Abstract
T cell recognition of autoantigens is critical to progressive immune-mediated destruction of islet cells, which leads to autoimmune diabetes. We identified a naturally presented autoantigen from the human islet antigen glutamic acid decarboxylase, 65-kDa isoform (GAD65), by using a combination of chromatography and mass spectrometry of peptides bound by the type I diabetes (insulin-dependent diabetes mellitus, IDDM)-associated HLA-DR4 molecule. Peptides encompassing this epitope-stimulated GAD65-specific T cells from diabetic patients and a DR4-positive individual at high risk for developing IDDM. T cell responses were antagonized by altered peptide ligands containing single amino acid modifications. This direct identification and manipulation of GAD65 epitope recognition provides an approach toward dissection of the complex CD4+ T cell response in IDDM.
Type I diabetes (insulin-dependent diabetes mellitus, IDDM), like many autoimmune diseases, exhibits exquisite target organ specificity. IDDM is characterized by immune-mediated destruction of beta cells in the pancreatic islet, coincident with the sparing of neighboring alpha and delta cells. The precise target-cell specificity in this disease implies the existence of antigenic self proteins derived from beta cells that are recognized by autoimmune T lymphocytes. Extensive analysis of serum antibodies in patients with IDDM has documented several self proteins that are candidates for this role (1, 2). The 65-kDa isoform of human glutamic acid decarboxylase (hGAD65) is expressed in pancreatic beta cells at high levels, and antibodies to hGAD65 are present in up to 70% of newly diagnosed diabetics. These antibodies are also often present for several years before the development of clinical diabetes, providing a useful serum marker for prediction of disease onset (1, 3, 4). Recently, it has been shown that the suppression of GAD expression in nonobese diabetic (NOD) mice prevents autoimmune diabetes (5), directly implicating GAD as a likely participant in IDDM progression.
Studies of T cell reactivity to autoantigens in diabetics have confirmed the immunogenicity of hGAD65 with reports of both CD4+ and CD8+ T cell responses (6–14). Approximately 70% of Caucasoid diabetics express the DRB1*0401, *0404, and *0405 MHC class II alleles. These DR4+ alleles are in linkage disequilibrium with the DQB1*0302 gene, the HLA-DQ marker most highly associated with IDDM (15). T cell responses to the hGAD65 proteins that are restricted by HLA-DR molecules predominate in patients with these HLA disease-susceptibility haplotypes, and previous studies using overlapping synthetic peptides identified approximately 10 peptides that were capable of efficient binding to DR4 molecules. Indeed, three of these candidate autoantigen epitopes, corresponding to residues 115–127, 274–286, and 554–566 of hGAD65, were immunogenic when used to immunize mice transgenic for HLA-DR4 (16). A separate study, also using DR4-transgenic mice, found that these same three epitopes also were included in immunodominant regions (116–130, 271–285, and 551–565) when the GAD65 protein, rather than the peptides, was used as the immunogen (17). We now report the direct identification of one of these epitopes by liquid chromatography–electrospray ionization mass spectrometric (LC/MS) analysis of peptides bound by DR4 molecules on human antigen-presenting cells (APCs). Alterations in this peptide epitope result in a potent antagonist that interferes with antigen-specific human T cell clones responding to this diabetes-associated autoantigen.
Materials and Methods
Patient Selection.
Patients, between the ages of 14 and 25 who were recently diagnosed with IDDM and who were being treated for diabetes at the Virginia Mason Medical Center Section of Endocrinology, were asked to participate in this study. All participating patients were typed for HLA class II DR and DQ alleles, and serum was tested for autoantibodies to hGAD65, insulin, and IA2, by using standard protocols described previously. Patients who were DR4-positive and who had autoantibodies to GAD65 were selected for T cell analysis. A nondiabetic individual, initially identified as positive for autoantibodies to ICA, GAD65, and IA2 in an ongoing serum-screening project, HLA-DR4 [HLA-DRB1*0404, *0405; HLA-DQB1*0302, *0302], also was studied. By using our criteria of a high-risk HLA genotype and two or more antiislet autoantibodies, we define this individual as at-risk for IDDM. Intravenous glucose tolerance tests were performed at the same time the T cell studies were initiated, and the test results were within normal limits. This patient continues to be followed in a prediabetes screening program at the Virginia Mason Research Center.
In Vitro T Cell Assays.
For assays, 105 thawed and irradiated DRB1*0404, DRB1*0405 or control (non DR4) peripheral blood lymphocytes (PBL) in a volume of 100 μl were added to wells of 96-well V-bottom plates containing peptide or medium and were allowed to incubate for 2–3 h, at which time 4 × 104 T cells were added for a total volume of 200–250 μl. At 20–24 h of coculture, supernatants were harvested for cytokine determination and the wells were replenished with fresh medium. At 48 h, wells were radiolabeled with 1 μCi of [3H]thymidine and cultured for an additional 18 h. The plates were harvested on a Tomtec (Orange, CT) manual mach III harvester and cpm were determined by liquid spectroscopy on a Wallac (Gaithersburg, MD) Microbeta LSC.
Traditional sandwich ELISAs were performed to test the supernatants for human IFN-γ, by using matched antibody sets obtained from Endogen (Cambridge, MA). The plates were read at 405 nm on a Microplate reader (Bio-Tek, Burlington, VT). The concentration of cytokine was estimated from standard curves by using linear regression.
Derivation of Human T Cell Clones.
PBL were primed for 10 days with a 10 μg/ml pool of peptides spanning the C terminus of human GAD65. At day 10 of culture, T cells were plated at 0.3, 3, and 10 cells per well together with 104 irradiated, autologous, GAD-pulsed PBL in 10 μl of IL2- and IL7-supplemented conditioned medium in sterile Terasaki plates. Conditioned medium consisted of RPMI medium 1640 supplemented with 2 mM l-glutamine, 100 μg/ml penicillin/streptomycin, 1 mM sodium pyruvate, and 15% (vol/vol) pooled human serum obtained from 20–25 healthy, nontransfused male donors. After 10–14 days of incubation in a 37° C, 5% (vol/vol) CO2 atmosphere, wells having positive growth were transferred to 96-well flat-bottom plates containing 105 irradiated, autologous, GAD-(555–567)-pulsed PBL, 10 units/ml IL2 (Intergen, Purchase, NY), 10 ng/ml IL7 (PharMingen), and 0.4 μg/ml phytohemagglutinin (Sigma). After another 14 days of culture, all wells were assayed for specificity to GAD-(555–567) by measuring both 3H uptake and IFN-γ production. Wells of interest were further expanded with autologous PBL plus supplemented conditioned medium as described above. The restriction elements were determined by testing an APC panel of BLS-1 cells transfected with HLA class II genes representative of the donor's DR type—i.e., BLS DRB1*0404, *0405, and DRB4*0101. All T cell clones in this study were found to recognize GAD-(555–567) in the context of both DRB1*0404 and *0405 gene products and not DRB4-encoded molecules.
Peptide Response and Antagonism Assays.
For prepulse assays, APCs were preincubated for 2–3 h with suboptimal concentrations of the agonist peptide, washed 3 times, then cultured with the antagonist peptides as indicated in Fig. 5. Peptides used for T cell stimulation and MHC-binding studies were synthesized with an Applied Biosystems 432 Peptide Synthesizer and competition-binding assays were performed as described (16).
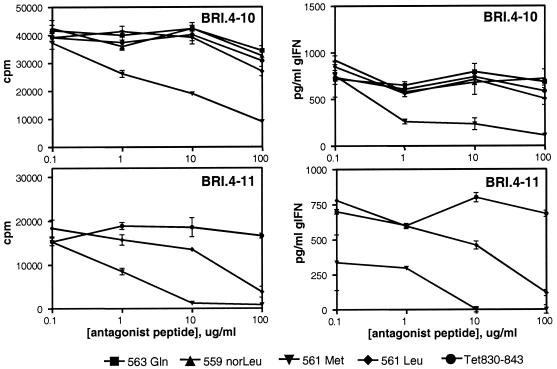
Figure 5.
T cell response to GAD-(555–567) in the presence of altered peptide ligands. Proliferative responses (Left) and INF-γ release (Right) are shown for T cell clones BRI.4–10 (Upper) and BRI.4–11 (Lower).
MHC Class II-Restricted Peptide Isolation.
The GAD65-transfected PREISS cell line (HLA-DRB1*0401) was grown up to 6 × 108 cells, washed twice in PBS, snap frozen in liquid nitrogen, and held at −80°C. Cells were lysed in 5 ml of lysis buffer [20 mM Tris⋅HCl (pH 8.0)/150 mM NaCl/1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)/5 μg/ml aprotinin/10 μg/ml leupeptin/10 μg/ml pepstatin A/5 mM EDTA/0.04% sodium azide/1 mM PMSF] for 1 h at 4°C. Cell lysate was centrifuged in an Eppendorf tube at 16,000 × g for 30 min at 4°C. The supernatant was transferred to a clean 15-ml conical tube and 100 μl of recombinant protein A Sepharose beads (Amersham Pharmacia) was added, followed by tube rotation for 4 h at 4°C. After removal of the beads, the supernatant was incubated overnight at 4°C with 100 μl of recombinant protein A Sepharose beads to which 2 mg of LB3.1 antibody had been bound. The beads were washed subsequently two times in lysis buffer, four times in 20 mM Tris⋅HCl (pH 8.0)/150 mM NaCl, two times in 20 mM Tris (pH 8.0)/1 M NaCl, and three times in 20 mM Tris⋅HCl (pH 8.0). Peptides were eluted in acid and separated from beads, antibody, and class II MHC molecules by passage through a 10,000-Da cutoff ultrafiltration unit (Millipore).
Liquid Chromatography/Tandem MS (LC/MS/MS) Parameters on the LCQ Ion Trap Mass Spectrometer.
Approximately 3 × 107 cell equivalents (corresponding to 5% of the sample) was loaded onto a C18 microcapillary column and gradient eluted directly into a Finnigan LCQ ion trap mass spectrometer (ThermoQuest, San Jose, CA). Nanoflow HPLC columns with integrated electrospray emitter tips were constructed as described (18). The LCQ ion trap mass spectrometer was operated in data-dependent mode (19), in which an initial MS scan recorded mass to charge (m/z) ratios of ions over the range 300–2,000. Then the five most abundant ions were selected for subsequent collision-activated dissociation to yield sequence-specific peptide fragment ions. The MS/MS spectra were searched by using the sequest algorithm (20) against both the single-protein database of interest and the nonredundant database maintained at the National Center for Biotechnology Information.
LC/MS Parameters on the Fourier-Transform Mass Spectrometer.
Approximately 1 × 107 cell equivalents were loaded onto a C18 column and gradient eluted directly into a home-built Fourier-transform ion cyclotron resonance mass spectrometer (18). HPLC gradient conditions were 0–70% acetonitrile in 0.1% acetic acid in 40 min. Full-scan mass spectra (300 ≤ m/z ≤ 5,000) were collected at approximately one scan per second, with typically 5,000–10,000 mass resolving power. The LC/MS data were searched manually for masses corresponding to the +2, +3, or +4 charge states of the possible nested set species.
Selected Reaction Monitoring Parameters on the TSQ7000 Triple Quadrupole Mass Spectrometer.
Approximately 3 × 107 cell equivalents were loaded onto a C18 microcapillary column and gradient eluted directly into a Finnigan TSQ7000 triple quadrupole mass spectrometer (ThermoQuest). HPLC gradient conditions were 0–70% acetonitrile in 0.1% acetic acid in 40 min. The +4 charge state of the peptide hGAD65-(552–572) (m/z 605.0) was selected in quadrupole 1 for subsequent collision-activated dissociation in quadrupole 2; the +3 charge states of the b19 (m/z 725.0), the b20 (m/z 762.5), and the b14 fragment ions (m/z 540.5) were monitored sequentially in quadrupole 3. The y7 fragment ion (m/z 370.0) from dissociation of the +3 charge state of hGAD65-(552–570) (m/z 650) was used as a reference. Retention times of fragment ions from hGAD65-(552–572) were recorded relative to this reference ion. In the coelution study, roughly 100 amol of the synthetic peptide hGAD65-(552–572) was spiked into a subsequent run of approximately 2.2 × 107 cell equivalents.
Peptide Synthesis.
The synthetic hGAD65 peptides used for mass spectrometric analyses were synthesized by standard fluorenylmethoxycarbonyl chemistry by using a Gilson model AMS422 peptide synthesizer.
Results and Discussion
Recognition of hGAD65 epitopes in the context of the DR4 restriction element was studied by using T cells from eight patients who were HLA-DR4-positive with recently onset IDDM (less than 15 months postdiagnosis). One of the most immunodominant of these epitopes corresponded to a region near the C terminus of hGAD65, which was represented in our antigen panel by peptides encompassing residues 553–585 (Table 1). Five of eight HLA-DR4 patients tested showed robust IFN-γ output after stimulation with peptide-pulsed autologous APCs. The specificity of this response was verified by lack of stimulation with other GAD65 peptides in the same experiment for each patient. Also shown in Table 1 is the T cell response for patient no. 6211, a nondiabetic HLA-DR4 individual at risk for IDDM, who also had strong IFN-γ cytokine responses to peptides from this GAD65 region. The observed variability in response between patients is concordant with previous similar studies and likely represents both genetic heterogeneity and the natural history of disease.
Table 1.
T cell reactivity to peptides from GAD65–(553–585) in DR4-positive patients
| Subject I.D. no. | HLA-DRB1 typing | Time after diagnosis, mo | IFN-γ, pg/ml | IL4, pg/ml | GAD65 Ab index* |
|---|---|---|---|---|---|
| 6118 | 0301, 0404 | 5 | 0 | 0 | 0.24 |
| 6544 | 0401, 0401 | 1 | 6 | 0 | 1.1 |
| 6616 | 0301, 0401 | 14 | 79 | 0 | 0 |
| 6545 | 0301, 0404 | 1.5 | 120 | 0 | 0.94 |
| 6862 | 0301, 0404 | 1.5 | 176 | 0 | 0.37 |
| 6815 | 0301, 0401 | 1.5 | 264 | 0 | 0.07 |
| 6434 | 0401, 0404 | 0.8 | 376 | 0 | 0.2 |
| 7417 | 0401, 0101 | 0.5 | 4 | 0 | 0.55 |
| 6211 | 0404, 0405 | – | 261 | 0 | 0.18 |
Ratio of autoantibody levels in patient sera relative to standardized index sera; positive Ab index > .03.
T cells from patient no. 6211 were expanded in serial culture by restimulation with GAD65 peptides incubated with autologous APCs. Specific T cell responses were present for both peptides 553–572 (53 pg/ml IFN-γ) and 555–567 (51 pg/ml IFN-γ), but not peptide 569–585 (5 pg/ml IFN-γ), localizing the minimal epitope to the 555–567 region. T cell clones were derived by expansion of this culture. Proliferation and cytokine response profiles for CD4+ T cell clones BRI.4–10 and BRI.4–11 are shown in Fig. 1. The presence of such T cells from an individual identified as “at-risk” for diabetes is a likely reflection of the preclinical stage of an autoimmune response to islet-related antigens.
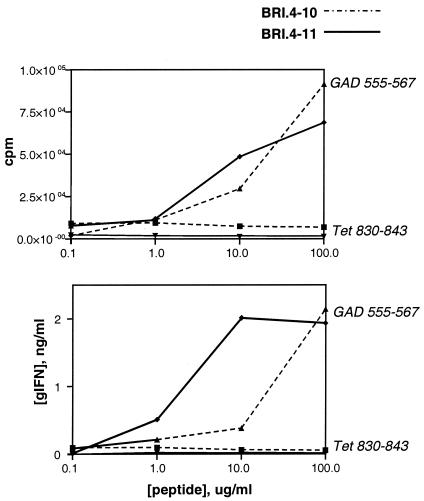
Figure 1.
T cell response profiles for human CD4+ T cell clones BRI.4–10 and BRI.4–11. Proliferation was measured by thymidine uptake (Upper) and IFN-γ release was determined by specific ELISA (Lower). Clones were stimulated with specific GAD peptides or with control peptides derived from tetanus toxoid (Tet 830–843).
Mass spectrometry provides for the direct sequence analysis and identification of naturally processed peptide antigens associated with class I and class II MHC molecules. The presence of posttranslational modifications on some MHC-associated peptides (21) underscores the importance of direct identification of the naturally presented species. The combination of offline HPLC fractionation, an epitope reconstitution assay, and LC/MS has been used previously to identify successfully a number of MHC class I-restricted peptide antigens (22). However, this approach has been more difficult to exploit for the identification of peptide epitopes associated with class II MHC molecules. Although the reasons for this difficulty are not entirely clear, it is generally recognized that class II peptides often occur in nested sets (23–25). Thus, HPLC fractionation may temporally separate and dilute species that collectively constitute an epitope, decreasing the ability to detect them by bioassay. Recent advances in mass spectrometric instrument control software (26, 27) now allow more rapid and efficient (MS and MS/MS) analysis of increasingly complex mixtures, effectively eliminating the need for fractionation.
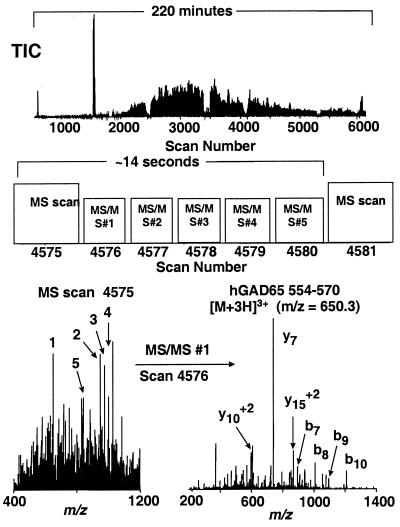
We sought to identify a DR4-restricted hGAD65 epitope from an unfractionated sample of class II-associated peptides. Previous studies have shown that transfection of GAD65 into the human B-LCL line PRIESS [DRB1*0401, *0401] resulted in an APC capable of stimulating T cell hybridomas derived from mice transgenic for HLA-DR4 that were immunized with GAD65 peptides (16). To determine the structure of the naturally processed hGAD65 peptides presented by this transfectant, the HLA-DR4-associated peptides were purified from 6 × 108 cells. An aliquot of this peptide extract was analyzed by nanoflow HPLC micro electrospray ionization mass spectrometry on a quadrupole ion trap mass spectrometer (19). The total ion chromatogram recorded in data-dependent mode indicates the enormous complexity of this sample (Fig. 2 Top). Approximately 5,000 MS/MS spectra were recorded, of which approximately 2,000 showed fragmentation features characteristic of peptides. These spectra were searched by using the sequest algorithm (20) against both an hGAD65 single-protein database and the nonredundant protein database maintained at the National Center for Biotechnology Information. Approximately 700 spectra were assigned tentatively to protein sequences in the nonredundant database. The MS/MS spectra of ions with m/z values of 974.8+2 and 650.3+3 matched to residues 554–570 of the hGAD65 protein. These matches were confirmed (i) by manual interpretation of the MS/MS spectra recorded on the ions at m/z 650.3+3 (Fig. 2 Bottom Right) and m/z 974.8+2 (Fig. 3); (ii) comparison with MS/MS spectra obtained for a synthetic peptide corresponding to this sequence (data not shown); and (iii) coelution of the naturally processed and synthetic peptides (data not shown). Based on a comparison with known amounts of synthetic peptide, we estimated that the observed signal intensity for naturally processed hGAD65-(554–570) corresponded to approximately 2 fmol or 30–50 peptide copies per cell.
Figure 2.
Identification of GAD65-derived peptides by using data-dependent MS/MS analysis of an unfractionated HLA-DR4-restricted peptide extract. (Top) Total ion chromatogram (TIC). (Middle) Data acquisition scheme. For MS scan no. 4575 m/z values for all ions in the 300–2,000 range were recorded (MS mode), and the instrument control computer automatically selected the five most abundant ions. Each of these ions was subjected to collision-activated dissociation to yield peptide sequence-specific fragment ions (MS/MS analysis) over the next five scans. The m/z values for these five ions were ignored by the instrument for a time equal to the observed chromatographic peak width (2 min for the data shown herein) to minimize redundant MS/MS analyses (e.g., dissociating the same peptide multiple times). After the next MS scan (no. 4581), the instrument selected the five most abundant ions, exclusive of those already identified in scan no. 4575. In this manner, ions having abundances over a wide dynamic range were automatically subjected to MS/MS analysis. (Bottom Left) MS scan no. 4575, showing ions that were selected for subsequent MS/MS analysis (numbered in order of abundance). (Bottom Right) MS/MS spectrum (scan no. 4576) for m/z = 650.3, the +3 charge state of the hGAD65 peptide 554–570 (corresponding to the first ion selected in MS scan no. 4575). All MS/MS spectra were searched against both an hGAD65 single-protein database and the nonredundant protein database maintained at the National Center for Biotechnology Information by using the SEQUEST search algorithm. The hGAD65 peptide sequence identified by this analysis was confirmed by manual interpretation of the MS/MS spectrum and by comparison with a synthetic peptide.
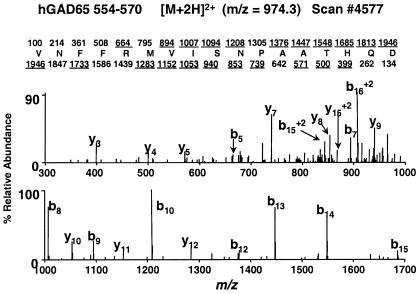
Figure 3.
MS/MS spectrum (scan no. 4578) for m/z = 974.8, the +2 charge state of hGAD65-(554–570). Note that all abundant fragment ions observed correspond to b-type (fragment ions containing the N terminus) and y-type (fragment ions containing the C terminus) sequence-specific ions. Predicted type b- and y-fragment ions, respectively, for hGAD-(554–570) are shown above and below the sequence. Ions observed in the mass spectrum are underlined.
Identification of a single peptide species of interest during the data-dependant LC/MS/MS analysis on a quadrupole ion trap mass spectrometer (QIT) was followed by an LC/MS analysis of an aliquot corresponding to 1 × 107 cell equivalents of the sample on a Fourier-transform ion cyclotron resonance mass spectrometer (18). The combination of high mass accuracy, high mass resolving power, and high sensitivity afforded by this technique allowed for identification of candidate nested-set peptides at less than 10 copies per cell. Candidate nested-set peptides were confirmed subsequently by comparison with synthetic peptides either through targeted MS/MS analysis on a QIT or through selected reaction monitoring analysis on a triple quadrupole mass spectrometer. This methodology allowed for the identification of two additional overlapping hGAD65 peptides [hGAD65-(552–570) and ()] present at approximately 80% and 10%, respectively, of the amount observed for hGAD65-(554–570).
The GAD-(554–570) sequence (VNFFRMVISNPAATHQD) contains a prototypic DR4-binding motif (28–31) in which F-557, V-560, S-562, and A-565 correspond to the P1, P4, P6, and P9 anchors—corresponding to the four main binding pockets in the DR4 molecule. Fig. 4 shows the binding properties of hGAD65-(554–570) analogs to HLA-DR4 molecules. As predicted, both hGAD65-(554–570) and the minimal epitope hGAD65-(555–567) [identified in the murine DR4 transgenic studies (16)] are strong binders; radical substitution (F → R) at the P1 anchor abrogates binding, consistent with the predicted motif (Fig. 4A).
Figure 4.
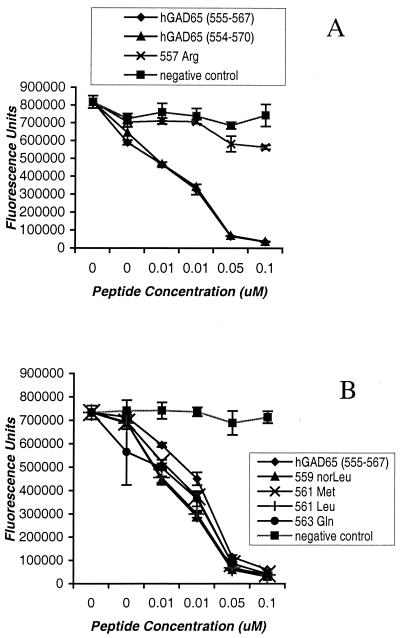
Competitive peptide binding to HLA-DR4 (DRB1*0101/DRB1*0401) molecules. Purified DR4 molecules were incubated with 0.1 μM biotinylated peptide standard (IAFTSEHSHFSLK) in the presence of various concentrations of GAD65 analogs, compared with hGAD65-(555–567) (NFFRMVISNPAAT). A nonbinding peptide standard (KSAVLEGTLTAEK) was used as a negative control.
Based on this motif, additional peptide analogs were synthesized, in which putative T cell contact residues on the peptide were modified. Conservative substitutions were introduced at P3 (methionine to norleucine at GAD559), P5 (isoleucine to methionine or leucine at GAD561), and P7 (asparagine to glutamine at GAD563) to alter the strength of the antigenic signal delivered for T cell antigen receptor (TCR) recognition without changing the class II-binding profile. As expected, binding each of these substituted peptides to DR4 class II molecules was comparable to that of the unmodified sequence (Fig. 4B). Each of these modified peptides also was tested for the ability to trigger proliferation or IFN-γ release from T cell clones BRI.4–10 and BRI.4–11. No T cell stimulation was observed, consistent with the predicted loss of agonist activity by changes at TCR contact residues of the peptide epitope (data not shown).
A number of studies have suggested the possibility for rational design of peptide antagonists by altering amino acid residues at TCR contact sites within an immunogenic epitope, to subtly alter the overall avidity of the TCR–MHC-peptide interaction (32, 33). Mechanistically, this altered interaction appears to interfere with the duration of TCR signaling events and therefore the efficiency of substrate phosphorylation and subsequent intracellular signaling (34–36). To determine whether the modified GAD65 peptides were capable of antagonizing the T cell response to hGAD65, APCs were prepulsed with the agonist hGAD-(555–567) peptide and then incubated with each of the modified peptides. Methionine substitution at P5 resulted in significant antagonism of the antigen-specific T cell response (Fig. 5). The T cell proliferative response of T cell clone BRI.4–10 was reduced by 80% when APCs were incubated with the Met-561 antagonist peptide. A control peptide derived from tetanus toxin 830–843, as well as the P3 and P7 substituted peptides, had no effect. The IFN-γ cytokine response of clone BRI.4–10 was antagonized similarly, with a much greater sensitivity to the Met-561 altered peptide ligand (APL; Fig. 5 Right). The Met-561 APL also antagonized T cell responses of clone BRI.4-11 by more than 90% (Fig. 5 Lower). No antagonistic affect was observed with human T cell clones specific for influenza virus hemagglutinin, an unrelated antigen (data not shown).
Another APL, Leu-561, partially antagonized the proliferative and cytokine responses of T cell clone BRI.4–11 only when used at high concentrations; however, no antagonism of clone BRI.4–10 was observed with this peptide. Interestingly, peptide Leu-561, like Met-561, represents a relatively conservative structural change at the P5 position, suggesting a crucial role for this central TCR contact site in the recognition of hGAD65-(555–567).
These data demonstrate the rational design of potential peptide antagonists for the GAD-(555–567) epitope, in which substitutions at the peptide P5 residue, a predicted TCR contact site, resulted in APLs that interfere with T cell activation in response to an immunodominant IDDM-associated autoantigen. The GAD-(555–567) epitope is one of several antigenic targets in the human T cell response to islets that occurs in IDDM. Approximately 70% of Caucasians with IDDM carry DRB1*04/DQB1*0302 haplotypes. Although only a very small number of human CD4+ T cell clones to diabetes autoantigens restricted by this haplotype have been studied, all use the DR molecule as the restricting element. This restriction presumably reflects the predominant expression of DR relative to DQ and other human class II molecules, or may indicate some preferential role in disease progression. The T cell clones used in this report were expanded initially in culture by using APCs consisting of peripheral blood mononuclear cells incubated with the intact GAD65 protein. This cloning strategy thus ensured that the epitope we report by direct structural determination using a human B cell line is also expressed as a normal product of antigen processing in nontransformed human cells.
Two clinical trials have recently reported the testing of an APL in multiple sclerosis (MS), where the APL was derived by modification of an immunodominant peptide from myelin basic protein (MBP; refs. 37 and 38). Notably, the immune and clinical responses to APL administration were complex and variable. In the report by Kappos et al. (37), patients receiving a high APL dose showed hypersensitivity reactions, some patients developed TH2 responses to the native MBP peptide, and only patients receiving a low APL dose showed partial reduction in lesions defined by magnetic resonance imaging. Bielekova et al. (38) report on a smaller group similarly treated, in which three of eight patients developed exacerbations of MS after APL administration, possibly because of cross-recognition of native MBP induced by treatment. These clinical studies indicate the potential for variance between in vitro antagonism studies and in vivo use in a complex autoimmune setting, and emphasize a cautionary note for applications in human diabetes which, like MS, is a progressive autoimmune disorder with intricate T cell interactions.
In the past, identification of peptide epitopes potentially contributing to human autoimmune disease has relied on analysis of MHC-binding properties and T cell response profiles, generally by using synthetic peptides corresponding to arbitrary or predictive motifs within an antigenic protein. The direct identification of the naturally processed peptides corresponding to autoantigens is more difficult because of the rarity of specific peptide–MHC complexes that are sufficient to stimulate autoreactive antigen-specific T cells. In this report, we describe the resolution of this barrier in the identification of an hGAD65 target epitope as an IDDM-associated autoantigen presented on HLA-DR4 molecules. T cell responses to this epitope are present in a majority of our patients who are DR4-positive and have IDDM, indicating that this epitope represents one of the determinants recognized by CD4+ T cells during the autoimmune events associated with IDDM. Identification of synthetic peptide analogs that act as functional antagonists of specific T cell activation for hGAD65-specific T cells will facilitate analysis of autoantigen recognition in the complex T cell response associated with IDDM.
Acknowledgments
We thank Laura Tsaknaridis and Lori Moriarity for technical assistance and Nicky Ducommun and Janice Abbas for preparation of the manuscript. This work was supported by U.S. Public Health Service Grants DK49841 (to G.T.N.), AI45199 (to V.H.E.), and AI33993 (to D.F.H.), and by a grant from the Juvenile Diabetes Foundation International (to G.T.N.).
Abbreviations
- IDDM
type I diabetes (insulin-dependent diabetes mellitus)
- GAD
glutamic acid decarboxylase
- hGAD65
65-kDa isoform of human GAD
- LC/MS
liquid chromatography–electrospray ionization mass spectrometric
- APC
antigen-presenting cell
- PBL
peripheral blood lymphocytes
- TCR
T cell antigen receptor
- APL
altered peptide ligand
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gianani R, Eisenbarth G S. In: Type I Diabetes. Molecular, Cellular, and Clinical Immunology. Eisenbarth G S, Lafferty K J, editors. New York: Oxford Univ. Press; 1996. pp. 209–229. [Google Scholar]
- 2.Nepom G T. Curr Opin Immunol. 1995;7:825–830. doi: 10.1016/0952-7915(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 3.Mehta V, Palmer J P. In: Prediction, Prevention and Genetic Counseling in IDDM. Palmer J P, editor. Chichester, PA: Wiley; 1996. pp. 3–16. [Google Scholar]
- 4.Lernmark A. J Intern Med. 1996;240:259–277. doi: 10.1046/j.1365-2796.1996.27859000.x. [DOI] [PubMed] [Google Scholar]
- 5.Yoon J W, Yoon C S, Lim H W, Huang Q Q, Kang Y, Pyun K H, Hirasawa K, Sherwin R S, Jun H S. Science. 1999;284:1183–1187. doi: 10.1126/science.284.5417.1183. [DOI] [PubMed] [Google Scholar]
- 6.Atkinson M A, Bowman M A, Campbell L, Darrow B L, Kaufman D L, Maclaren N K. J Clin Invest. 1994;94:2125–2129. doi: 10.1172/JCI117567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong N W, Jones D B. Lancet. 1994;344:406–407. doi: 10.1016/s0140-6736(94)91432-x. [DOI] [PubMed] [Google Scholar]
- 8.Lohmann T, Leslie R D G, Hawa M, Geysen M, Rodda S, Londei M. Lancet. 1994;343:1607–1608. doi: 10.1016/s0140-6736(94)93061-9. [DOI] [PubMed] [Google Scholar]
- 9.Panina-Bordignon P, Lang R, van Endert P M, Benazzi E, Felix A M, Pastore R M, Spinas G A, Sinigaglia F. J Exp Med. 1995;181:1923–1927. doi: 10.1084/jem.181.5.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worsaae A, Hejnaes K, Moody A, Ludvigsson J, Pociot F, Lorenzen T, Dyrberg T. Autoimmunity. 1995;22:183–189. doi: 10.3109/08916939508995315. [DOI] [PubMed] [Google Scholar]
- 11.Schloot N C, Roep B O, Wegmann D R, Yu L, Wang T B, Eisenbarth G S. Diabetologia. 1997;40:332–338. doi: 10.1007/s001250050683. [DOI] [PubMed] [Google Scholar]
- 12.Weiss U, Manfras B J, Terjung D, Eiermann T, Wolpl A, Loliger C, Kuhnl P, Boehm B O. Scand J Immunol. 1995;42:673–678. doi: 10.1111/j.1365-3083.1995.tb03710.x. [DOI] [PubMed] [Google Scholar]
- 13.Bach J M, Otto H, Nepom G T, Jung G, Cohen H, Timsit J, Boitard C, van Endert P M. J Autoimmun. 1997;10:375–386. doi: 10.1006/jaut.1997.0143. [DOI] [PubMed] [Google Scholar]
- 14.Endl J, Otto H, Jung G, Dreisbusch B, Donie F, Stahl P, Elbracht R, Schmitz G, Meinl E, Hummel M, et al. J Clin Invest. 1997;99:2405–2415. doi: 10.1172/JCI119423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nepom G T, Erlich H. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- 16.Wicker L S, Chen S-L, Nepom G T, Elliott J F, Freed D C, Bansal A, Zheng S, Herman A, Lernmark Å, Zaller D M, et al. J Clin Invest. 1996;98:2597–2603. doi: 10.1172/JCI119079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel S D, Cope A P, Congia M, Chen T T, Kim E, Fugger L, Wherrett D, Sonderstrup-McDevitt G. Proc Natl Acad Sci USA. 1997;94:8082–8087. doi: 10.1073/pnas.94.15.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin S E, Shabanowitz J, Hunt D F, Marto J A. Anal Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 19.Burlingame A L, Carr S A, Baldwin M A, editors. Mass Spectrometry in Biology and Medicine. Totowa, NJ: Humana; 1999. [Google Scholar]
- 20.Yates J R., III Electrophoresis. 1998;19:893–900. doi: 10.1002/elps.1150190604. [DOI] [PubMed] [Google Scholar]
- 21.Meadows L, Wang W, den Haan J M, Blokland E, Reinhardus C, Drijfhout J W, Shabanowitz J, Pierce R, Agulnik A I, Bishop C E, et al. Immunity. 1997;6:273–281. doi: 10.1016/s1074-7613(00)80330-1. [DOI] [PubMed] [Google Scholar]
- 22.Hunt D F, Henderson R A, Shabanowitz J, Sakaguchi K, Michel H, Sevilir, Cox A L, Appella E, Engelhard V H. Science. 1992;255:1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 23.Rudensky A Y, Rath S, Preston-Hurlburt P, Murphy D B, Janeway C A., Jr Nature (London) 1991;353:660–662. doi: 10.1038/353660a0. [DOI] [PubMed] [Google Scholar]
- 24.Hunt D F, Michel H, Dickinson T A, Shabanowitz J, Cox A L, Sakaguchi K, Appella E, Grey H M, Sette A. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 25.Chicz R M, Urban R G, Gorga J C, Vignali D A, Lane W S, Strominger J L. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis M T, Stahl D C, Hefta S A, Lee T D. Anal Chem. 1995;67:4549–4556. doi: 10.1021/ac00120a019. [DOI] [PubMed] [Google Scholar]
- 27.McCormack A L, Schieltz D M, Goode B, Yang S, Barnes G, Drubin D, Yates J R. Anal Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 28.Hammer J, Belunis C, Bolin D, Papadopoulos J, Walsky R, Higelin J, Danho W, Sinigaglia F, Nagy Z A. Proc Natl Acad Sci USA. 1994;91:4456–4460. doi: 10.1073/pnas.91.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill C M, Liu A, Marshall K W, Mayer J, Jorgensen B, Yuan B, Cubbon R M, Nichols E A, Wicker L S, Rothbard J B. J Immunol. 1994;152:2890–2898. [PubMed] [Google Scholar]
- 30.Sette A, Sidney J, Oseroff C, del Guercio M-F, Southwood S, Arrhenius T, Powell M F, Colon S M, Gaeta F C A, Grey H M. J Immunol. 1993;151:3163–3170. [PubMed] [Google Scholar]
- 31.Dessen A, Lawrence C M, Cupo S, Zaller D M, Wiley D C. Immunity. 1997;7:473–481. doi: 10.1016/s1074-7613(00)80369-6. [DOI] [PubMed] [Google Scholar]
- 32.Lamont A G, Grey H M, Powell M F, Sette A. In: Molecular Autoimmunity. Talal N, editor. San Diego: Academic; 1991. pp. 425–435. [Google Scholar]
- 33.Evavold B D, Sloan-Lancaster J, Allen P M. Immunol Today. 1993;14:602–609. doi: 10.1016/0167-5699(93)90200-5. [DOI] [PubMed] [Google Scholar]
- 34.Racioppi L, Matarese G, D'Oro U, De Pascale M, Masci A M, Fontana S, Zappacosta S. Proc Natl Acad Sci USA. 1996;93:10360–10365. doi: 10.1073/pnas.93.19.10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madrenas J, Wange R L, Wang J L, Isakov N, Samelson L E, Germain R N. Science. 1995;267:515–518. doi: 10.1126/science.7824949. [DOI] [PubMed] [Google Scholar]
- 36.La Face D M, Couture C, Anderson K, Shih G, Alexander J, Sette A, Mustelin T, Altman A, Grey H M. J Immunol. 1997;158:2057–2064. [PubMed] [Google Scholar]
- 37.Kappos L, Comi G C, Panitch H, Oger J, Antel J, Conlon P, Steinman L The Altered Peptide Ligand in Relapsing MS Study Group. Nat Med. 2000;6:1176–1182. doi: 10.1038/80525. [DOI] [PubMed] [Google Scholar]
- 38.Bielekova B, Goodwin B, Richert N, Cortese I, Kondo T, Afshar G, Gran B, Eaton J, Antel J, Frank J A, et al. Nat Med. 2000;6:1167–1175. doi: 10.1038/80516. [DOI] [PubMed] [Google Scholar]