Abstract
Purpose
The dismal outcome of esophageal cancer patients highlights the need for novel prognostic biomarkers, such as microRNAs (miRNAs). While recent studies have established the role of miRNAs in esophageal carcinoma, a comprehensive multi-center study investigating different histological types, including squamous cell carcinoma (SCC) and adenocarinoma (ADC) with or without Barrett's, is still lacking.
Experimental Design
MiRNA expression was measured in cancerous and adjacent non-cancerous tissue pairs collected from 100 ADC and 70 SCC patients enrolled at 4 clinical centers from the US, Canada, and Japan. Microarray-based expression was measured in a subset of samples in two cohorts and was validated in all available samples.
Results
In ADC patients, miR-21, miR-223, miR-192, and miR-194 expression was elevated, while miR-203 expression was reduced in cancerous compared to non-cancerous tissue. In SCC patients, we found elevated miR-21 and reduced mir-375 expression levels in cancerous compared to non-cancerous tissue. When comparing cancerous tissue expression between ADC and SCC patients, mir-194 and mir-375 were elevated in ADC patients. Significantly, elevated mir-21 expression in non-cancerous tissue of SCC patients and reduced levels of mir-375 in cancerous tissue of ADC patients with Barrett's were strongly associated with worse prognosis. Associations with prognosis were independent of tumor stage or nodal status, cohort type, and chemoradiation therapy.
Conclusions
Our multi-center-based results highlight miRNAs involved in major histological types of esophageal carcinoma and uncover significant associations with prognosis. Elucidating miRNAs relevant to esophageal carcinogenesis is potentially clinically useful for developing prognostic biomarkers and identifying novel drug targets and therapies.
Keywords: microRNA, esophageal cancer, prognosis, Barrett's, expression profiling
INTRODUCTION
Esophageal cancer is the 8th most common cancer and the 6th most common cause of cancer deaths worldwide (1). Often diagnosed at later stages, the survival rate for affected patients is very low, ranging from 10% in Europe (2) to 16% in the United States (3). The incidence of esophageal cancer varies greatly by geographical location, where it is most common in China, South East Africa, and Japan, and by gender, where males are affected more than females (7:1 ratio). In recent years, the incidence of Barrett's associated adenocarcinoma (ADC), mainly caused by gastric reflux and obesity, has been rising, while the incidence of squamous cell carcinoma (SCC), mainly caused by cigarette smoking and alcohol consumption, has been decreasing in the United States (4). Barrett's results from chronic gastric reflux and is characterized by the replacement of normal esophageal squamous cell epithelium by metaplastic columnar epithelium. This chronic inflammatory condition is a well recognized precursor of esophageal ADC (5).
MiRNAs are small (20 – 24 nucleotides), well-conserved, non-coding RNA molecules that regulate the translation of mRNAs (6). Since the discovery of the first miRNA, lin-4, in C. elegans in 1993 (7), the miRNA registry has housed sequences from 218 miRNAs in 2002 to 9539 as of March 2009, including miRNAs in primates, rodents, birds, fish, worms, flies, plants and viruses (8). Each miRNA is thought to play a role in the post-transcriptional regulation of hundreds of genes, and translational blockade of a given gene may require binding of more than one miRNA (6). The role of miRNAs has been well established in various human cancers (9) and altered miRNA expression has been reported in most tumor types (10,11). In addition, miRNAs are often located in fragile sites or cancer-associated genomic regions (12). Recently, we and others reported the involvement of let-7 and miR-155 in lung cancer diagnosis and prognosis (13–15) and high expression of miR-21 was associated with poor survival and therapeutic outcome in colon cancer (16). Other expression profiling studies identified miRNA signatures in hepatocellular carcinoma (17,18), pancreatic cancer (19), breast cancer (20), papillary thyroid cancer (21), and chronic lymphocytic leukemia (22). Importantly, the successful use of antagomirs to silence miRNAs in mice (23) and non-human primates (24) suggests the possible therapeutic use of miRNAs. In the context of esophageal carcinoma, increased expression of RNASEN, a miRNA processing enzyme, in tumor samples of SCC patients suggests the role of miRNA in esophageal tumor progression (25). Recently, miRNA expression profiles unique to esophageal cancer histological types, Barrett's status, and survival have been uncovered (26–28), albeit limited samples sizes.
In this study, we utilized four independent clinical centers to confirm and find novel miRNAs involved in the pathogenesis of human esophageal cancers and Barrett's , and to explore their utility as predictors of prognosis. Using 100 ADC and 70 SCC patients split into training (32 ADC and 44 SCC) and validation sets (68 ADC and 26 SCC), we first generated miRNA microarray (29) profiles and confirmed expression differences of relevant miRNAs using real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) in all samples. Furthermore, we analyzed associations between miRNA expression and survival to explore the possible utility of miRNAs as prognostic biomarkers of esophageal carcinoma.
MATERIALS AND METHODS
Cohorts and clinico-pathological data
A total of 170 patients with available cancerous and adjacent non-cancerous tissue from surgical resection were divided into training and validation sets. The training set includes 44 SCC cases and 32 ADC cases, of which 18 were also diagnosed with Barrett's, while the validation set comprises 26 SCC cases and 68 ADC cases, including 45 patients also diagnosed with Barrett's. Patients were recruited from 4 different cancer centers: 1) University of Maryland (UMD) Medical System in Baltimore, MD, 2) Nippon Medical School in Tokyo, Japan, 3) New York Presbyterian-Weill Cornell Medical Center in NY, US, 4) Queen Elizabeth II Health Science Centre, Halifax NS, Canada (Table 1). Samples collected from the UMD cohort were divided such that 32 ADC and 11 SCC patients with sufficient RNA for microarray and qRT-PCR analysis were included in the training set, and 41 ADC and 13 SCC were included in the validation set. Disease stage and survival were obtained from medical records, pathology reports, State of Maryland records, and the National Death Index. These studies were approved by the Institutional Review Boards of the participating institutions. Clinico-pathological data relevant to this study were provided from their respective sources and include gender, age, histology, presence/absence of Barrett's, neo-adjuvant chemoradiation therapy administration (prior to surgery), alcohol consumption, smoking status, and pathologic staging (Table 1).
Table 1.
Patient clinical, pathological, and demographic characteristics
| Sample Type | Characteristics | UMD | Canada | Japan | Cornell | P* | |
|---|---|---|---|---|---|---|---|
| Adenocarcinoma | Total | 73 | 27 | NA | NA | ||
|
|
|||||||
| Gender | Male | 63 | 26 | 0.3 | |||
| Female | 10 | 1 | |||||
|
|
|||||||
| Age | < 62 | 39 | 12 | 0.5 | |||
| >= 62 | 34 | 15 | |||||
|
|
|||||||
| Barrett's Esophagus (Adeno only) | Yes | 48 | 15 | 0.4 | |||
| No | 25 | 12 | |||||
|
|
|||||||
| Chemoradiation | Yes | 45 | 0 | 1.00E-09 | |||
| Therapy | No | 27 | 27 | ||||
|
|
|||||||
| Alcohol Consumption | Yes | 44 | 26 | 0.01 | |||
| No | 17 | 1 | |||||
| Unknown | 12 | 0 | |||||
|
|
|||||||
| Smoking | Yes | 45 | 21 | 0.8 | |||
| No | 11 | 6 | |||||
| Unknown | 17 | 0 | |||||
|
|
|||||||
| Nodal Status | Yes | 57 | 18 | 0.3 | |||
| No | 16 | 9 | |||||
|
|
|||||||
| Pathologic Staging † | 0 | 20 | 0 | 3.00E-05 | |||
| I | 18 | 6 | |||||
| II | 20 | 3 | |||||
| III | 11 | 16 | |||||
| IV | 4 | 2 | |||||
|
| |||||||
| Squamous Cell | Total | 24 | NA | 33 | 13 | ||
|
|
|||||||
| Carcinoma | Gender | Male | 12 | 30 | 10 | 0.002 | |
| Female | 12 | 3 | 3 | ||||
|
|
|||||||
| Age | < 62 | 11 | 14 | 3 | 0.4 | ||
| >= 62 | 13 | 19 | 10 | ||||
|
|
|||||||
| Chemoradiation | Yes | 14 | 15 | 4 | 0.2 | ||
| Therapy | No | 9 | 18 | 9 | |||
| Unknown | 1 | 0 | 0 | ||||
|
|
|||||||
| Alcohol Consumption | Yes | 18 | 26 | 0 | 0.3 | ||
| No | 1 | 5 | 0 | ||||
| Unknown | 5 | 2 | 13 | ||||
|
|
|||||||
| Smoking | Yes | 19 | 24 | 11 | 0.4 | ||
| No | 2 | 7 | 2 | ||||
| Unknown | 3 | 2 | 0 | ||||
|
|
|||||||
| Nodal Status | Yes | 5 | 18 | 10 | 0.0005 | ||
| No | 18 | 14 | 3 | ||||
| Unknown | 1 | 0 | 0 | ||||
|
|
|||||||
| Pathologic Staging † | 0 | 10 | 2 | 0 | 0.002 | ||
| I | 2 | 5 | 1 | ||||
| II | 9 | 12 | 3 | ||||
| III | 1 | 6 | 4 | ||||
| IV | 1 | 8 | 5 | ||||
| Unknown | 1 | 0 | 0 | ||||
P calculated from Fisher's exact Test
Determined at the time of surgery
RNA Isolation and quantification of miRNA
Total RNA used for quantification of miRNA levels was extracted from esophageal tissue using TRIZOL (Invitrogen, cat. no. 15596-026), according to the manufacturer's procedures. MiRNA expression levels were measured using miRNA microarray chips version 3 (Ohio State University) containing 329 human miRNAs and 249 mouse miRNA probes in duplicate (29). Five μg of total RNA were converted to biotin-labeled first strand cDNA, hybridized onto the chips, and processed by direct detection of the biotin-containing transcripts by streptavidin-Alexa 647 conjugate. Slides were subsequently scanned with the Axon 4000B Scanner (Molecular Device, Inc) and spot intensities were quantified with Genepix (version Pro 6.0.1.00). Microarray data is currently being submitted to the Gene Expression Omnibus, in compliance with MIAME guidelines.
Validation of miRNA altered levels was performed by qRT-PCR using Taqman miRNA reverse transcription assays (Applied Biosystems, cat. no. 4366596) and appropriate primers, following the manufacturer's instructions. In brief, 10 ng of total RNA was used as a template for a 15 μl reverse transcription reaction using probes specially designed for specific mature miRNAs. For each miRNA, reactions were performed in triplicate using the 7500 RT-PCR system (Applied Biosystems) and RNU66 (Applied Biosystems, cat. no. 4373382) was used as a normalization control.
Statistical Analysis
Pre-processing and normalization of miRNA microarray expression values were performed in R (version 2.6.0), and differential expression analysis was carried out in BRB ArrayTools (version 3.5.0) developed by Dr. Richard Simon and Amy Peng Lam (http://linus.nci.nih.gov/BRB-ArrayTools.html). (See Supplemental Data for more detail). Differential expression analysis was restricted to human miRNA probes with non-missing intensity values in at least 25% of the samples. Altered expression of miRNA probes was determined using the Class Comparison Tool, which performs t-tests, and expression changes with a P < 0.05 and corresponding False Discovery Rate < 10% were considered to be statistically significant. A paired t-test was performed when comparing cancerous and adjacent non-cancerous tissue expression, while a t-test with a random block design by date was applied for all other comparisons.
qRT-PCR measurements were considered concordant with microarray expression when both measurements showed statistically significant and same direction fold changes. qRT-PCR expression counts were normalized to RNU66 counts and two-sided paired or unpaired t-tests (for comparing cancerous and adjacent non-cancerous tissue, and all other comparisons, respectively) were performed.
Survival analysis was performed on dichotomized miRNA expression values using the median expression value within each cohort as a cutoff. Kaplan-Meier curves were constructed and survival differences were assessed using the log rank test. The proportional hazards assumption was tested using Schoenfeld residuals and was met for all models reported. Univariate and multivariate Cox analysis was performed to assess associations between clinical variables and prognosis, and to adjust for relevant clinical variables (see Supplemental Data). Multivariate Cox models included cohort type to account for the different cohorts that were used in the study, and staging or nodal involvement as they were associated with survival in the univariate analysis.
To ensure a sufficient number of events per group in the multivariate analysis, samples from different cohorts were combined. To make sure that cohort type did not bias associations with prognosis, we verified that there was no association between cohort type and survival (see Supplemental Figure 1). In addition, miRNA expression was dichotomized into high and low within each cohort, using a median cutoff. Finally, all multivariate Cox models were also adjusted for cohort type to ensure independence of miRNA prognosis associations from cohort type associations. Of note, hazard ratios showed the same trend in each cohort for a given miRNA although in some cases, P values exceeded 0.05 (data not shown).
Importantly, the administration or lack thereof of neo-adjuvant chemoradiation therapy was not associated with survival. Statistical significance of expression validation and survival analysis was achieved when P < 0.005 (corresponding to P < 0.05 after applying the stringent Bonferroni correction for 9 multiple comparisons) and borderline statistical significance was achieved when 0.005 < P < 0.05.
Sample classifications were performed using the R package “pamr” (version 1.34.0), Prediction Analysis of Microarrays (PAM) (30). Robustness of the models was evaluated using bootstrap techniques. P values describing the accuracy level (i.e. is an 80% accuracy significantly different from random) and the difference in accuracies observed between models built using all probes and models built after removing “persistent probes” were calculated (see Supplemental Data). Finally, we performed Receiving Operating Curves analysis to select optimal model parameters (see Supplemental Data).
RESULTS
Cancerous and adjacent non-cancerous tissue resected from 170 patients were utilized in this study, which was conducted in three steps: (a) Analysis of ADC patients (N = 32 in training set, N = 68 in validation set), including Barrett's associated and sporadic ADC; (b) Analysis of SCC patients (N = 44 in training set, N = 26 in validation set); and (c) Comparison of ADC and SCC patients. Clinical characteristics of all patients used in this study are summarized in Table 1. In ADC patients, patients in each cohort differed by administration of neoadjuvant chemoradiation therapy, alcohol consumption, and pathologic staging while differences in gender, nodal status, and stage were observed between cohorts of SCC patients.
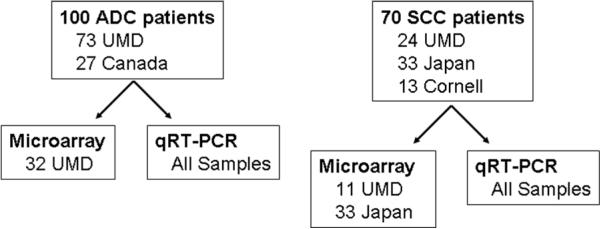
A flow chart of the analysis performed is shown in Figure 1. MicroRNA microarray expression values were first evaluated in training set samples (Supplemental Table 1) and expression of select miRNAs was subsequently confirmed using qRT-PCR in all samples. The predictive capacity miRNAs was first evaluated by inputting microarray expression values in Prediction Analysis of Microarrays and classifying samples by tumor status and types (Supplemental Table 2). When classifying ADC samples, 71% accuracy (P = 0.005) was obtained when discerning cancerous from adjacent non-cancerous tissue. Diagnostic prediction of ADC patients with Barrett's increased the accuracy to 77% (P = 0.006) while the prediction of patients with sporadic ADC yielded near random class assignment (58% accuracy). Analogously, there were no differentially expressed miRNAs identified in patients with sporadic ADC using standard t-tests. Furthermore, 78% accuracy (P = 0.003) was obtained when classifying cancerous tissue expression of Barrett's associated or sporadic ADC patients. Non-cancerous tissue expression was not predictive of Barrett's associated ADC. Classification of SCC samples into cancerous and non-cancerous tissue yielded 86% accuracy (P < 1e-4). Finally, classification of samples by histology yielded 82% and 85% accuracies using cancerous and non-cancerous tissue expression profiles, respectively. Importantly, there is a large overlap between miRNA probes that contribute most to the classifications (Supplemental Table 3), and those that show differential expression using ANOVA. These miRNAs are of interest as they may play a role in esophageal carcinogenesis.
Figure 1.
Study design flow chart. Differential expression analysis and associations with prognosis were performed in patients with ADC and SCC, separately. Microarray analysis was performed on a subset of samples as a hypothesis-generating tool to subsequently focus further validation on a subset of miRNAs that are potentially relevant to esophageal carcinogenesis. Validation using qRT-PCR was performed on all samples available.
MicroRNA Differential Expression in ADC Patients
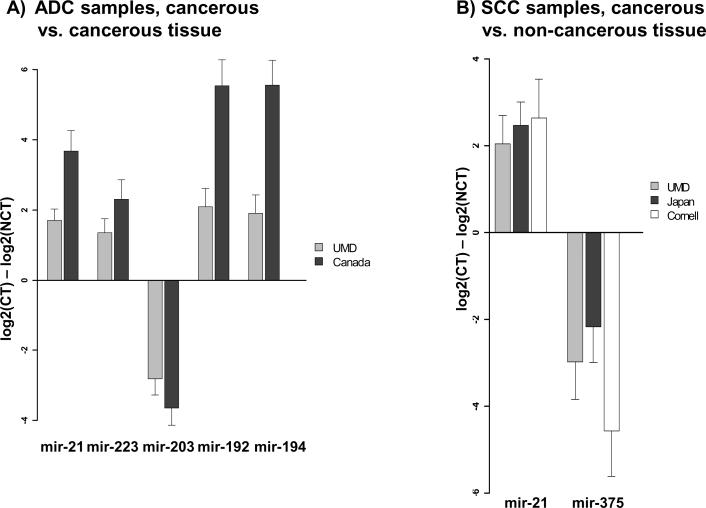
Alterations in miRNA microarray expression levels specific to ADC patients were evaluated in 32 cancerous and adjacent non-cancerous tissue pairs (Supplemental Table 4–5). Interestingly, many of the differentially expressed probes are located in fragile sites and Cancer Associated Genomic Regions. Expression measurements of select miRNAs (P<0.05, FDR < 10%, and largest fold changes) were validated using qRT-PCR. Elevated expression of miR-21, miR-223, mir-192, mir-194, and reduced expression of miR-203 in ADC cancerous compared to adjacent non-cancerous tissue was confirmed in training and validation set samples, including those from the UMD and Canada cohorts (Figure 2A). These 5 miRNAs were also differentially expressed between ADC patients with or without Barrett's. Of note, mir-21, mir-223, mir-192, and mir-194 were also differentially expressed in patients that have received neo-adjuvant chemoradation therapy, compared to those that have not (Supplemental Table 6).
Figure 2.
qRT-PCR validation of differentially expressed miRNAs. A) Altered expression between cancerous tissue (NCT) and non-cancerous tissue (NCT) in ADC patients is shown. Mir-21, mir-223, mir-192, and mir-194 are over-expressed in tumors while mir-203 is under-expressed in tumors. B) Differential expression between CT and NCT in SCC patients demonstrates that mir-21 is up-regulated and mir-375 is down-regulated in SCC tumors. All expression values are normalized to RNAU66, and differential expression is statistically significant (P<0.005).
In addition, altered expression of miR-192 (Barrett's:non-Barrett's fold change: 6.94, pval = 0.0008) and miR-194 (Barrett's:non-Barrett's fold change: 5.78, pval = 0.0009) in cancerous tissue between Barrett's associated and sporadic ADC patients was confirmed by qRT-PCR in all samples of the UMD cohort. This association was not confirmed in the Canada cohort (mir-192 Barrett's:non-Barrett's fold change: 0.41, pval = 0.17; mir-194 Barrett's:non-Barrett's fold change: 0.46, pval = 0.22). ADC patients from the UMD and Canada cohort differ by stage and neoadjuvant chemoradiation therapy (none of the Canada cohort patients have undergone therapy). Nonetheless, mir-192 and mir-194 were not differentially expressed between patients with low stage (TNM = 0, I) and high stage (TNM = II, III, IV). Due to an insufficient number of UMD cohort patients that have not undergone chemoration therapy and do not have Barrett's (N=2), we were unable to assess whether the Barrett's associated differential expression is affected by therapy. However, mir-192 and mir-194 expression is increased in Barrett's associated patients that have undergone chemoradiation therapy. More detailed differential expression by cohort type of all miRNAs evaluated is detailed in Supplemental Table 7.
Association between miRNA expression and survival of ADC patients
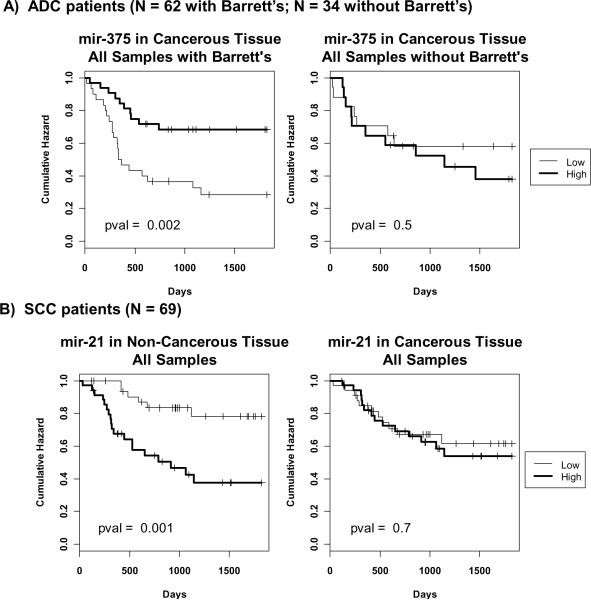
MiRNA expression values derived from qRT-PCR were dichotomized based on a within cohort median cutoff (see Methods). Associations between miRNA expression and survival were not observed in ADC patients. However, when evaluating ADC patients with Barrett's, low expression of miR-375 (HR = 0.31; 95% confidence interval [CI] = 0.15 − 0.67) in cancerous tissue was strongly associated with worse prognosis. While mir-375 expression is altered between ADC patients with Barrett's that have and have not received neoadjuvant chemoradiation therapy (Supplemental Table 6), multivariate analysis shows that the association between mir-375 and survival is independent of cohort type, stage, and therapy (HR = 0.29; 95% CI = 0.13 − 0.64). In addition, low expression of miR-223 (HR = 0.45; 95% confidence interval [CI] = 0.22 − 0.93) in cancerous tissue was borderline associated with poor prognosis, independent of cohort type, stage, and therapy (HR = 0.46; 95% CI = 0.22 − 0.94) (Figure 3A, Table 2, Supplemental Table 8). Expression of mir-223 is not altered in ADC patients with Barrett's that have or have not received therapy (Supplemental Table 6). There was no significant association between miRNA and survival in ADC patients without Barrett's.
Figure 3.
Kaplan-Meier Analysis depicting associations with qRT-PCR miRNA expression and survival. MiRNA expression values were dichotomized into low and high groups, using the within cohort median expression value as a cutoff. A) Associations observed in adenocarcinoma (ADC) patients with and without Barrett's. Reduced expression of mir-375 in cancerous tissue of ADC patients with Barrett's is associated with worse prognosis. This association is not observed in ADC patients without Barrett's. Survival profiles were compared using the log rank test. B) Associations observed in squamous cell carcinoma (SCC) patients. Elevated expression of mir-21 in non-cancerous tissue is associated with worse prognosis. This association is not found when cancerous tissue expression is evaluated.
Table 2.
Univariate and multivariate Cox modeling of all available samples to assess associations between miRNA qRT-PCR expression levels and survival.
| Comparison | Univariate | Multivariate* | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| ADC patients with BE | mir-375 in CT (N=62) | 0.31 | 0.15–0.67 | 0.003 | 0.29 | 0.13–0.66 | 0.001 |
|
| |||||||
| SSC Patients | mir-21 in NCT (N=69) | 4.23 | 1.68–10.61 | 0.002 | 4.71 | 1.74–12.79 | 0.002 |
| mir-155 in NCT (N=69) | 2.69 | 1.12–6.46 | 0.03 | 3.08 | 1.21–7.87 | 0.02 | |
| mir-146b in NCT (N=69) | 2.4 | 1.03–5.56 | 0.04 | 2.7 | 1.1–6.6 | 0.03 | |
| mir-181b in NCT (N=69) | 2.76 | 1.15–6.62 | 0.02 | 3.52 | 1.37–9.04 | 0.009 | |
| mir-223 in CT (N=69) | 0.43 | 0.19–0.97 | 0.04 | 0.25 | 0.09–0.69 | 0.007 | |
Abbreviations: CT- cancerous tissue; NCT- non-cancerous tissue
Adjusted for stage (0-I vs. II-IV), cohort type, and neo-adjuvant chemoradiation therapy in ADC patients with BE; adjusted for nodal status, cohort type, and neo-adjuvant chemoradiation therapy in SCC patients
MicroRNA Differential Expression in SCC Patients
Altered miRNA expression specific to SCC was first analyzed using microarrays in 44 patients (Supplemental Tables 1, 9). Thirty-five percent of the probes differentially expressed between cancerous and non-cancerous tissue are located in Cancer Associated Genomic Regions. Expression measurements of miRNAs with largest fold changes (P<0.05, FDR < 10%) were confirmed using qRT-PCR in all available cases, including 26 additional validation set samples. Elevated expression levels of miR-21 and reduced levels of miR-375 were confirmed in patients from the UMD, Japan, and Cornell cohorts when comparing cancerous and adjacent non-cancerous tissue (Figure 2B). Neither mir-21 nor mir-375 expression was associated with administration of neo-adjuvant chemoradiation therapy (Supplemental Table 6). Interestingly, elevated levels of miR-21 in cancerous tissue were also observed in ADC samples, suggesting it might participate in the different histological types of esophageal tumorigenesis. Detailed differential expression results by cohort type are shown in Supplemental Table 7.
Association between miRNA expression and survival in SCC patients
Similar to the analysis of ADC patients, qRT-PCR expression values were dichotomized based on a median cutoff within each cohort. Kaplan-Meier analysis revealed a statistically significant association between high expression of miR-21 in non-cancerous tissue (HR = 4.23; 95% CI = 1.68 − 10.61) and worse prognosis (Figure 3B, Table 2, Supplemental Table 8). Elevated levels of miR-155 (HR = 2.69; 95% CI = 1.12 − 6.46), miR-146b (HR = 2.4; 95% CI = 1.03 − 5.56), and miR-181b (HR = 2.76; 95% CI = 1.15 − 6.62) in non-cancerous tissue showed a borderline significant association with worse prognosis. Furthermore, reduced miR-223 expression in cancerous tissue (HR = 0.43; 95% CI = 0.19 − 0.97) was borderline associated with poor prognosis. Mir-21, mir-155, mir-146b, and mir-181b expression is not associated with neo-adjuvant chemoradiation therapy in SCC patients (Supplemental Table 6). Multivariate Cox modeling revealed that all associations between the expression of these miRNAs and survival are independent of cohort type, nodal involvement, and therapy.
MicroRNA Differential Expression Between ADC and SCC patients
Microarray analysis results are shown in Supplemental Table 10 and demonstrate that differential expression by histological type is only detected in cancerous tissue, suggesting that adjacent non-cancerous tissue in both histological subtypes have similar miRNA profiles. Elevated expression levels in cancerous tissue of miR-194 (ADC:SCC fold change: 4.59, pval = 0.005) and miR-375 (ADC:SCC fold change: 6.41, pval = 0.009) in ADC compared to SCC patients were observed using qRT-PCR in patients without Barrett's. qRT-PCR expression differences between ADC and SCC patients was evaluated in the UMD cohort only, since it is the only cohort that includes a sufficient amount of ADC and SCC patients. In addition, only patients without Barrett's were evaluated to avoid confounding of differential expression by Barrett's status. Altered expression in these miRNAs underlies differences in biological mechanisms involved in the two different histological types of esophageal cancerous cells.
DISCUSSION
To our knowledge, we have performed the largest study to date that assesses the potential diagnostic and prognostic utility of miRNAs in esophageal cancer. MiRNA expression was evaluated in 170 cancerous and adjacent non-cancerous tissue pairs and we identified miRNAs important for classification of samples into diagnostic and Barrett's categories. Elevated miR-21 levels were observed in both SCC and ADC samples indicating that miR-21 involvement in esophageal carcinogenesis is independent of histological type. Furthermore, increased expression of miR-194, miR-192, miR-223 and reduced expression of miR-203 are observed in ADC patients from two independent clinical centers while decreased expression of miR-375 is detected in SCC patients from three clinical centers. Altered expression of these miRNAs is specific to histological type, which may provide clues as to the different molecular progression undertaken by these cancers and suggests a potential avenue for histology-specific therapy to improve prognosis.
Expression levels of miRNAs mentioned above were validated in samples from the testing and validation cohorts using qRT-PCR. Over-expression of miR-21 is of interest since it is ubiquitously induced in solid tumors, including esophagus, liver, lung, breast, stomach, prostate, colon, pancreas (10,11,13,16–21) and in chronic lymphocytic leukemia (22). MiR-21 targets tumor and metastasis suppressor genes, including phosphatase and tensin homolog PTEN (31), tumor suppressor gene tropomyosin 1 TPM1 (32), programmed cell death 4 PDCD4 (33,34), and Sprouty2 (35), thereby demonstrating its involvement in tumor growth, invasion, and metastasis. Our laboratory has also revealed that elevated miR-21 in tumors are associated with poor prognosis and therapeutic outcome in colon cancer (16).
Importantly, we demonstrate here that altered levels of miR-21 in non-cancerous tissue of SCC patients are associated with survival, suggesting that miR-21 may have an effect in SCC tumors via the stroma. Knowing that inflammatory responses are implicated in all stages of tumorigenesis (36), targeting mir-21 in stromal cells provides a possible avenue for chemoprevention. We have previously established that the combination of cytokine expression in non-cancerous and cancerous tissue of lung ADC patients are predictors of survival, suggesting a possible interaction between the tumor and its surrounding lung environment (37). A similar association between cytokine expression and survival was observed in esophageal cancer (unpublished results). Furthermore, there is growing evidence for the role of miRNAs in regulating innate and acquired immune response (38–40). Specifically, mir-21 expression has been associated with immune-related diseases, including B-cell lymphoma (41) and chronic lymphocytic leukemia (22). Furthermore, a recent study demonstrated the Stat3-dependent effect of interleukin-6 on miR-21 induction, which contributed to the oncogenic potential of Stat3 (42). Consequently, our finding that increased levels of miR-21 are associated with worse prognosis in non-cancerous tissue is possibly a reflection of an immune response that is associated with tumorigenesis.
In addition, our study shows that mir-375 levels in cancerous tissue are associated with prognosis in Barrett's associated ADC patients. A recent study has demonstrated that mir-375-deficient mice are hyperglycemic and that the pancreatic alpha-cell mass is decreased, a sign of impaired proliferation (43). Furthermore, the same group performed a combined analysis of putative targets and microarray expression showing that putative mir-375 targets control cellular growth and proliferation (43). Experimentally validated targets of mir-375, as described in Tarbase (44), include Mxi1, JAK2, and Ahr. MXi1 is a c-MYC antagonist previously found to be over-expressed in esophageal ADC (45). A direct association between JAK2 and PI3-kinase upon glycine-extended gastrin stimulation has been shown in Barrett's associated ADC cells (46). The suppression of Ahr signaling as a chemopreventive approach was supported by a study that showed a reduction of DNA adducts in esophageal SCC cell lines by blocking the transcription factor Ahr (47). These results combined with our finding that reduced levels of mir-375 are associated with worse prognosis suggest that mir-375 may be a good marker for cell proliferation.
In concordance with our observations, a recent study based on a cohort of 7 patients reported that miR-21 is over-expressed in ADC, miR-143 is under-expressed in ADC, and miR-194 is over-expressed in Barrett's (28) . The study also reported over-expression of miR-203, miR-205, miR-143, and miR-215 in Barrett's. Another study reported the analysis of 20 cases and 9 normal epithelial tissue and revealed an over-expression of miR-21 and under-expression of miR-203 and miR-205 in cancerous tissue in both histological subtypes (26), which is concordant with our microarray results. In a previous study evaluating miRNA expression in SCC patients, high expression of miR-103 and miR-107 correlated with poor survival in 30 patients, a finding confirmed in an independent set of 22 SCC patients (27). These results were not in concordance with our analysis, perhaps due to their use of a different microarray platform and more limited sample size.
The administration of neo-adjuvant chemoradiation therapy (prior to surgery) in 54% of patients used in this study and complete pathologic response in 22% of patients limits our ability to negate the role of therapy on associations between miRNA expression and diagnosis/prognosis. Differential expression in cancerous tissue and non-cancerous tissue was observed in patients that had or had not undergone neo-adjuvant chemoradiation therapy (Supplemental Table 10) although associations were not consistent across different cohort types. Because therapy was administered prior to tissue collection and no pre-therapy tissue is available, it was not possible to directly predict therapeutic response.
Of note, patients with complete pathologic response are not necessarily cured, perhaps due to remaining systemic processes or the inability to detect small metastatic disease (48). It is still a debate whether such patients have longer survival than patients without complete pathologic response (49). This observation further demonstrates the importance of identifying molecular biomarkers, such as miRNAs, that would help refine staging and predict treatment response. Furthermore, while chronic alcohol consumption and smoking may adversely affect survival of esophageal cancer patients (50), we were unable to adequately assess the influence of those covariates in our multivariate Cox analysis due to missing values (16% and 23% missing values for smoking and alcohol consumption, respectively). Although preliminary analysis of available data did not show an association between smoking, alcohol consumption and survival (data not shown), further studies are warranted to confirm the association of miRNAs with survival, independent of these two factors.
In conclusion, we identified miRNAs whose expression is altered in and between SCC and ADC cancerous tissue, and in cancerous tissue between Barrett's associated and sporadic ADC cancerous tissue. Furthermore, we have established a strong association between elevated miR-21 levels in the non-cancerous tissue of SCC patients with worse prognosis, thereby suggesting a possible association between miR-21 in the stromal environment, immune response, and SCC. Prognostic association of miRNA expression in non-cancerous tissue is of particular interest because altered levels of these miRNAs may be evident prior to advanced disease stage and the occurrence of symptoms. In Barrett's associated ADC patients, high levels of mir-375, which targets genes associated with tumor cell proliferation, in the cancerous tissue was associated with worse prognosis. The ability to block miRNA transcription may open avenues for the possible use of miRNAs in identifying novel drug targets and therapies for esophageal carcinoma.
Supplementary Material
Acknowledgements
We thank Dr. Raymond Jones, John Cottrell, and Audrey Salabes at the University of Maryland and Baltimore Veterans Administration for tissue and data collection, and Dr. Leoni Leondaridis (Advance Medical Systems Consultants) for coordination of data from the National Death Index. We also thank Dr. Stefan Ambs at the National Cancer Institute and Dr. Joe Y. Chang at the University of Texas MD Anderson Cancer Center, Houston, TX for helpful discussion, and David B. Chou at the Howard Hughes Medical Institute for his technical assistance. This research was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institutes of Health. Giang Huong Nguyen was funded by the Howard Hughes Medical Institute Grant for Graduate Medical Education. Dr. Rosemary Braun is supported by the Cancer Prevention Fellowship Program, National Cancer Institute, National Institutes of Health, Bethesda, MD. Dr. Alan Casson was supported by the Nova Scotia Health Research Foundation and by the National Cancer Institute of Canada with funds from the Canadian Cancer Society. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Md. (http://biowulf.nih.gov).
Footnotes
Disclosure of Potential conflicts of Interests: No potential conflicts of interest are disclosed.
STATEMENT OF TRANSLATIONAL RELEVANCE Developing novel prognostic biomarkers and identifying novel drug targets and therapies in esophageal cancer is of great interest given the dismal survival rates and the debate over whether patients that have undergone chemoradiation therapy have longer survival rates than those that have not. Our results provide preliminary evidence for the potential clinical utility of microRNAs (miRNAs) as prognostic biomarkers for adenocarinoma and squamous cell carcinoma of the esophagus. Importantly, the ability to use antagomirs to block specific miRNA transcription opens avenues for the use of miRNAs in novel personal drug therapies for esophageal carcinoma. Furthermore, miRNAs predictive of prognosis could help to stratify patients that would benefit most from particular treatment regimens. The comprehensive multi-center study of esophageal cancer presented in this study provides a basis for potential future applications of miRNAs in the clinic.
Reference List
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Sant M, Aareleid T, Berrino F, et al. EUROCARE-3: survival of cancer patients diagnosed 1990-94--results and commentary. Ann Oncol. 2003;14(Suppl 5):v61–118. doi: 10.1093/annonc/mdg754. [DOI] [PubMed] [Google Scholar]
- 3.Surveillance and Epidemiology and End Results (SEER) 2007.
- 4.Crew KD, Neugut AI. Epidemiology of upper gastrointestinal malignancies. Semin Oncol. 2004;31:450–64. doi: 10.1053/j.seminoncol.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 5.Maley CC, Rustgi AK. Barrett's esophagus and its progression to adenocarcinoma. J Natl Compr Canc Netw. 2006;4:367–74. doi: 10.6004/jnccn.2006.0031. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 15.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 18.Ji J, Wang XW. New kids on the block: Diagnostic and prognostic microRNAs in hepatocellular carcinoma. Cancer Biol Ther. 2009;8 doi: 10.4161/cbt.8.18.8898. [DOI] [PubMed] [Google Scholar]
- 19.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.lorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 21.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulci V, Chiaretti S, Goldoni M, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–51. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 23.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with `antagomirs'. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 24.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008 doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 25.Sugito N, Ishiguro H, Kuwabara Y, et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–8. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- 26.Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255–60. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y, Chen Z, Zhang L, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 28.Watson DI, Wijnhoven BP, Michael MZ, Mayne GC, Hussey DJ. Hp24 microrna expression profiles in barrett's oesophagus. ANZ J Surg. 2007;77(Suppl 1):A45. [Google Scholar]
- 29.Liu CG, Calin GA, Meloon B, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–4. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–72. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 33.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 34.Hiyoshi Y, Kamohara H, Karashima R, et al. MicroRNA-21 Regulates the Proliferation and Invasion in Esophageal Squamous Cell Carcinoma. Clin Cancer Res. 2009 doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- 35.Sayed D, Rane S, Lypowy J, et al. MicroRNA-21 Targets Sprouty2 and Promotes Cellular Outgrowths. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surh YJ, Kundu JK, Na HK, Lee JS. Redox-sensitive transcription factors as prime targets for chemoprevention with anti-inflammatory and antioxidative phytochemicals. J Nutr. 2005;135:2993S–3001S. doi: 10.1093/jn/135.12.2993S. [DOI] [PubMed] [Google Scholar]
- 37.Seike M, Yanaihara N, Bowman ED, et al. A cytokine gene signature of the lung adenocarcinoma and its tissue environment predicts prognosis. J Natl Cancer Inst. 2007;99:1257–69. doi: 10.1093/jnci/djm083. [DOI] [PubMed] [Google Scholar]
- 38.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–30. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 39.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008 doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–80. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 41.Lawrie CH, Soneji S, Marafioti T, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–61. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 42.Loffler D, Brocke-Heidrich K, Pfeifer G, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 43.Poy MN, Hausser J, Trajkovski M, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813–8. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boult JK, Taniere P, Hallissey MT, Campbell MJ, Tselepis C. Oesophageal adenocarcinoma is associated with a deregulation in the MYC/MAX/MAD network. Br J Cancer. 2008;98:1985–92. doi: 10.1038/sj.bjc.6604398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogunwobi OO, Beales IL. Glycine-extended gastrin stimulates proliferation via. Mol Cell Endocrinol. 2008;296:94–102. doi: 10.1016/j.mce.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Hughes D, Guttenplan JB, Marcus CB, Subbaramaiah K, Dannenberg AJ. Heat shock protein 90 inhibitors suppress aryl hydrocarbon receptor-mediated activation of CYP1A1 and CYP1B1 transcription and DNA adduct formation. Cancer Prev Res (Phila Pa) 2008;1:485–93. doi: 10.1158/1940-6207.CAPR-08-0149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Chang EY, Smith CA, Corless CL, Thomas CR, Jr., Hunter JG, Jobe BA. Accuracy of pathologic examination in detection of complete response after chemoradiation for esophageal cancer. Am J Surg. 2007;193:614–7. doi: 10.1016/j.amjsurg.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Mooney MM. Neoadjuvant and adjuvant chemotherapy for esophageal adenocarcinoma. J Surg Oncol. 2005;92:230–8. doi: 10.1002/jso.20364. [DOI] [PubMed] [Google Scholar]
- 50.Sundelof M, Lagergren J, Ye W. Patient demographics and lifestyle factors influencing long-term survival of oesophageal cancer and gastric cardia cancer in a nationwide study in Sweden. Eur J Cancer. 2008 doi: 10.1016/j.ejca.2008.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.