Abstract
Consensus HIV-1 genes can decrease the genetic distances between candidate immunogens and field virus strains. To ensure the functionality and optimal presentation of immunologic epitopes, we generated two group-M consensus env genes that contain variable regions either from a wild-type B/C recombinant virus isolate (CON6) or minimal consensus elements (CON-S) in the V1, V2, V4, and V5 regions. C57BL/6 and BALB/c mice were primed twice with CON6, CON-S, and subtype control (92UG37_A and HXB2/Bal_B) DNA and boosted with recombinant vaccinia virus (rVV). Mean antibody titers against 92UG37_A, 89.6_B, 96ZM651_C, CON6, and CON-S Env protein were determined. Both CON6 and CON-S induced higher mean antibody titers against several of the proteins, as compared with the subtype controls. However, no significant differences were found in mean antibody titers in animals immunized with CON6 or CON-S. Cellular immune responses were measured by using five complete Env overlapping peptide sets: subtype A (92UG37_A), subtype B (MN_B, 89.6_B and SF162_B), and subtype C (Chn19_C). The intensity of the induced cellular responses was measured by using pooled Env peptides; T-cell epitopes were identified by using matrix peptide pools and individual peptides. No significant differences in T-cell immune-response intensities were noted between CON6 and CON-S immunized BALB/c and C57BL/6 mice. In BALB/c mice, 10 and eight nonoverlapping T-cell epitopes were identified in CON6 and CON-S, whereas eight epitopes were identified in 92UG37_A and HXB2/BAL_B. In C57BL/6 mice, nine and six nonoverlapping T-cell epitopes were identified after immunization with CON6 and CON-S, respectively, whereas only four and three were identified in 92UG37_A and HXB2/BAL_B, respectively. When combined together from both mouse strains, 18 epitopes were identified. The group M artificial consensus env genes, CON6 and CON-S, were equally immunogenic in breadth and intensity for inducing humoral and cellular immune responses.
Introduction
Since its discovery in 1981, human immunodeficiency virus type I (HIV-1) has exploded into a global pandemic. More than 60 million people have been infected, and 33 million are currently living with HIV-1.45 Because of the high level of genetic variation and the rapid increase in viral population, HIV-1 has evolved into nine defined genetically distinct viral subtypes.26 Regions, countries, and even cities have multiple HIV-1 subtypes cocirculating that give rise to recombinant circulating viruses. It has been determined that >20% of viral sequences are intersubtype recombinants.31,36 Previous vaccine studies have shown that a small amount of genetic divergence between the vaccine strain and the challenge strain will negate any protective immunity; therefore, it is unlikely that a single subtype will be effective at inducing immunity against natural challenge in such a diverse population.2,5,22,42
Several approaches have been investigated to overcome the challenge of genetic diversity. First, conserved T- and B-cell epitopes were explored, and many cross-subtype T-cell responses have been identified.9,15,17,18,43 Although T-cell epitopes are more easily defined than B = cell epitopes, several cross-subtype neutralizing antibodies have been identified and mapped.7,10,35,44,48–50 However, further experiments have failed to induce antibody responses to these epitopes, and passive transfer is not a practical prophylactic.21,27,32,46 Epitope vaccines are limited because viral-escape mutants are easily selected for during infection.1,4,8,39
Second, a multisubtype immunization has been investigated. Cocktails of peptides, proteins, DNA expression plasmids, and recombinant viral vectors have been used to increase the breadth of the anti–HIV-1 immune responses.6,14,23 Kong23 and Seaman41 have shown that T- and B-cell immune responses to polyvalent vaccines are equivalent to the immune responses induced by monovalent vaccines.
Finally, to minimize the genetic diversity between the immunogen and challenge strain, several investigators have proposed the use of a centralized HIV-1 gene as an immunogen. These centralized sequences can be established by using several methods: consensus, ancestral, mosaic, and center of tree (COT).16,19,20,24,25,37,38 All of these methods result in a sequence that localizes to the central polytomic node of an HIV-1 phylogenetic tree. Analysis of the synthetic centralized sequences indicates that many experimentally defined T-cell epitopes from many subtype HIV-1 viruses are preserved, indicating that centralized genes may induce broadly cross-reactive T-cell immune responses.20,40
Previous studies reported the generation of a group M consensus env gene (CON6). CON6 was biologically functional, used the CCR5 coreceptor, induced T-cell immune responses and neutralizing antibody against HIV-1 primary isolates.20 CON6 was compared with a multisubtype immunogen as well as three subtype immunogens (subtypes A, B, and C). T-cell data showed that the CON6 immunogen induced broader T-cell immune responses, as compared with single-subtype immunogens, and equally broad responses, as compared with the multisubtype immunogen.47
With the intention to preserve as much biologic function as possible, CON6 was generated without consensus variable loops, V1, V2, V4, and V5, because of variation, insertions, and deletions. Instead, these variable loops were replaced with subtype C env sequence from a CRF07_BC virus (Fig. 1). It is important to note that subtype C viruses, in general, have shorter variable regions as compared with the other subtypes. Later, a complete consensus env, CON-S, gene was made by using a minimal variable element approach (Fig. 1). This was accomplished by selecting a sequence restricted to the minimal-length env sequence in the alignment without completely removing the variable regions during phylogenetic analysis. In this study, we characterized the CON-S immunogen and compared the immune responses induced by CON6 immunogen and the fully consensus CON-S immunogen. Both CON6 and CON-S are biologically functional in that they form infectious pseudovirions and use the CCR5 coreceptor for entry (29). No significant differences in CON6- or CON-S–induced antibody titers or T-cell immune responses against Env peptide pools were found, and little difference was noted in the induced T-cell epitopes. These data indicate that CON6 and CON-S are comparable immunogens and have the potential to be used as a broadly cross-reactive immunogen for most HIV-1 subtypes.
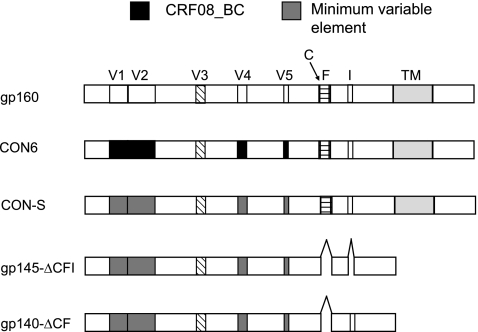
FIG. 1.
CON6 and CON-S were generated by using two group M consensus env genes that contain variable regions either from a wild-type B/C recombinant virus isolate (CON6) or minimal consensus elements (CON-S) in the V1, V2, V4, and V5 regions. The minimal variable elements and the subtype C sequence positions are shown as gray and black boxes, respectively. C, F, and I indicate the location of the cleavage, fusion, and intervening regions, respectively. gp145CFI and gp140CFI indicate the location of the deletions and truncations.
Methods and Materials
Env gp140 immunogens
To generate the gp145CFI membrane-bound envelope immunogens, CON6, CON-S, 92UG37_A and HXB2/Bal_B gp140CF plasmids were constructed by introducing a stop codon after the membrane-spanning domain (YIKIFIMIVGGLIGLRIVFAVLSIVN), a deletion of gp120/gp41 cleavage site and fusion domain of gp41, as previously described, and a deletion between HR1 and HR2 in gp41.6,10,12 The env genes were cloned into the DNA expression vector pCMV/R at XbaI and BamHI sites for use as DNA immunogens. The same genes, with the exception of an introduced stop codon before the membrane-spanning domain (gp140CFI), were also cloned into the shuttle plasmid pSC65 at SalI and KpnI sites to generate recombinant vaccinia viruses (rVVs), as previously described13,47 (Fig. 1).
Animal immunization
Female BALB/c and C57BL/6 mice (6–8 weeks old) were purchased from Charles River Laboratories (Wilmington, MA) and housed in the Duke University Animal Facility under the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC) guidelines with animal-use protocols approved by the Duke University Animal Use and Care Committee. Five mice per group were immunized intramuscularly (i.m.) in the quadriceps with gp145CFI and empty vector control DNA (50 μg) twice at 3-week intervals. Three weeks after the last DNA immunization, the mice were boosted with rVVs and wild-type VV (107 pfu) intraperitoneally. Two weeks after the boost, the mice were killed, and spleens were collected. Pre- and post-bleeds were obtained by retrorbital bleeding with a Natellson heparinized collection tube.
Enzyme-linked immunospot (ELISpot) assay
Three overlapping Env peptide sets from subtypes A, B, and C were used for T-cell epitope mapping. Subtype type A (92UG037_A) gp140 overlapping peptides were synthesized through SynPep Corporation (Dublin, CA). It consisted of 168 peptides that were 15 amino acids long with an 11-amino-acid overlap. Subtype B (SF162P3 _B) and subtype C (Chn19_C) peptides were obtained from the NIH AIDS Research and Reference Reagent Program (Bethesda, MD). The SF162P3 peptide set consisted of 182 peptides that were 15 amino acids long with an 11-amino = acid overlap. The Chn19 peptide set consisted of 72 peptides that were 20 amino acids long with a 10-amino-acid overlap. Spleens from individual mice were minced and then forced through a 70-μm nylon cell strainer (BD Labware, Franklin Lakes, NJ). ELISPOTS were performed according to manufacturer protocol (Mabtech, Stockholm, Sweden). In brief, single-cell suspensions of splenocytes were plated in 96-well polyvinylidene difluoride–backed plates (MultiScreen-IP; Millipore, Billerica, MA) coated with 50 μl of anti-mouse IFN-γ mAb AN18 (5 μg/ml; Mabtech) overnight at 4°C. The plates were blocked with Hepes-buffered complete RPMI medium at 37°C for 2 h. Equal volumes (50 μl) of each peptide pool and splenocytes (107 cells/ml) were added to the wells in duplicate. Plates were incubated overnight (14–16 h) at 37°C with 5% CO2. After the plates were washed 6 times with PBS, 50 μl of 1:1,000-diluted biotinylated anti-mouse IFN-γ mAb (Mabtech) was added to each well. Plates were incubated at RT for 2 h and then washed 3 times with PBS. Fifty microliters of streptavidin-alkaline phosphatase conjugate (1:1,000 dilution; Mabtech) was added to each well. After incubation at RT for 1 h, the plates were washed 5 times with PBST. Finally, 100 μl of BCIP/NBT (Plus) alkaline phosphatase substrate (Moss, Pasadena, MD) were added to each well. The plates were incubated at RT for 10 min. After washing with water, plates were air-dried. Spots were counted by using an automated ELISpot plate reader (Immunospot counting system, CTL Analyzers) and expressed as spot-forming cells (SFCs) per 106 splenocytes. Responses were considered positive if the numbers of the spots were 4 times more than those of the negative control and ≥50 SFC/106 cells per well. CD4 and CD8 T-cell depletion and enrichment were performed by using immunomagnetic depletion beads according to the manufacturer's instruction (Dynal Biotech ASA, Oslo, Norway). ELISpot assays were performed with CD4 or CD8-depleted cells to determine whether the T-cell epitopes were either MHC class I or II restricted.
Antibody titers
ELISA plates were coated overnight at 4°C with 100 μl/well HIV-1 Env protein at a concentration of 5 μg/ml in PBS. The plates were washed twice with PBS containing 0.1% Tween 20 (PBST) and blocked with 300 μl blocking buffer (4% BSA and 0.01% NaN3 in PBS) for 2 h at RT. Diluted mouse sera (100 μl) was added into the antigen-coated microtiter wells and incubated for 1 h at RT. After the plate was washed 3 times with PBST, it was incubated with the 1:2,000 diluted goat anti-mouse IgG alkaline phosphatase conjugate (Sigma, St. Louis, MO) for 1 h at RT. Substrate p-nitrophenyl phosphate disodium (PIERCE, Rockford, IL) was added to each well for color development. Absorbance was measured at 405 nm with the Wallac 1420 Multilabel Counter (Perkin Elmer Life Sciences, Boston, MA). The titer was determined as the highest dilution that was twofold higher than the prebleed sera.
Statistical analysis
Antibody and pooled T-cell immune responses were analyzed by using a Student's t test (Microsoft Excel) and are considered significant if p ≤ 0.05. Error bars in all figures represent standard error (n = 5).
Results
Induced humoral immune responses
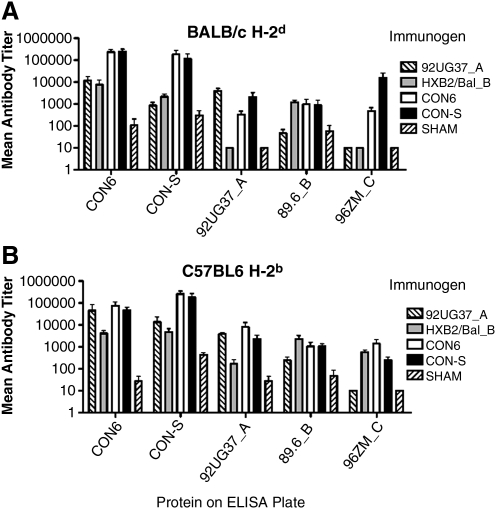
No statistically significant differences in the mean antibody titers induced in CON6 and CON-S immunized animals against all proteins in both BALB/c and C57BL/6 mice (Fig. 2). The SHAM group represents animals immunized with empty DNA expression plasmid and boosted with vaccinia virus WR that did not express any HIV envelope immunogen. Undetectable antibody titers were assigned a value of 10 for statistical evaluation. Figure 2 indicates some level of anti-envelope antibody in the SHAM group; however, this is due to the inability to assay titers >10 and the small amount of nonspecific binding typical at low serum dilutions. This low level of anti-envelope antibody was not significant and was consistently several orders of magnitude less than that of immunized mice. In BALB/c mice, CON6 and CON-S induced significantly higher GMTs than both subtype controls against CON-S Env protein. CON6 induced significantly higher mean antibody titers against 96ZM651_C protein than did 92UG37_A and HXB2/Bal_B (p ≤ 0.02). 92UG37_A induced a higher GMT than HXB2/Bal_B and CON6 against 92UG37_A_B Env protein, whereas CON6 had a higher mean antibody titer than HXB2/Bal_B against 92UG37_A Env protein (p ≤ 0.05) (Fig. 2A). In C57BL/6 mice, the subtype control immunized mice had a higher mean antibody titer against the homologous Env protein, whereas no significant differences between CON6 and CON-S immunogens were found, as compared with the homologous immunogen against the same Env protein (p ≤ 0.05) (Fig. 2B). HXB2/Bal_B, CON-S, and CON6 Env protein induced greater mean antibody titers against 96ZM651_C Env protein; however, the data were not statistically significant (Fig. 2B). Data in this and other studies consistently showed that the consensus immunogens induce higher anti-envelope immune responses as compared with wild-type immunogens.29,40,47 Although the data presented here only indicate anti-envelope immune responses, they do not measure effective HIV-neutralizing antibodies. However, Liao et al.29 studied differences between CON6 and CON-S and showed that both of the consensus immunogens induced greater cross-reactive HIV-neutralizing antibodies as compared with wild-type immunogens.
FIG. 2.
Mean antibody titers induced in BALB/c (A) and C57BL/6 (B) mice by 92UG37_A, HXB2/Bal_B, CON6, and CON-S. Mice were immunized twice i.m. with 50 μg of DNA expression plasmid followed by boosting with 107 pfu envelope expressing recombinant vaccinia virus. Two weeks after boosting, sera were used to measure anti-HIV-1 envelope humoral immune responses, as measured by ELISA assay. Undetectable antibody was reported as a titer of 10 for statistical purposes. Error bars indicate standard error (n = 5).
Cellular Immune responses to three pooled Env peptide sets
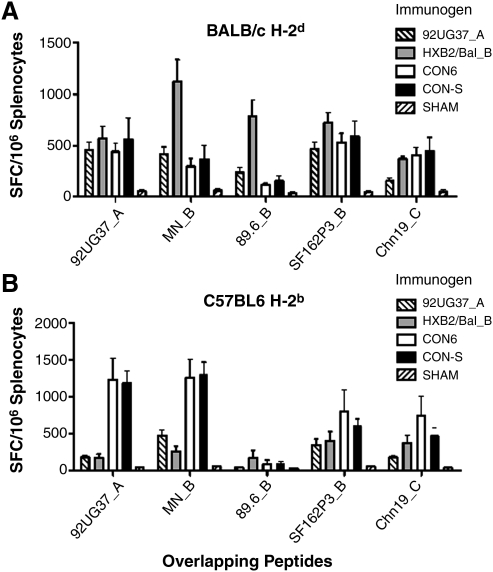
To determine the relative intensity of induced T-cell immune responses, individual mouse splenocytes were assayed for the ability to recognize peptide pools that contained all of the overlapping Env peptides for each set. No significant differences were seen between CON6 and CON-S induced T-cell responses to Env peptide pools in both BALB/c and C57BL/6 mouse strains (Fig. 3). In BALB/c mice, CON6, CON-S, and HXB2/Bal_B induced significantly greater T-cell immune responses against Chn19_C Env peptides than did 92UG37_A (p ≤ 0.05) (Fig. 3A). In C57BL/6 immunized mice, CON6 and CON-S induced significantly greater T-cell immune responses against subtype A peptides, UG37_A, than did 92UG37_A and HXB2/Bal_B. CON6 and CON-S also induced greater immune responses against subtype C, Chn19_C, Env peptides than did 5304_A (p ≤ 0.05) (Fig. 3B).
FIG. 3.
The intensity of T-cell immune responses against overlapping envelope peptide pools in BALB/c (A) and C57BL/6 (B) mice. Mice were immunized twice i.m. with 50 μg of DNA expression plasmid followed by boosting with 107 pfu envelope expressing recombinant vaccinia virus. Two weeks after boosting, the mice were killed, and splenocytes were used to measure total T-cell responses to overlapping envelope peptide pools, as measured by ELISPOT assay. Error bars indicate standard error (n = 5).
Breadth of T-cell epitopes
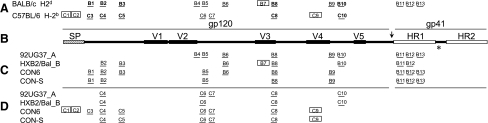
To determine the relative breadth of CON6 and CON-S immunogens, the T-cell epitopes were identified by using matrix peptide pools and individual peptides. Because of the large number of overlapping Env peptide sets, the epitopes were screened and identified by using pooled splenocytes from all five mice in each group. In BALB/c and C57BL/6 mice, 13 and 10 T-cell epitopes were identified, respectively. The epitopes for BALB/c and C57BL/6 were designated B1–B13 and C1–C10, respectively (Fig. 4A). In BALB/c mice, CON6 and CON-S induced responses to 10 and eight epitopes, respectively. Both subtype controls, 92UG37_A and HXB2/Bal_B, induced responses to eight epitopes (Table 1 and Fig. 4C). In C57BL/6 mice, CON6 and CON-S induced responses to nine and six T-cell epitopes, respectively. The subtype controls, 92UG37_A and HXB2/Bal_B, induced responses to four and three T-cell epitopes, respectively. Three epitopes were found to be cross-reactive: C4, C6, and C8. CON6 and CON-S induced responses to several epitopes (C1, C2, C3, C5, and C9) that the subtype controls did not; whereas the subtype controls only induced responses to one epitope, C10, that was not induced by CON6 or CON-S (Table 1 and Fig. 4D). These results were confirmed in previous and successive studies.
FIG. 4.
T-cell epitope mapping of CON6, CON-S, and subtype Env immunogens. All of the epitopes identified in both MHC haplotypes are shown in (A). A schematic representation of the HIV-1 envelope relative structural elements is represented in (B). The approximate location of the epitopes induced by the envelope immunogens in BALB/c and C57BL/6 mice are shown in (C) and (D), respectively. Lines represent the epitope positions relative to the signal peptide, variable regions, gp120, gp41, and heptad repeats. Boxed epitopes are MHC class 1 restricted CTL epitopes, and bolded epitopes are common in both MHC haplotypes. BALB/c epitopes are labeled B1–B13, and C57BL/6 epitopes are labeled C1–C10.
Table 1.
The Number of T Cell Epitopes Induced by HIV Envelope Immunogens
| |
Immunogen |
|||
|---|---|---|---|---|
| H-2d | 92UG37_A | HXB2/Bal_B | CON6 | CON-S |
| No. of epitopes | ||||
| Subtype A | 6 | 5 | 7 | 7 |
| Subtype B | 8 | 5 | 6 | 5 |
| Subtype C | 1 | 4 | 5 | 3 |
| Total | 8 | 6 | 10 | 8 |
| H-2b | ||||
| No. of epitopes | ||||
| Subtype A | 2 | 1 | 5 | 4 |
| Subtype B | 3 | 3 | 6 | 5 |
| Subtype C | 1 | 2 | 5 | 2 |
| Total | 4 | 4 | 9 | 6 |
Mice were immunized with subtype A, Subtype B, CON6 and CON-S immunogens. The mice were immunized i.m. 2X with 50 mg of DNA expression plasmid followed by boosting with 107 pfu of envelope expressing vaccinia virus. Two weeks after boosting the splenocytes were used to map epitopes induced by the immunogens using the ELISPOT assay and overlapping envelope peptide sets corresponding to subtype A (92UG37_A), subtype B (MN, 89.6, SF162) and subtype (Chn19P) wild type HIV-1 envelope sequences.
Location of T cell epitopes and identity of conserved epitopes
Because CON6 uses subtype C variable loops, it was possible that CON6 would be biased toward subtype C immunogenicity. However, few T-cell epitopes were found to localize in the variable regions. Epitope C9 localized to the V4 region, whereas epitopes B4 and B9 overlapped with V2 and V4, respectively (Fig. 4). Three epitopes, B7, B8, and C8, localized to V3; however, the V3 regions in CON6 and CONS are identical. CON6 induced two CTL epitopes in the signal peptide of the envelope, and both CON6 and CON-S induced the CTL epitope C9 in the V4 variable region (Fig. 4). Epitopes B8, B11, and B12 were found to be broadly cross-reactive, in that they were induced by all four immunogens. Epitopes B4, B7, and B10 were not detected by either CON6 or CON-S immunized animals, whereas both CON6- and CON-S–induced epitope B1 and 92UG37_A and HXB2/Bal_B did not (Fig. 4). Supplemental figures show the relative position and peptide sequences to map the T-cell epitopes in both BALB/c and C57BL mice. The numbers flanking the peptide sequences indicate the catalog number of the peptide that was received from the NIH AIDS Reagent and Repository.
Determination of MHC class I or II restriction on T-cell epitopes
To determine whether responding T cells were either CD8 or CD4, we performed ELISpot assays with either CD4- or CD8-depleted cells for all positive peptides in both mouse strains. T-cell responses that were generated by CD8 T cells were considered MHC I–restricted CTL epitopes, and responses that were derived from CD4 T cells were considered MHC II–restricted Th cells. Four epitopes (B7, C1, C2, and C9) were recognized by CD8 T cells in BALB/c or C57BL/6 mice. All other epitopes were recognized by CD4 T cells (Fig. 4 and Supplemental Figures 1 and 2).
Discussion
One of the most difficult obstacles to overcome in developing an HIV vaccine is the high level of genetic diversity. In this study, we showed that no significant differences exist between CON6 and CON-S in inducing humoral and cellular immunity and that both immunogens have advantages over subtype-specific immunogens, in that they induce more T-cell epitopes at a greater intensity and at equivalent or greater humoral responses. These data suggest that both CON6 and CON-S have the potential to be further developed as a broadly reactive AIDS vaccine.
When investigating the humoral immune responses induced by the consensus and subtype immunogens, we found no differences between CON6 and CON-S against all Env proteins in both mouse strains. In general, consensus immunized animals had higher or equivalent mean antibody titers than did subtype immunized animals. Also, HXB2/Bal_B–immunized animals had higher mean antibody titers against the consensus Env proteins than homologous protein, indicating that these antibodies had a higher affinity for B-cell epitopes presented by CON6 and CON-S. The subtype immunogens were biased toward homologous Env protein, whereas CON6 and CON-S showed little variation against all three subtype Env proteins. Also, CON6 and CON-S were the only immunogens to induce anti-96ZM651_C antibodies consistently. The ability of both CON6 and CON-S to induce neutralizing antibodies against subtypes B and C primary HIV-1 isolates has been reported (29). Liao et al.29 showed that the variable regions played a significant role in the induction of neutralizing antibodies and showed that the minimal variable-region approach resulted in more neutralization, especially in regard to the V3 region.
When the cellular immune responses were examined, again, we found that CON6 and CON-S were equivalent at inducing responses against subtype epitopes in both mouse strains. Neither the subtype nor the consensus immunogens showed a bias toward a particular subtype. An explanation for a greater intensity of cellular immune responses in CON6 and CON-S immunized C57BL/6 mice could be that the 3 CTL epitopes were induced only by the consensus immunogens. Two of the CTL epitopes reacted to Chn19_C peptides, and one reacted to UG37 peptides. The two CTL epitopes in the signal peptide of the CON6 were not in CON-S, even though these sequences were the same. This may be due to differing proteolytic processing of the genes because of the differing variable regions. Because these peptides are synthetic and overlap by 10 or 11 amino acids, they do not realistically represent a processed and presented antigen. Therefore, the in vitro immune responses may vary from those in vivo.
Relatively few differences were found in the T-cell responses induced by CON6 and CON-S. In both mouse strains, CON6 induced more responses than CON-S. This is interesting because the epitopes that were absent in CON-S were not in the variable regions, and the sequence homology between the two immunogens is identical in the conserved regions. This may be a result of deleted or mutated proteolytic cleavage sites, sequence effects on presentation, or alterations in ubiquination. However, in both mouse strains, CON6 and CON-S induced more T-cell responses than did the subtype immunogens. Although this study is not designed to evaluate the protective nature of the T-cell epitopes, theoretically it is better to have more epitopes, because HIV can lose epitopes by mutation, as shown in previous studies.1,3–5,8,32,39 Also, CTL epitopes have been shown to provide additional suppressive activity, either through direct control of HIV infected cells or perhaps through secreted soluble anti-HIV factors, as shown in several CD8+ T-cell depletion studies.1,3–5,8,32,39 Interestingly, the Th1 and Th2 biased backgrounds of C57BL6 and BALB/c mice, respectively, were evident in the immune responses induced by the consensus immunogens, whereas both CON6 and CON-S induced greater anti-HIV humoral responses in the BALB/c mice as compared with the C57BL6 mice. However, both CON6 and CON-S induced greater anti-HIV cellular responses in the C57BL6 mice as compared with the BALB/c mice. This may be due simply to the increased numbers of CTL and Th epitopes induced in the consensus immunogens or may be the result of synergistic immune stimulation induced by cooperative Th and CTL cytokine secretion and regulation.
In this study, we showed that little difference exists between consensus immunogens with wild-type or subtype C variable regions and that both consensus immunogens are better at inducing cross-clade immunity than are subtype-specific immunogens. However, it must be noted that this study examined the induction of humoral and cellular immune responses against wild-type and consensus immunogens in two mouse strains and indicates only the possibility of what could be seen in a more diverse MHC, HLA, or Mamu genetic background; this provides evidence that further investigation is warranted.
Supplementary Material
Acknowledgments
We would like to thank the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH for HIV-1 overlapping envelope peptides. This work was supported by the NIH grants AI055386, AI061734, and AI064518. E.A.W. and Z.T.C. were supported by a NIH training grant 5T32 AI07392.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Allen TM. O'Connor DH. Jing P, et al. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 2.Amara RR. Villinger F. Altman JD, et al. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Vaccine. 2002;20:1949–1955. doi: 10.1016/s0264-410x(02)00076-2. [DOI] [PubMed] [Google Scholar]
- 3.Barouch DH. Kunstman J. Glowczwskie J, et al. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J Virol. 2003;77:7367–7375. doi: 10.1128/JVI.77.13.7367-7375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch DH. Kunstman J. Kuroda MJ, et al. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature. 2003;415:335–339. doi: 10.1038/415335a. [DOI] [PubMed] [Google Scholar]
- 5.Barouch DH. Santra S. Schmitz JE, et al. Control of viremia, prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett JA. Wasserman SS. Hicks CB, et al. Safety and immunogenicity of an HLA-based HIV envelope polyvalent synthetic peptide immunogen. DATRI 010 Study Group. Division of AIDS Treatment Research Initiative. Aids. 1998;12:1291–1300. doi: 10.1097/00002030-199811000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Binley JM. Wrin Korber B, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:3232–3252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borrow P. Lewicki H. Wei X, et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 9.Braibant M. Brunet S. Costagliola D, et al. Antibodies to conserved epitopes of the HIV-1 envelope in sera from long-term non-progressors: prevalence and association with neutralizing activity. Aids. 2006;20:1923–1930. doi: 10.1097/01.aids.0000247113.43714.5e. [DOI] [PubMed] [Google Scholar]
- 10.Burton DR. Pyati J. Koduri R, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 11.Castro BA. Homsy J. Lennette E. HIV-1 expression in chimpanzees can be activated by CD8+ cell depletion or CMV infection. Clin Immunol Immunopathol. 1992;65:227–233. doi: 10.1016/0090-1229(92)90151-d. [DOI] [PubMed] [Google Scholar]
- 12.Catanzaro AT. Roederer M. Koup RA, et al. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine. 2007;25:4085–4092. doi: 10.1016/j.vaccine.2007.02.050. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarti S. Sisler JR. Moss BB, et al. Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques. 1997;23:1094–1097. doi: 10.2144/97236st07. [DOI] [PubMed] [Google Scholar]
- 14.Cho MW. Kim YB. Lee MK. Polyvalent envelope glycoprotein vaccine elicits a broader neutralizing antibody response but is unable to provide sterilizing protection against heterologous Simian/human immunodeficiency virus infection in pigtailed macaques. J Virol. 2001;75:2224–2234. doi: 10.1128/JVI.75.5.2224-2234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Groot AS. Rivera DS. McMurry JA, et al. Identification of immunogenic HLA-B7 “Achilles' heel” epitopes within highly conserved regions of HIV. Vaccine. 2008;26:3059–3071. doi: 10.1016/j.vaccine.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellenberger DL. Li B. Lupo LD, et al. Generation of a consensus sequence from prevalent and incident HIV-1 infections in West Africa to guide AIDS vaccine development. Virology. 2002;302:155–163. doi: 10.1006/viro.2002.1577. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari G. Humphrey W. McElrath MJ, et al. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci U S A. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari G. Kostyu DD. Cox J, et al. Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines. AIDS Res Hum Retroviruses. 2000;16:1433–1443. doi: 10.1089/08892220050140982. [DOI] [PubMed] [Google Scholar]
- 19.Gao F. Korber BT. Weaver E, et al. Centralized immunogens as a vaccine strategy to overcome HIV-1 diversity. Expert Rev Vaccines. 2004;3:S161–S618. doi: 10.1586/14760584.3.4.s161. [DOI] [PubMed] [Google Scholar]
- 20.Gao F. Weaver EA. Lu Z, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol. 2005;79:1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauduin MC. Parren PW. Weir R, et al. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat Med. 1997;3:1389–1393. doi: 10.1038/nm1297-1389. [DOI] [PubMed] [Google Scholar]
- 22.Jin X. Bauer DE. Tuttleton SE, et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong WP. Huang Y. Yang ZY, et al. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J Virol. 2003;77:12764–12772. doi: 10.1128/JVI.77.23.12764-12772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong WP. Wu L. Wallstrom TC, et al. Expanded breadth of the T-cell response to mosaic human immunodeficiency virus type 1 envelope DNA vaccination. J Virol. 2009;83:2201–2215. doi: 10.1128/JVI.02256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korber B. Muldoon M. Theiler J. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 26.Leitner T, et al. HIV sequence compendium 2003. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; Los Alamos, New Mexico: 2004. [Google Scholar]
- 27.Letvin NL. Montefiori DC. Yasutomi Y, et al. Potent, protective anti-HIV immune responses generated by bimodal HIV envelope DNA plus protein vaccination. Proc Natl Acad Sci U S A. 1997;94:9378–9383. doi: 10.1073/pnas.94.17.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letvin NL. Schmitz JE. Jordan HL, et al. Cytotoxic T lymphocytes specific for the simian immunodeficiency virus. Immunol Rev. 1999;170:127–134. doi: 10.1111/j.1600-065x.1999.tb01334.x. [DOI] [PubMed] [Google Scholar]
- 29.Liao HX. Sutherland LL. Xia SM, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lifson JD. Rossio JL. Piatak M, Jr, et al. Role of CD8(+) lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J Virol. 2001;75:10187–10199. doi: 10.1128/JVI.75.21.10187-10199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machuca A. Tang S. Hu J, et al. Increased genetic diversity and intersubtype recombinants of HIV-1 in blood donors from urban Cameroon. J Acquir Immune Defic Syndr. 2007;45:361–363. doi: 10.1097/QAI.0b013e318053754c. [DOI] [PubMed] [Google Scholar]
- 32.Mascola JR. Stiegler G. VanCott TC, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 33.Matano T. Shibata R. Siemon C, et al. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol. 1998;72:164–169. doi: 10.1128/jvi.72.1.164-169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metzner KJ. Jin X. Lee FV, et al. Effects of in vivo CD8(+) T cell depletion on virus replication in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J Exp Med. 2000;191:1921–1931. doi: 10.1084/jem.191.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore JP. Cao Y. Conley AJ, et al. Studies with monoclonal antibodies to the V3 region of HIV-1 gp120 reveal limitations to the utility of solid-phase peptide binding assays. J Acquir Immune Defic Syndr. 1994;7:332–229. [PubMed] [Google Scholar]
- 36.Munoz-Nieto M. Perez-Alvarez L. Thomson M, et al. IV type 1 intersubtype recombinants during the evolution of a dual infection with subtypes B and G. AIDS Res Hum Retroviruses. 2008;4:337–343. doi: 10.1089/aid.2007.0230. [DOI] [PubMed] [Google Scholar]
- 37.Nickle DC, et al. Consensus and ancestral state HIV vaccines (Letter) Science. 2003;24:299. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- 38.Novitsky V. Smith UR. Gilbert P, et al. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J Virol. 2002;76:5435–5451. doi: 10.1128/JVI.76.11.5435-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peyerl FW. Barouch DH. Yeh WW, et al. Simian-human immunodeficiency virus escape from cytotoxic T-lymphocyte recognition at a structurally constrained epitope. J Virol. 2003;77:12572–12578. doi: 10.1128/JVI.77.23.12572-12578.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santra S. Korber BT. Muldoon M, et al. A centralized gene-based HIV-1 vaccine elicits broad cross-clade cellular immune responses in rhesus monkeys. Proc Natl Acad Sci U S A. 2008;105:89–94. doi: 10.1073/pnas.0803352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seaman MS. Xu L. Beaudry K, et al. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J Virol. 2005;79:2956–2963. doi: 10.1128/JVI.79.5.2956-2963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiver JW. Fu TM. Chen L, et al. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature. 2002;415:331–335. doi: 10.1038/415331a. [DOI] [PubMed] [Google Scholar]
- 43.Sreepian A. Srisurapanon S. Horthongkham N, et al. Conserved neutralizing epitopes of HIV type 1 CRF01_AE against primary isolates in long-term nonprogressors. AIDS Res Hum Retroviruses. 2004;20:531–542. doi: 10.1089/088922204323087787. [DOI] [PubMed] [Google Scholar]
- 44.Trkola A. Pomales AB. Yuan H, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.UNAIDS/WHO. Report on the global AIDS epidemic. 2008. 2008. UNAIDS/WHO.
- 46.Veazey RS. Shattock RJ. Pope M, et al. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat Med. 2003;9:34–36. doi: 10.1038/nm833. [DOI] [PubMed] [Google Scholar]
- 47.Weaver EA. Lu Z. Camacho ZT, et al. B-subtype T-cell immune responses induced by a human immunodeficiency virus type 1 group m consensus env immunogen. J Virol. 2006;80:6745–6756. doi: 10.1128/JVI.02484-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwick MB. Bonnycastle LL. Menendez A, et al. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J Virol. 2001;75:6692–6699. doi: 10.1128/JVI.75.14.6692-6699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zwick MB. Labrijn AF. Wang W, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2002;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zwick MB. Wang M. Poignard P, et al. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J Virol. 2001;75:12198–12208. doi: 10.1128/JVI.75.24.12198-12208.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.