Abstract
Human immunodeficiency virus type 1 (HIV) infection of the central nervous system frequently causes HIV-associated neurocognitive disorders (HAND). The role of HIV Nef and other accessory proteins in HAND pathogenesis is unclear. To determine whether HIV nef undergoes adaptive selection in brain, we cloned 100 nef sequences (n = 30 brain and n = 70 lymphoid) from four patients with AIDS and HIV-associated dementia (HAD). Normalized nonsynonymous substitutions were more frequent at the divergence of lymphoid and brain sequences, indicating stronger adaptive selection in brain compared to lymphoid tissue. Brain-specific nonsynonymous substitutions were found within an NH3-terminal CTL epitope, the PACS1 binding motif, or positions predicted to be important for activation of the myeloid-restricted Src family tyrosine kinase Hck. These results suggest that adaptive selection of HIV nef in brain may reflect altered requirements for efficient replication in macrophages and brain-specific immune selection pressures.
Human immunodeficiency virus type I (HIV) infects macrophages and microglia in the central nervous system (CNS) and causes HIV-associated neurocognitive disorders (HAND) in 10–30% of AIDS patients.1 Env and nef, two genes likely to influence HAND pathogenesis,2,3 undergo divergent evolution between brain and lymphoid tissue,3–5 suggesting viral adaptation to the unique brain environment. Likewise, divergence of cerebrospinal fluid (CSF) and lymphoid env sequences correlates with an increase in adaptive selection.6,7 A functional consequence is likely to be an increased capacity to replicate in macrophages and microglia, the main target cells for HIV infection in brain.1,8 Nef expression enhances HIV replication in macrophages, influences the release of proinflammatory cytokines from infected macrophages,3,9 and induces neurological disease in a murine model when expressed in the absence of other HIV proteins.10 If Nef undergoes functional adaptation in the brain environment, phylogenetic analysis of brain and lymphoid nef sequences may reveal evidence of adaptive selection.
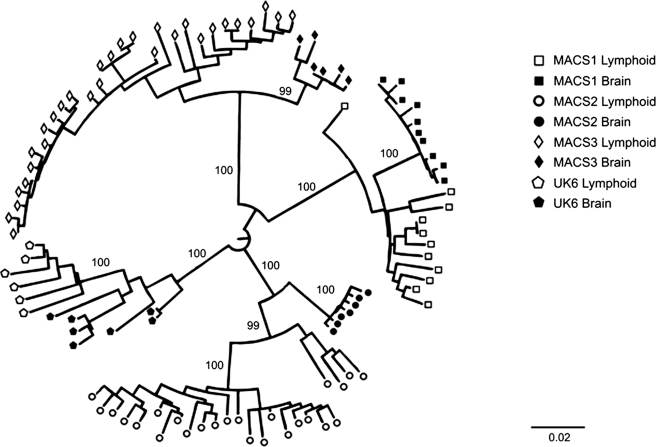
To generate a dataset of nef sequences for phylogenetic analysis, we used single HIV genome copies to polymerase chain reaction (PCR) amplify nef from autopsy tissues from four AIDS patients with HIV-associated dementia (HAD) and moderate to severe encephalitis (Table 1). Nef sequences from patients MACS2 and MACS3 were previously reported by Agopian et al.5,11 The sequences from patients MACS1 and UK6 were not previously described. Viral loads in the brain were ∼2- to 150-fold lower than in spleen or lymph node (Table 1). PCR amplified nef genes were sequenced and 100 unique sequences from all four patients were used to construct a neighbor-joining phylogenetic tree based on Maximum Composite Likelihood distances (Fig. 1).12 The consensus tree out of 10,000 bootstrapped trees is shown. Sequences from each patient clustered together, forming patient-specific nodes. Furthermore, sequences from each tissue clustered together, forming two tissue-specific nodes within each patient-specific node. Using the Slatkin–Maddison-based Panmixis test,13 distinct compartmentalization of Nef sequences by tissue was highly significant (p-values <0.0001 for all four patient sets).
Table 1.
Clinical Characteristics of Study Subjects and Tissue Sample Viral Loads
| |
|
|
|
Viral load (HIV copy #/105cells)b |
|
|---|---|---|---|---|---|
| Patient | Last CD4 count (cells/μl) | Clinical dementiaa | HIV encephalitisa | Brainc | Lymphoidd |
| MACS1e | 2 | Yes | Severe | 194 | 254 (SP) |
| MACS2e | 52 | Yes | Moderate | 156 | 1080 (LN) 300 (SP) |
| MACS3e | 95 | Yes | Moderate | 80 | 12600 (LN) |
| UK6 | 54 | Yes | Severe | 82 | 291 (SP) |
Determined by clinical history prior to and neuropathological examination at autopsy.
Real time PCR quantitation of HIV genomes/(Real time PCR quantitation of CCR5 copies/2 copies of CCR5 per cell × 105 cells).
Brain samples were collected from the frontal lobe.
Lymphoid tissue samples were collected from spleen (SP) and/or lymph node (LN).
FIG. 1.
Phylogenetic analysis of brain- and lymphoid-derived Nef sequences. The consensus neighbor joining tree based on 10,000 reconstructions was created using MEGA 4.0 software using the Maximum Composite Likelihood method. The input CLUSTAL W nucleotide alignment included sequences from all patients and tissues. The scale bar (lower right) represents the length of a branch where 2 nucleotide substitutions occurred out of 100 total nucleotides. Significant bootstrap values (>95) representing the frequency out of 100 trees in which the node occurs are shown at the indicated positions.
Brain nefs were more closely related to each other than were lymphoid nefs in all four patients. Indeed, the average genetic distance between brain nef sequences was 1.3- to 5-fold closer than that between lymphoid nef sequences (Table 2). Reduced genetic distance in brain nef sequences was significant for MACS1 and MACS2 (Table 2, Student's t test p-value <0.0001) but not for MACS3 and UK6 (p-values = 0.0574 and 0.1080, respectively). Therefore, reduced genetic diversity, as measured by genetic distance within the brain, is not a conserved feature of brain nef sequences.
Table 2.
Diversity and Selection within and between Tissue Compartments
| |
|
Within lymphoid |
Within brain |
Between brain and lymphoid |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Sequence comparison | Min.a | Avg.a | Max.a | Min. | Avg. | Max. | Min. | Avg. | Max. |
| MACS1 | Genetic distanceb | 0.3 | 2.0 | 3.8 | 0.0 | 0.4c | 1.0 | 2.40 | 3.4c | 4.03 |
| dS/dNd | 1.23 | 9.14 | 42.8 | 1.00 | 1.48c | 3.75 | 3.49 | 5.53c | 7.92 | |
| MACS2 | Genetic distanceb | 0.3 | 2.4 | 5.6 | 0.00 | 1.1c | 2.0 | 4.4 | 5.4c | 6.4 |
| dS/dNd | 1.23 | 4.59 | 46.9 | 1.00 | 1.62c | 3.68 | 0.95 | 1.71c | 2.48 | |
| MACS3 | Genetic distanceb | 0.0 | 1.7 | 4.2 | 0.33 | 1.1 | 1.8 | 2.0 | 3.4c | 6.1 |
| dS/dNd | 0.91 | 3.77 | 22.3 | 1.00 | 1.23c | 1.82 | 1.38 | 2.81c | 4.52 | |
| UK6 | Genetic distanceb | 0.8 | 2.7 | 1.4 | 0.2 | 2.1 | 4.5 | 3.3 | 4.7c | 6.0 |
| dS/dNd | 0.74 | 8.12 | 21.4 | 0.72 | 3.94c | 5.32 | 0.98 | 2.76c | 6.36 | |
Min., minimum observed value; Avg., average observed value; Max., maximum observed value.
Genetic distance is the number of nucleotide substitutions per 100 sites calculated with the maximum composite likelihood method in Mega 4.0 software. The average pairwise genetic distances for all lymphoid sequences (Within lymphoid), for all brain sequences (Within brain), and between each brain and lymphoid sequence (Between brain and lymphoid) are shown.
Significantly different than within lymphoid (Mann–Whitney test p-value <0.0001).
SNAP (Synonymous Nonsynonymous Analysis Program) was used to calculate the Jukes–Cantor correction of the observed proportion of synonymous substitutions relative to the number of total possible synonymous substitutions (dS) and the Jukes–Cantor correction of the observed proportion of nonsynonymous substitutions relative to the number of total possible nonsynonymous substitutions (dN). The ratio of dS to dN was subsequently calculated. Average values for the groups described above are shown.
To determine whether nef is under brain-specific selection pressures in late stage disease, the SNAP analysis program (available at http://www.hiv.lanl.gov) was used to compare the average ratio of normalized synonymous to nonsynonymous substitutions (dS/dN). In all four patients, dS/dN values were consistently higher in lymphoid tissue than in brain, implying that nef in lymphoid tissue is under stronger negative selection pressure (Table 2). To determine what type of selection occurred at the divergence of lymphoid and brain sequences, the average dS/dN values within each tissue or between brain and lymphoid sequences were generated for each patient (Table 2). The dS/dN value between brain and lymphoid tissue was 1.3- to 2.9-fold lower than the dS/dN value within lymphoid tissue (Mann–Whitney p-value <0.0001). This finding suggests stronger adaptive selection occurred at the divergence between brain and lymphoid sequences.
The dS/dN ratio is an index of the cumulative number of substitutions but does not identify the codons where these substitutions occurred. To identify codons undergoing positive selection at the divergence between lymphoid and brain, we employed SLAC analysis14 (Single Likelihood Ancestor Counting, HyPhy software) to calculate the probability that codon variation is due to either negative or positive selection. SLAC identifies codons under positive (adaptive) selection when significantly greater than expected amounts of nonsynonymous substitutions are found. When significantly greater than expected amounts of synonymous substitutions are found, the codon is categorized as being under negative (purifying) selection. Analysis of brain and lymphoid sequences from all four patients identified five codons under positive selection (Table 3). At codons 32 and 158, positive selection is a result of variation between patients rather than between tissues. Codons 10 and 194 were conserved within each tissue, but varied between brain and lymphoid tissue in 4/4 or 3/4 patient data sets, respectively. Codon 22 varied between brain and lymphoid tissue only for patient UK6. Substitutions at codons 10 and 194 may therefore be important for nef adaptation to the brain microenvironment.
Table 3.
SLACa Predicted Sites of Positive Selection in Brain and Lymphoid Tissue
| |
|
Consensus amino acid (frequency)b |
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Codon (NL4-3 residue #) | Tissue | MACS1 | MACS2 | MACS3 | UK6 | p Valuec | ||||
| 10 (10) | Brain | L | (10/10) | V | (7/7) | V | (6/6) | M | (4/7) | 0.0049 |
| Lymphoid | S | (7/7) | G | (25/26) | L | (25/28) | K | (5/6) | ||
| 22 (22) | Brain | R | (10/10) | R | (7/7) | R | (6/6) | R | (7/7) | 0.0935 |
| Lymphoid | R | (7/7) | R | (23/26) | R | (25/28) | Q | (4/6) | ||
| 32 (28) | Brain | D | (10/10) | D | (6/7) | A | (6/6) | D | (6/7) | 0.0672 |
| Lymphoid | D | (6/10) | E | (26/26) | A | (27/28) | D | (6/6) | ||
| 158 (152) | Brain | E | (10/10) | E | (7/7) | Q | (4/6) | K | (7/7) | 0.0868 |
| Lymphoid | E | (6/10) | K | (26/26) | Q | (15/28) | K | (6/6) | ||
| 194 (188) | Brain | R | (10/10) | R | (7/7) | R | (6/6) | L | (7/7) | 0.0932 |
| Lymphoid | L | (10/10) | H | (12/26) | R | (27/28) | T | (5/6) | ||
| Q | (11/26) | |||||||||
Single Likelihood Ancestor Counting (HyPhy software package) was used to reconstruct ancestral sequences at each node of the phylogenetic tree shown in Fig. 1. Substitutions are calculated at each branch and terminal node. Wherever a variation occurs, the dS and dN were calculated.
Consensus amino acid is the residue at that position in >60% of sequences. Where no consensus residue was apparent, the two most frequent residues are shown. Frequency indicates the fraction of total sequences from the tissue of each patient in which the residue occurred.
p Values are based on a two-tailed binomial distribution that measures how likely the observed proportion of synonymous substitutions differs from the expected value derived from a global dN/dS ratio fitted to reconstructed ancestral states at each patient-specific node of the maximum likelihood phylogenetic tree. p-values < 0.1 were considered significant.
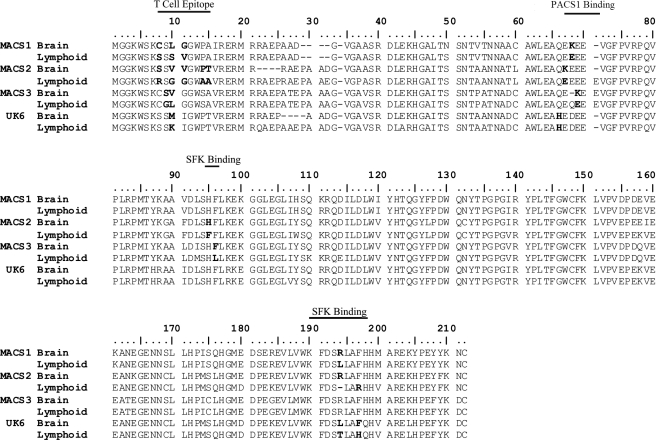
Codon 10 did not have a preferred amino acid for brain or lymphoid sequences (Table 3 and Fig. 2). This codon is located within an epitope recognized by both cytotoxic T lymphocytes and CD4 helper T cells15; as such, different residues may be required to escape immune surveillance within specific tissues. Tissue-specific changes were also observed in five other codons contained in these T cell epitopes (Fig. 2). SLAC analysis of lymphoid nef sequences alone indicated that positions 10 and 14 were under negative selection in patients UK6 and MACS2, respectively (Table 4). At codon 10, there was evidence for stronger adaptive selection and weaker negative selection at the divergence between lymphoid and brain sequences.
FIG. 2.
Alignment of predicted brain- and lymphoid-derived Nef amino acid sequences. Consensus nucleotide sequences were generated using Bioedit software from CLUSTAL W alignments of each patient-specific tissue set of sequences in which the representative base occurred in at least 60% of sequences in the set. Consensus nucleotide sequences associated with each patient-specific tissue were used to generate a predicted amino acid sequence. The resulting amino acid sequences were aligned with CLUSTAL W. Residues of interest are in bold. “T Cell Epitope” indicates residues within reported CTL or CD4+ T helper epitopes. “PACS1 Binding” indicates residues interacting with the PACS1 protein. “SFK Binding” indicates residues that may influence the association of Nef with Src family kinases.
Table 4.
SLACa Predicted Sites of Lymphoid-Specific Negative Selection
| Codon (NL4-3 residue #) | Patient | p Valueb | Codon description |
|---|---|---|---|
| 10 (10) | UK6 | 0.0257 | NH2-terminal T cell epitope |
| 14 (14) | MACS2 | 0.0167 | NH2-terminal T cell epitopes |
| 67 (62) | MACS2 | 0.0041 | MHC-I downmodulation |
| 68 (63) | MACS1 | 0.0904 | MHC-I downmodulation |
| 69 (insertion at 63) | MACS3 | 0.0709 | MHC-I downmodulation |
Single Likelihood Ancestor Counting (HyPhy software package) was used to reconstruct ancestral sequences at each node of a lymphoid-only phylogenetic tree. Substitutions are calculated at each branch and terminal node. Wherever a variation occurs, the dS and dN were calculated.
p Values are based on a two-tailed binomial distribution that measures how likely the observed proportion of synonymous substitutions differs from the expected value derived from a global dN/dS ratio fitted to reconstructed ancestral states at each node of patient-specific maximum likelihood phylogentic trees.
Tissue-specific substitutions were dispersed throughout the PACS1 binding motif, a motif essential for the ability of Nef to downmodulate MHC Class I (Fig. 2). As we previously reported,5 the glutamate-to-lysine change in all MACS2 brain Nefs at codon 67 and in the majority of MACS3 brain Nefs at codon 69 diminishes MHC Class I downmodulation.5 MACS3 brain sequences without the glutamate-to-lysine change contain a proline-to-serine change at position 122, which also diminishes the ability of Nef to downmodulate MHC Class I.5 Here, we extend these findings to include an analogous amino acid substitution of a glutamate-to-lysine at codon 68 in MACS1 brain Nefs (Fig. 2). SLAC analysis of lymphoid sequences indicates that negative selection likely occurred at these positions in patients MACS1, MACS2, and MACS3 (Table 4). In the PACS1 binding motif, as at codon 10, there is evidence for stronger adaptive selection and weaker negative selection at the divergence between lymphoid and brain.
Codon 194 (position 188 relative to the NL4-3 sequence) is under positive selection at the divergence between lymphoid and brain sequences in patients MACS1, MACS2, and UK6 but not patient MACS3. However, MACS3 brain sequences contain a substitution at codon 96 (NL4-3 residue 90, Fig. 2), a site that is predicted to interact directly with the Src family kinase (SFK) SH3 domain.16 Codon 194 (188) along with residues 91 (85), 95 (89), 96 (90), and 197 (191) form a motif containing determinants of Nef's ability to associate with activated PAK2 kinase.11 Mutation of this motif has been shown to disrupt the ability of Nef to activate Hck, a myeloid-restricted Src family kinase (SFK).17 Therefore, residues within the PAK2 association motif and those that interact with the SKF SH3 domain may act in concert to activate Hck. Thus, all four patient sequence datasets contain tissue-specific changes at positions that may influence the ability of Nef to activate Hck.
In summary, we showed that the divergence between lymphoid and brain nef sequences in late stage disease is characterized by stronger adaptive selection and weaker negative selection. These adaptive mutations, which likely accrued following negative selection at an earlier stage of disease,18 occurred in three distinct regions in HIV Nef. Adaptive mutations in the CTL and CD4+ T helper epitopes surrounding position 10 are likely tissue-specific escape mutations caused by the differential frequency of HIV-specific CTLs in brain versus peripheral blood.19 Brain-specific, adaptive mutations in the PACS1 binding domain abrogate the ability of Nef to downmodulate MHC Class I.5 This may reflect a decrease in fitness advantage for downmodulating MHC Class I in brain, perhaps due to its lower expression on ramified microglia20 or reduced CTL selection pressure in the brain compared to lymphoid tissue, and possibly an increase in the fitness advantage for altering the composition of the T cell epitope in this domain. Adaptive mutations also occurred within a motif potentially involved in Nef-mediated Hck activation, which may enhance replication and pathogenesis by increasing the promoter activity of the HIV LTR,21 inducing proliferation of infected microglia22 and/or dysregulating the immune response by altering macrophage phagocytosis and chemotaxis.23 Evidence of viral adaptive selection at the divergence between lymphoid and brain nef sequences implies that brain-derived nef sequences adaptively evolve to enhance viral replication in brain macrophages and microglia and escape brain-specific immune surveillance.
Sequence Data
GenBank accession numbers for sequences used in this study are as follows: GU049621–GU049630 (MACS1 brain), GU049631–GU049641 (MACS1 spleen), DQ885391–DQ885408 (MACS2 brain), DQ358037–DQ358047 and GU049642–GU049646 (MACS2 spleen), DQ358015–DQ358036 (MACS2 lymph node), DQ885409–DQ885420 (MACS3 brain), DQ885421–DQ885454 (MACS3 lymph node), GU049653–GU049659 (UK6 brain), GU049647–GU049652 (UK6 spleen).
Acknowledgments
This work was supported by NIH Grants AI73415 and MH83588. K.O. was supported in part by the Dana-Farber AIDS training grant (NIH #T32 AI007386). K.O. and J.M. were supported in part by the Harvard Medical School Neurodegeneration and Repair training grant (NIH #T32 AG00222).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Gonzalez-Scarano F. Martin-Garcia J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5(1):69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- 2.Ohagen A. Ghosh S. He J, et al. Apoptosis induced by infection of primary brain cultures with diverse human immunodeficiency virus type 1 isolates: Evidence for a role of the envelope. J Virol. 1999;73(2):897–906. doi: 10.1128/jvi.73.2.897-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Marle G. Henry S. Todoruk T, et al. Human immunodeficiency virus type 1 Nef protein mediates neural cell death: A neurotoxic role for IP-10. Virology. 2004;329(2):302–318. doi: 10.1016/j.virol.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 4.Ohagen A. Devitt A. Kunstman KJ, et al. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J Virol. 2003;77(22):12336–12345. doi: 10.1128/JVI.77.22.12336-12345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agopian K. Wei BL. Garcia JV. Gabuzda D. CD4 and MHC-I downregulation are conserved in primary HIV-1 Nef alleles from brain and lymphoid tissues, but Pak2 activation is highly variable. Virology. 2007;358(1):119–135. doi: 10.1016/j.virol.2006.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillai SK. Pond SL. Liu Y, et al. Genetic attributes of cerebrospinal fluid-derived HIV-1 env. Brain. 2006;129(Pt 7):1872–1883. doi: 10.1093/brain/awl136. [DOI] [PubMed] [Google Scholar]
- 7.Kosakovsky Pond SL. Poon AF. Zarate S, et al. Estimating selection pressures on HIV-1 using phylogenetic likelihood models. Stat Med. 2008;27(23):4779–4789. doi: 10.1002/sim.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorry PR. Taylor J. Holm GH, et al. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol. 2002;76(12):6277–6292. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahlknecht U. Deng C. Lu MC, et al. Resistance to apoptosis in HIV-infected CD4+ T lymphocytes is mediated by macrophages: Role for Nef and immune activation in viral persistence. J Immunol. 2000;165(11):6437–6446. doi: 10.4049/jimmunol.165.11.6437. [DOI] [PubMed] [Google Scholar]
- 10.Mordelet E. Kissa K. Cressant A, et al. Histopathological and cognitive defects induced by Nef in the brain. FASEB J. 2004;18(15):1851–1861. doi: 10.1096/fj.04-2308com. [DOI] [PubMed] [Google Scholar]
- 11.Agopian K. Wei BL. Garcia JV. Gabuzda D. A hydrophobic binding surface on the human immunodeficiency virus type 1 Nef core is critical for association with p21-activated kinase 2. J Virol. 2006;80(6):3050–3061. doi: 10.1128/JVI.80.6.3050-3061.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 13.Slatkin M. Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123(3):603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosakovsky Pond SL. Frost SD. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 15.HIV Molecular Immunology 2006/2007. Los Alamos, New Mexico: Los Alamos National Laboratory, Theoretical Biology and Biophysics; 2007. [Google Scholar]
- 16.Arold S. O'Brien R. Franken P, et al. RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry. 1998;37(42):14683–14691. doi: 10.1021/bi980989q. [DOI] [PubMed] [Google Scholar]
- 17.Trible RP. Emert-Sedlak L. Wales TE. Ayyavoo V. Engen JR. Smithgall TE. Allosteric loss-of-function mutations in HIV-1 Nef from a long-term non-progressor. J Mol Biol. 2007;374(1):121–129. doi: 10.1016/j.jmb.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeve AB. Patel K. Pearce NC, et al. Reduced genetic diversity in lymphoid and central nervous system tissues and selection-induced tissue-specific compartmentalization of neuropathogenic SIVsmmFGb during acute infection. AIDS Res Hum Retroviruses. 2009;25(6):583–601. doi: 10.1089/aid.2008.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadagopal S. Lorey SL. Barnett L, et al. Enhancement of human immunodeficiency virus (HIV)-specific CD8+ T cells in cerebrospinal fluid compared to those in blood among antiretroviral therapy-naive HIV-positive subjects. J Virol. 2008;82(21):10418–10428. doi: 10.1128/JVI.01190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aloisi F. Immune function of microglia. Glia. 2001;36(2):165–179. doi: 10.1002/glia.1106. [DOI] [PubMed] [Google Scholar]
- 21.Kim MO. Suh HS. Si Q. Terman BI. Lee SC. Anti-CD45RO suppresses human immunodeficiency virus type 1 replication in microglia: Role of Hck tyrosine kinase and implications for AIDS dementia. J Virol. 2006;80(1):62–72. doi: 10.1128/JVI.80.1.62-72.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suh HS. Kim MO. Lee SC. Inhibition of granulocyte-macrophage colony-stimulating factor signaling and microglial proliferation by anti-CD45RO: Role of Hck tyrosine kinase and phosphatidylinositol 3-kinase/Akt. J Immunol. 2005;174(5):2712–2719. doi: 10.4049/jimmunol.174.5.2712. [DOI] [PubMed] [Google Scholar]
- 23.Ernst M. Inglese M. Scholz GM, et al. Constitutive activation of the SRC family kinase Hck results in spontaneous pulmonary inflammation and an enhanced innate immune response. J Exp Med. 2002;196(5):589–604. doi: 10.1084/jem.20020873. [DOI] [PMC free article] [PubMed] [Google Scholar]