-Prot ID: P37288), human κ-type opioid receptor (Swiss-Prot ID: P41145), and human proton-sensing receptor GPR4 (Swiss-Prot ID: P46093). These receptors belong to the β-, γ-, and δ-groups of class A GPCRs, respectively; and they may be structurally more divergent from the templates than any receptors in the α-group. The superposition of the predicted models to their respective template structures shows that there may be numerous differences in the way the TM helices are packed in these receptors (Supplementary Figure 1). For instance, helix II in the vasopressin V1a receptor model lacks a kink despite the P1072.60 residue, and allows the extracellular segment of helix I, that is known to participate in agonist-binding,64 to be positioned closer to the binding site. Similarly, the extracellular segment of helix II in the κ-type opioid receptor model is slightly rotated and positions V1182.63, a residue known to participate in agonist-binding,65 closer to the binding pocket. Helix V in the proton-sensing GPR4 receptor model does not have a bulge at the helix center, and consequently the extracellular segment of helix V is slightly rotated.
Taken together, the main advantage of the foldGPCR protocol is that it has the ability to accurately model the structural divergence between the template and target, and capture some of the structural features unique to the target. A caveat is in the case of modeling receptors that are close homologs to the template structure, for instance in modeling β1AR based on β2AR. The structures of turkey β1AR and human β2AR differ only by 0.7 Å Cα RMSD in the TM region.18 The foldGPCR models for the human β1AR based on the β2AR structure is at best ~1.7 Å Cα RMSD from the turkey β1AR structure, and these models would be expected to be worse than homology models based on β2AR. Nevertheless, for template-target pairs with significant structural divergence such as β2AR and rhodopsin, the foldGPCR protocol seems to be able to generate better models than the homology modeling approach.
Another advantage of the foldGPCR protocol is that it is very flexible in incorporating additional or different sets of distance restraints. The stepwise assembly and GB/REX refinement approach is effective in generating models of reasonable accuracy given certain template- and experimentally-derived distance restraints. The protocol provides a framework to generate receptor-ligand complex models for GPCRs using any arbitrary set of sparse distance restraints that define the topology of the helical bundle, inter-residue contacts, and receptor-ligand interactions. Restraint sets derived from another template, multiple templates, or other biochemical or biophysical experiments may be incorporated or substituted given that there is a sufficient number of restraints to yield converged models. For example, one interesting application would be to generate an agonist-bound receptor model using the recent crystal structures of opsin, which show structural features of an active state,16,17 together with distance measurements suggested from EPR experiments.66
CONCLUSIONS
We have developed a novel modeling protocol (foldGPCR) for predicting the structure of the class A GPCR 7-TM helical bundle in complex with a ligand. The protocol aims to accurately model the structural divergence between the template and target in the TM helices, by using tertiary restraints and an all-atom implicit membrane GB force field to yield conformations capturing structural features unique to the target. It is implemented by a stepwise hierarchical approach, in which the TM helical bundle and the ligand are assembled by simulated annealing trials in the first step, and then the receptor-ligand complex is refined with replica exchange sampling in the second step. The protocol is applied to modeling the human β2AR bound to carazolol using contact restraints derived from the bovine rhodopsin template, and models with ~2.0 Å Cα RMSD from the β2AR crystal structure are obtained. Although the overall accuracy is comparable to homology models obtained with limited refinement, we find that some helical shifts can be modeled much closer to the β2AR structure than the rhodopsin template. As an increasing number of experimentally determined structures become available for the GPCR superfamily, template-based modeling methods with limited ability in refinement will become adequate for a larger proportion of the family. The de novo modeling protocol presented here is expected to be most useful in providing reliable models for those members of the family that require significant refinement from the best available template. Modeling the loops accurately and predicting the agonist-induced conformational changes remain a formidable challenge in GPCR structure prediction, and will require more experimental structures before reliable models can be built.
Supplementary Material
ACKNOWLEDGEMENTS
M.M. thanks Drs. Roger Armen and Jennifer Knight for help in developing the parameters for the ligands, and Drs. Wonpil Im and David Bostick for useful feedback in the early stages of this research. The authors are grateful to the Stevens group for helpful discussions. The authors thank Dr. Nagarajan Vaidehi for kindly sharing the MembStruk model. This research was supported by the NIH and Novartis through grants RR012255 and SFP_1562, respectively.
REFERENCES
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3(9):639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63(6):1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 3.Gloriam DE, Fredriksson R, Schioth HB. The G protein-coupled receptor subset of the rat genome. BMC Genomics. 2007;8:338. doi: 10.1186/1471-2164-8-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyndall JD, Sandilya R. GPCR agonists and antagonists in the clinic. Med Chem. 2005;1(4):405–421. doi: 10.2174/1573406054368675. [DOI] [PubMed] [Google Scholar]
- 5.Lefkowitz RJ. The superfamily of heptahelical receptors. Nat Cell Biol. 2000;2(7):E133–136. doi: 10.1038/35017152. [DOI] [PubMed] [Google Scholar]
- 6.Mirzadegan T, Benko G, Filipek S, Palczewski K. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry. 2003;42(10):2759–2767. doi: 10.1021/bi027224+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surgand JS, Rodrigo J, Kellenberger E, Rognan D. A chemogenomic analysis of the transmembrane binding cavity of human G-protein-coupled receptors. Proteins. 2006;62(2):509–538. doi: 10.1002/prot.20768. [DOI] [PubMed] [Google Scholar]
- 8.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 9.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342(2):571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343(5):1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 11.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103(44):16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318(5854):1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 14.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. A Specific Cholesterol Binding Site Is Established by the 2.8 A Structure of the Human beta(2)-Adrenergic Receptor. Structure. 2008;16(6):897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453(7193):363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 16.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454(7201):183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 17.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 18.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454(7203):486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322(5905):1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259(3):393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 21.Baldwin JM, Schertler GF, Unger VM. An alpha-carbon template for the transmembrane helices in the rhodopsin family of G-protein-coupled receptors. J Mol Biol. 1997;272(1):144–164. doi: 10.1006/jmbi.1997.1240. [DOI] [PubMed] [Google Scholar]
- 22.Herzyk P, Hubbard RE. Combined biophysical and biochemical information confirms arrangement of transmembrane helices visible from the three-dimensional map of frog rhodopsin. J Mol Biol. 1998;281(4):741–754. doi: 10.1006/jmbi.1998.1981. [DOI] [PubMed] [Google Scholar]
- 23.Pogozheva ID, Lomize AL, Mosberg HI. The transmembrane 7-alpha-bundle of rhodopsin: distance geometry calculations with hydrogen bonding constraints. Biophys J. 1997;72(5):1963–1985. doi: 10.1016/S0006-3495(97)78842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomize AL, Pogozheva ID, Mosberg HI. Structural organization of G-protein-coupled receptors. J Comput Aided Mol Des. 1999;13(4):325–353. doi: 10.1023/a:1008050821744. [DOI] [PubMed] [Google Scholar]
- 25.Bissantz C, Bernard P, Hibert M, Rognan D. Protein-based virtual screening of chemical databases. II. Are homology models of G-Protein Coupled Receptors suitable targets? Proteins. 2003;50(1):5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006;2(2):e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaidehi N, Floriano WB, Trabanino R, Hall SE, Freddolino P, Choi EJ, Zamanakos G, Goddard WA., III Prediction of structure and function of G protein-coupled receptors. Proc Natl Acad Sci U S A. 2002;99(20):12622–12627. doi: 10.1073/pnas.122357199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shacham S, Marantz Y, Bar-Haim S, Kalid O, Warshaviak D, Avisar N, Inbal B, Heifetz A, Fichman M, Topf M, Naor Z, Noiman S, Becker OM. PREDICT modeling and in-silico screening for G-protein coupled receptors. Proteins. 2004;57(1):51–86. doi: 10.1002/prot.20195. [DOI] [PubMed] [Google Scholar]
- 29.Tress M, Ezkurdia I, Grana O, Lopez G, Valencia A. Assessment of predictions submitted for the CASP6 comparative modeling category. Proteins. 2005;61(Suppl 7):27–45. doi: 10.1002/prot.20720. [DOI] [PubMed] [Google Scholar]
- 30.Archer E, Maigret B, Escrieut C, Pradayrol L, Fourmy D. Rhodopsin crystal: new template yielding realistic models of G-protein-coupled receptors? Trends Pharmacol Sci. 2003;24(1):36–40. doi: 10.1016/s0165-6147(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 31.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 32.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(Suppl 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 33.Im W, Feig M, Brooks CL., III An implicit membrane generalized born theory for the study of structure, stability, and interactions of membrane proteins. Biophys J. 2003;85(5):2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bu L, Im W, Brooks CL., III Membrane assembly of simple helix homo-oligomers studied via molecular dynamics simulations. Biophys J. 2007;92(3):854–863. doi: 10.1529/biophysj.106.095216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bu L, Michino M, Wolf RM, Brooks CL., III Improved model building and assessment of the Calcium-sensing receptor transmembrane domain. Proteins. 2008;71(1):215–226. doi: 10.1002/prot.21685. [DOI] [PubMed] [Google Scholar]
- 36.Bu L, Brooks CL., III De novo prediction of the structures of M. tuberculosis membrane proteins. J Am Chem Soc. 2008;130(16):5384–5385. doi: 10.1021/ja710213p. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Im W, Brooks CL., III Refinement of NMR structures using implicit solvent and advanced sampling techniques. J Am Chem Soc. 2004;126(49):16038–16047. doi: 10.1021/ja047624f. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Brooks CL., III Can molecular dynamics simulations provide high-resolution refinement of protein structure? Proteins. 2007;67(4):922–930. doi: 10.1002/prot.21345. [DOI] [PubMed] [Google Scholar]
- 39.Chopra G, Summa CM, Levitt M. Solvent dramatically affects protein structure refinement. Proc Natl Acad Sci U S A. 2008;105(51):20239–20244. doi: 10.1073/pnas.0810818105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobilka B, Schertler GF. New G-protein-coupled receptor crystal structures: insights and limitations. Trends Pharmacol Sci. 2008;29(2):79–83. doi: 10.1016/j.tips.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci U S A. 2004;101(20):7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohl CA, Strauss CE, Chivian D, Baker D. Modeling structurally variable regions in homologous proteins with rosetta. Proteins. 2004;55(3):656–677. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]
- 43.Michino M, Abola E, Brooks CL, 3rd, Dixon JS, Moult J, Stevens RC. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat Rev Drug Discov. 2009;8(6):455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy minimization and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 45.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10(1):59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 47.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 48.Momany F, Rone R. Validation of the general purpose QUANTA ®3.2/CHARMm® force field. J Comput Chem. 1992;13(7):888–900. [Google Scholar]
- 49.Roche O, Kiyama R, Brooks CL., III Ligand-protein database: linking protein-ligand complex structures to binding data. J Med Chem. 2001;44(22):3592–3598. doi: 10.1021/jm000467k. [DOI] [PubMed] [Google Scholar]
- 50.Zagrovic B, Pande VS. How does averaging affect protein structure comparison on the ensemble level? Biophys J. 2004;87(4):2240–2246. doi: 10.1529/biophysj.104.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 52.Mitsutake A, Sugita Y, Okamoto Y. Generalized-ensemble algorithms for molecular simulations of biopolymers. Biopolymers. 2001;60(2):96–123. doi: 10.1002/1097-0282(2001)60:2<96::AID-BIP1007>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 53.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 54.Mackerell AD, Jr., Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 55.Im W, Lee MS, Brooks CL., III Generalized born model with a simple smoothing function. J Comput Chem. 2003;24(14):1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 56.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22(5):623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 57.Feig M, Karanicolas J, Brooks CL., III MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph Model. 2004;22(5):377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Fleishman SJ, Harrington S, Friesner RA, Honig B, Ben-Tal N. An automatic method for predicting transmembrane protein structures using cryo-EM and evolutionary data. Biophys J. 2004;87(5):3448–3459. doi: 10.1529/biophysj.104.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michino M, Brooks CL., III Predicting structurally conserved contacts for homologous proteins using sequence conservation filters. Proteins. 2009;77(2):448–453. doi: 10.1002/prot.22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi L, Javitch JA. The binding site of aminergic G protein-coupled receptors: the transmembrane segments and second extracellular loop. Annu Rev Pharmacol Toxicol. 2002;42:437–467. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- 61.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768(4):794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhattacharya S, Hall SE, Li H, Vaidehi N. Ligand-stabilized conformational states of human beta(2) adrenergic receptor: insight into G-protein-coupled receptor activation. Biophys J. 2008;94(6):2027–2042. doi: 10.1529/biophysj.107.117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Skolnick J. Tertiary structure predictions on a comprehensive benchmark of medium to large size proteins. Biophys J. 2004;87(4):2647–2655. doi: 10.1529/biophysj.104.045385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hawtin SR, Wesley VJ, Simms J, Argent CC, Latif K, Wheatley M. The N-terminal juxtamembrane segment of the V1a vasopressin receptor provides two independent epitopes required for high-affinity agonist binding and signaling. Mol Endocrinol. 2005;19(11):2871–2881. doi: 10.1210/me.2005-0148. [DOI] [PubMed] [Google Scholar]
- 65.Vortherms TA, Mosier PD, Westkaemper RB, Roth BL. Differential helical orientations among related G protein-coupled receptors provide a novel mechanism for selectivity. Studies with salvinorin A and the kappa-opioid receptor. J Biol Chem. 2007;282(5):3146–3156. doi: 10.1074/jbc.M609264200. [DOI] [PubMed] [Google Scholar]
- 66.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274(5288):768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.