Abstract
FSH is a heterodimeric glycoprotein hormone secreted from the gonadotrope cell population of the anterior pituitary. Despite its crucial role in mammalian reproduction, very little is known about regulation of the FSH β-subunit gene at the molecular level. In this report, we examine the basis for cell-specific expression of FSHβ using the mouse LβT2 and αT3-1 gonadotrope-derived cell lines. Characterization of the hormonal content of LβT2 and αT3-1 cells at the protein level classifies these cells as relatively mature and immature gonadotropes, respectively. We studied LβT2 cell-specific expression of FSHβ using 398 bp of the mouse FSHβ regulatory region linked to a luciferase reporter gene in transient transfection assays. This mouse FSHβ promoter can direct reporter gene expression specifically to LβT2 cells when compared with other pituitary- and non-pituitary-derived cell lines, including αT3-1 cells. Furthermore, it is induced by activin, and interruption of the autocrine activin loop in LβT2 cells by the addition of follistatin reduces its expression. Truncation analysis indicates that several regions of the promoter are involved in this specificity and that these can be dissociated from activin regulation. We identify binding sites for the orphan nuclear receptor steroidogenic factor-1 and the heterotrimeric transcription factor nuclear factor Y and show that these elements functionally interact to regulate FSHβ gene expression in an LβT2 cell-specific manner. Moreover, steroidogenic factor-1 and nuclear factor Y are shown to physically interact with each other. This study is the first to demonstrate the presence of basal FSHβ protein in LβT2 cells and to identify specific elements within the FSHβ promoter that contribute to basal and cell-specific expression of the gene.
FSH and LH are both members of the glycoprotein hormone family that also includes TSH, produced in pituitary thyrotropes, and chorionic gonadotropin, produced in human and primate placenta (1). Each of these hormones shares a common α-subunit [α-glycoprotein subunit (α-GSU)] and contains a unique β-subunit that confers the physiological specificity to each hormone. Because the β-subunit genes of FSH and LH are expressed exclusively in the gonadotrope population of the anterior pituitary, the synthesis and secretion of FSH and LH are restricted to these cells.
Although some success has been achieved in understanding the molecular mechanisms governing basal and cell-specific expression of the α-GSU and LHβ genes (2–7), very little is understood about regulation of the FSHβ gene. This is due, in part, to the heterogeneity of the anterior pituitary gland in vivo and, until recently, the lack of an FSHβ-expressing gonadotrope-derived cell line in which to study its regulation. Use of transgenic mice has allowed large regulatory regions important for hormonal and gonadotrope-specific regulation of FSHβ to be defined. These studies demonstrated that a 10-kb region of the human FSHβ gene, including 4 kb of 5′-flanking sequence and 2 kb of 3′-flanking sequence, is sufficient to direct gonadotrope-specific expression (8). Similarly, a 5.5-kb fragment of the 5′-flanking sequence of the ovine FSHβ gene can confer pituitary-specific expression of a luciferase reporter gene (9). However, specific elements involved in tissue-specific expression have not been identified.
The LβT2 gonadotrope-derived cell line was recently shown to express endogenous FSHβ mRNA (10) and secrete substantial levels of FSH in response to activin (11), an important regulator of FSH synthesis in vivo (12, 13). Because LβT2 cells produce FSHβ, they must contain all of the factors necessary for its expression. These cells therefore present a model system in which to dissect details of the molecular basis for expression of FSHβ in the gonadotrope. Indeed, the molecular mechanisms governing GnRH and activin regulation of the ovine FSHβ gene are beginning to be elucidated using these cells (10, 14). In the present study, we examine the basis for cell-specific expression of FSHβ in LβT2 gonadotrope cells by comparison to a non-FSH-producing gonadotrope-derived cell line, αT3-1, and to a fibroblast-derived cell line, NIH3T3. We demonstrate the presence of endogenous FSHβ protein in LβT2 cells, even in the absence of treatment by activin or GnRH. Specific promoter elements that bind the orphan nuclear receptor steroidogenic factor-1 (SF-1), a known regulator of gonadotrope-specific genes, and the ubiquitous transcription factor, nuclear factor Y (NF-Y), are identified within 398 bp of the proximal mouse FSHβ regulatory region. These contribute to cell-specific expression but are not involved in repression by follistatin. Finally, NF-Y and SF-1 are shown to interact both physically as well as functionally; this functional interaction occurs in a manner that contributes to LβT2 cell-specific expression of the FSHβ gene.
Results
The FSH β-Subunit Is Specifically Expressed in LβT2 Cells
Synthesis and secretion of FSH are restricted to mature gonadotropes and are dependent on the presence of the β-subunit of the hormone. Although LβT2 cells have recently been shown to express FSHβ mRNA under non-hormone-stimulated conditions (10), a separate study detected FSH secretion only after treatment with activin (11); intracellular FSHβ protein levels have not been examined. To characterize the hormonal content of LβT2 cells under nonstimulated conditions, immunocytochemistry was performed using antibodies specific for several gonadotrope markers, including FSHβ, LHβ, α-GSU, and SF-1; the non-FSHβ-expressing cell line, αT3-1, was used for comparison (Fig. 1). All gonadotrope markers, including FSHβ, are detected in LβT2 cells under these basal conditions. In contrast to LβT2 cells, αT3-1 cells have been characterized as committed, but immature, gonadotrope-derived cells (15), expressing high levels of α-GSU and SF-1 mRNA but neither of the gonadotropin β-subunits. Examination of protein expression in these cells confirms their lack of the β-subunits. Consistent with the previously described translational control of α-GSU in αT3-1 cells (16), they contain very low levels of α-GSU protein despite the high levels of α-GSU message.
Fig. 1. The FSH β-Subunit Is Specifically Expressed in LβT2 Cells.
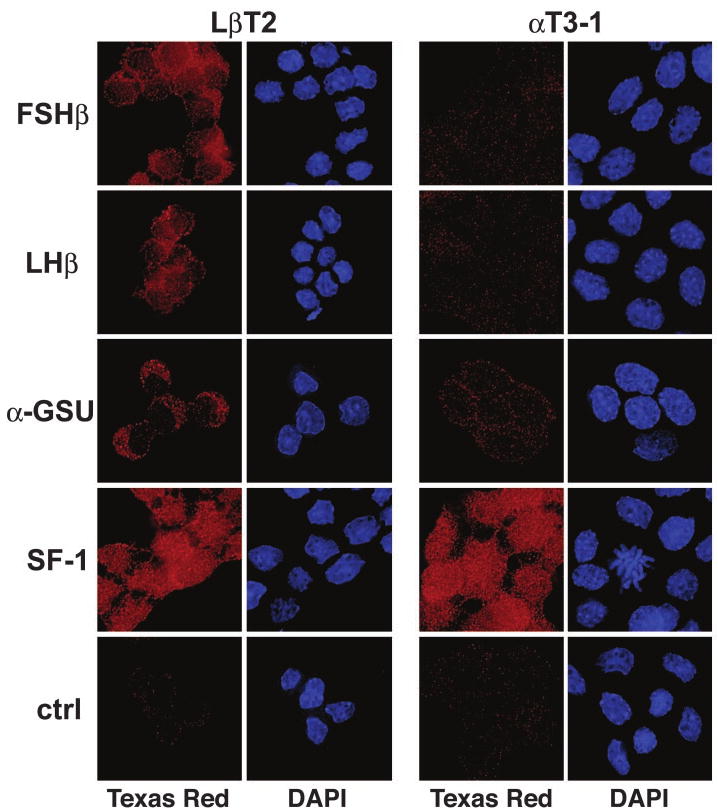
The β-subunits of FSH and LH are present in LβT2 cells but not αT3-1 cells. LβT2 and αT3-1 cells were grown on glass coverslips, fixed, and immunostained. The proteins were visualized using antibodies specific to FSH (FSHβ), LH (LHβ), α-GSU, or steroidogenic factor 1 (SF-1) and a Texas red-conjugated antirabbit IgG secondary antibody. Nuclei were stained with 4,6-diamidino-2-phenylindole. Representative fields are shown.
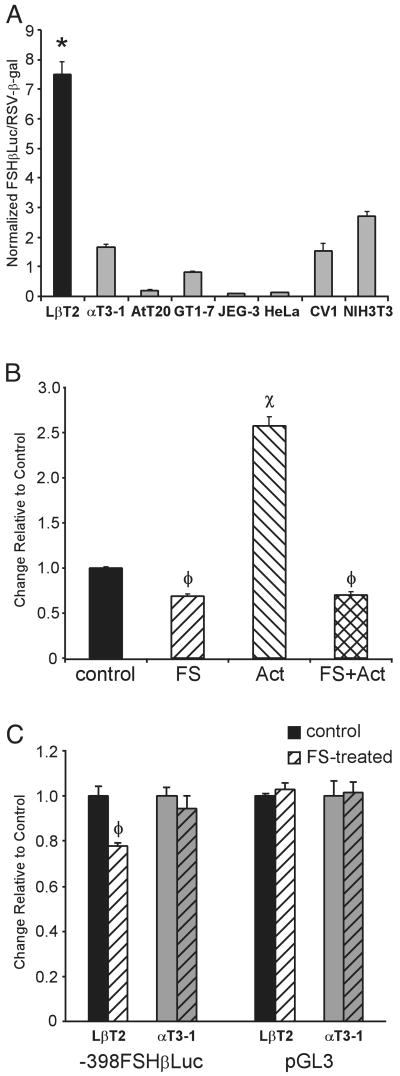
Previous work has shown that LβT2 cells specifically support transcriptional activation of the ovine FSHβ 5′-regulatory region (10). Because the endogenous FSHβ gene is expressed in LβT2 gonadotrope cells but not in αT3-1 cells, we postulated that these cell lines could be used as a model to identify factors involved in the induction of mouse FSHβ gene expression during gonadotrope maturation. To test this, a DNA fragment encompassing 398 bp of the mouse FSHβ 5′-regulatory region was linked to a luciferase reporter gene (−398FSHβLuc) and used in transient transfection studies. Activity of the FSHβ promoter was examined in LβT2 cells and compared with a variety of other pituitary and non-pituitary-derived cell lines (Fig. 2A). A Rous sarcoma virus (RSV) enhancer and promoter driving a β-galactosidase reporter gene (RSV-β-gal) was cotransfected as an internal control. Additionally, an RSV-luciferase (RSV-Luc) plasmid was transfected in parallel with FSHβLuc into each cell type to control for differences in transfection efficiency, transcription rates, reporter mRNA half-life, luciferase and β-galactosidase protein stability, and metabolic rates between distinct cell types. The ratio of RSV-Luc to RSV-β-gal was set to 100 for each cell line, and the FSHβLuc value was normalized to the RSV-Luc/RSV-β-gal value. To validate this approach, we tested the activities of the promoterless pUC18-luciferase and a thymidine kinase-Luc plasmid and found each to be expressed at equivalent levels in αT3-1 and LβT2 cells relative to the RSV controls (data not shown).
Fig. 2. The Mouse FSH β-Subunit Promoter Is Specifically Active and Responsive to Follistatin in LβT2 Cells.
A, The 398-bp mouse FSHβ promoter was linked to the luciferase reporter gene (−398FSHβLuc) and transiently transfected into pituitary-derived LβT2 (differentiated gonadotrope), αT3-1 (precursor gonadotrope), and AtT20 (corticotrope) cells and non-pituitary-derived GT1–7 (hypothalamic neuroendocrine), JEG-3 (placental), HeLa (cervical fibroblast), CV1 (kidney), and NIH3T3 (fibroblast) cells. RSV-β-gal was cotransfected as an internal control. To control for differences in transfection efficiencies and transcription rates between distinct cell lines, a parallel control, RSV-Luc, was transfected into each cell line with the RSV-β-gal internal control. The ratio of RSV-Luc divided by RSV-β-gal was set at 100 for each cell line (not shown). Results represent the mean ± sem of at least three independent experiments, each performed in triplicate (n ≥ 9). The bar marked with an asterisk (*) (LβT2 cells) is statistically significant from all other bars (P < 0.05). Statistically significant differences between other cell lines are not shown. B, The 398-bp mouse FSHβ promoter responds to follistatin and activin. LβT2 cells were transiently transfected with −398FSHβLuc, along with the RSV-β-gal internal control, and then treated with vehicle (control), 100 ng/ml follistatin (FS), 10 ng/ml activin (Act), or a combination of both follistatin and activin (FS+Act) for 24 h in DMEM with 10% fetal bovine serum. The value of the vehicle-treated sample was set at 1, and the follistatin- and activin-treated samples are shown relative to this control. Results represent the mean ± sem of three independent experiments, each performed in triplicate (n = 9). The bars marked with φ (FS and FS+Act) and x (Act) are statistically different from the untreated control and are also different from each other (P < 0.05). C, The mouse FSHβ promoter responds to follistatin in LβT2 cells, but not αT3-1 cells. LβT2 and αT3-1 cells were transiently transfected with either −398FSHβLuc or pGL3, along with the RSV-β-gal internal control, and then treated with either vehicle (control) or 100 ng/ml follistatin for 24 h (FS-treated, hatched bars). The values of the vehicle-treated samples were set at 1 and the FS-treated samples are shown relative to this control. Results represent the mean ± sem of at least three independent experiments, each performed in triplicate (n ≥ 9). The bar marked with φ is statistically different from the untreated control (P < 0.05).
Results from these experiments show that the transfected mouse FSHβ reporter gene is specifically expressed in the more differentiated LβT2 pituitary gonadotrope cell line (Fig. 2A). FSHβ promoter activity is approximately 4.5-fold higher in LβT2 cells than in the gonadotrope precursor αT3-1 cells and up to 80-fold higher than in some non-gonadotrope-derived cell lines such as AtT20 (corticotrope derived), JEG-3 (placental), and HeLa (cervical fibroblast).
Our previous study examining transcriptional regulation of the ovine FSHβ 5′-regulatory region demonstrated that activin specifically regulates FSHβ gene expression in LβT2 cells but not in αT3-1 or NIH3T3 cells (10). This LβT2 cell-specific response to activin includes activation of the ovine FSHβ promoter after the addition of exogenous activin as well as inhibition of ovine FSHβ promoter activity by neutralizing endogenous activin with follistatin, a potent activin-binding protein that blocks binding of activin to its receptors (17–19). Similarly, we now show that the mouse FSHβ promoter is responsive to both activin and follistatin in LβT2 cells (Fig. 2B). Activity of the 398-bp mouse FSHβ promoter in LβT2 cells is reduced approximately 20–25% in follistatin-treated cells when compared with untreated control cells. Treatment with activin stimulates −398FSHβLuc expression approximately 2.5-fold, an effect that is completely blocked by addition of follistatin. We used this ability of follistatin to disrupt autocrine activin activity to determine whether endogenous activin is contributing to LβT2 cell-specific expression of mouse FSHβ. LβT2 and αT3-1 cells were transiently transfected with mouse −398FSHβLuc and then treated with follistatin (Fig. 2C). The promoterless luciferase vector, pGL3, was used as a negative control. As with our previous work, follistatin treatment inhibits expression of FSHβLuc in LβT2 cells but not αT3-1 cells. Although this demonstrates that an endogenous activin autocrine loop contributes some degree to LβT2 cell-specific expression of FSHβ, it also reveals that the specificity to LβT2 cells is not solely due to the action of the activin autocrine loop and must reside in additional transcriptional regulators.
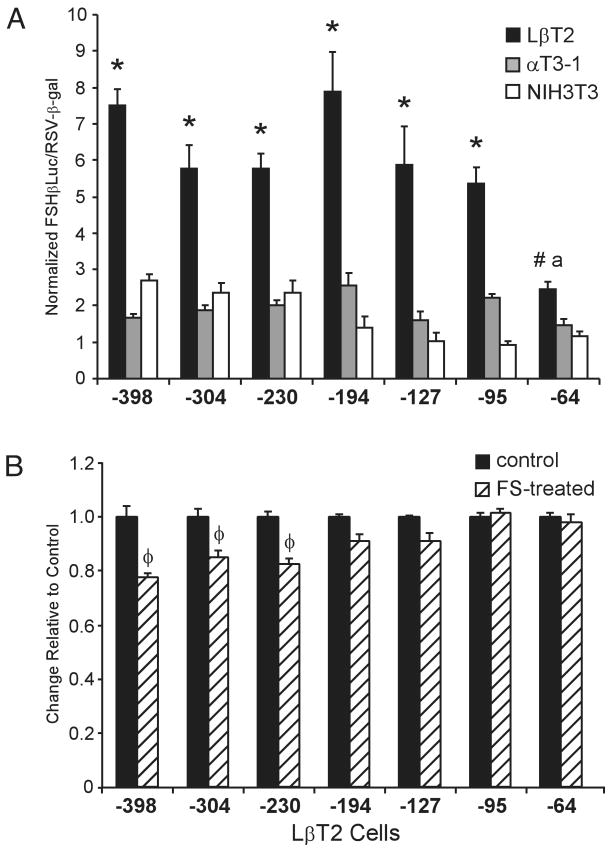
To delineate the regions of the mouse FSHβ promoter involved in basal expression and LβT2 cell specificity, 5′-truncations of mouse FSHβLuc were transiently transfected into LβT2, αT3-1, and NIH3T3 cells (Fig. 3A). Analyses of the transcriptional specificity of these truncations indicate that several regions of the FSHβ promoter contribute to LβT2 cell specificity; however, the largest decreases in specificity between the cell types are seen upon truncation of the proximal promoter regions from −127 bp (3.7-fold) to −95 bp (2.4-fold) and from −95 bp to −64 bp (1.7-fold). The only statistically significant decrease in basal expression within LβT2 cells alone is observed upon truncation of the sequences from −95 bp to −64 bp.
Fig. 3. Different Regions of the FSHβ Promoter Contribute to Basal Activity and Follistatin Responsiveness in LβT2 Cells.
A, Proximal promoter sequences are important for basal activity of the mouse FSHβ promoter. Truncations of the mouse FSHβ promoter linked to luciferase were transiently transfected into LβT2, αT3-1, and NIH3T3 cells. Results represent the mean ± sem of at least three independent experiments, each performed in triplicate (n ≥ 9). Asterisks (*) designate a statistically significant difference from αT3-1 and NIH3T3 cells (P < 0.05) # designates significant difference from NIH3T3 cells (P < 0.05) but not αT3-1 cells; the letter a designates a statistical difference between the −64-bp FSHβLuc truncation and all other truncations in LβT2 cells (P < 0.05). Statistical differences between αT3-1 and NIH3T3 cells are not shown. B, Follistatin responsiveness of the mouse FSHβ promoter is localized to a more distal region of the promoter. LβT2 cells were transiently transfected with mouse FSHβLuc truncations and treated with either vehicle (control) or 100 ng/ml follistatin for 24 h (FS-treated, hatched bars). To facilitate comparisons between promoter responses, the control samples were set at 1 and the FS-treated samples are shown relative to this control. Results represent the mean ± sem of at least three independent experiments, each performed in triplicate (n ≥ 9). Bars marked with φ are statistically different from the untreated controls (P < 0.05).
Because the endogenous activin autocrine loop might contribute to LβT2 cell specificity, we also sought to identify the region of the mouse FSHβ promoter regulated by endogenous activin, by localizing follistatin repression. LβT2 cells were transiently transfected with mouse FSHβLuc truncations and then treated with follistatin for 24 h (Fig. 3B). In contrast to basal expression, the follistatin responsiveness of the mouse FSHβ promoter localizes to more distal sequences of the promoter, between −230 bp and −194 bp. This demonstrates that the contribution of endogenous activin to FSHβ promoter activity in LβT2 cells is separate from the downstream factors contributing to basal and cell-specific activity.
The Mouse FSHβ Promoter Contains SF-1 and NF-Y Binding Elements
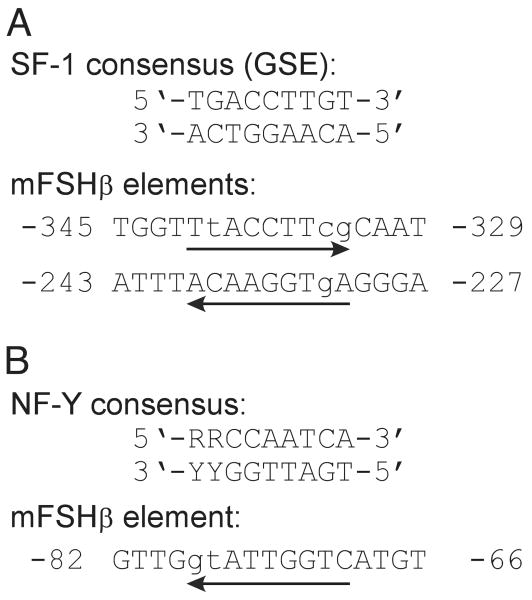
To identify specific elements that may contribute to basal expression of FSHβ in LβT2 cells, we analyzed the sequence of the mouse FSHβ regulatory region. Because of the known importance of SF-1 for gonadotrope function (20, 21), we began by perusing the sequence for SF-1 binding sites, also known as gonadotrope-specific elements, or GSEs. Two putative GSEs were identified at approximately −341 bp (6/9 match) and −239 bp (8/9 match) relative to the transcriptional start site based on sequence similarity to the GSEs from the LHβ and α-GSU promoters (Fig. 4A). The −239GSE is conserved in FSHβ regulatory regions from rat, human, bovine, and ovine; the −341GSE is conserved in rat. Due to the significant decrease in promoter activity observed upon deletion of proximal promoter regions, the sequence from −127 bp to −64 bp was searched for putative transcription factor binding sites using the TRANSFAC database (22). This identified a consensus-binding element, at approximately −76 bp, for the heterotrimeric nuclear factor-Y (NF-Y) transcription factor (Fig. 4B). Analysis of FSHβ promoter sequences from other species indicates that this NF-Y element is conserved in mouse, rat, and human.
Fig. 4. The Mouse FSHβ Promoter Contains Consensus SF-1 and NF-Y Binding Sites.
A, The mouse FSHβ promoter was scanned for regions of conservation with a consensus SF-1 binding site (GSE). Two putative SF-1 binding sites were identified at approximately −341 and −239 relative to the transcriptional start site and are shown aligned with the GSE consensus sequence from the LHβ and α-GSU promoters. B, The proximal region of the mouse FSHβ promoter was searched for possible transcription factor binding sites using the TRANSFAC database (http://transfac.gbf.de/TRANSFAC) (22). A putative NF-Y element was identified and is shown aligned with a consensus NF-Y binding sequence. Mismatches are shown in lower-case; arrows indicate the direction of the homology region, R denotes A or G, and Y denotes C or T. Note that homology for the −239 SF-1 and NF-Y binding sites is on the opposite strand of the consensus shown, as indicated by arrows.
SF-1 binding to the putative GSEs was confirmed using EMSAs. A specific complex is present when nuclear proteins from LβT2 and αT3-1 cells, but not NIH3T3 cells, are bound to both the −341SF-1 probe (Fig. 5A) and the −239SF-1 probe (Fig. 5B). This complex is competed by the consensus GSE from the α-GSU promoter (lanes 4 and 10, GSEcon), and binding is blocked by addition of a SF-1-specific antibody (lanes 5 and 11). The stronger binding of the SF-1-containing complex to the −239SF-1 probe is consistent with this site being a more highly conserved GSE.
Fig. 5. SF-1 Interacts with the FSHβ Promoter at Two Sites.
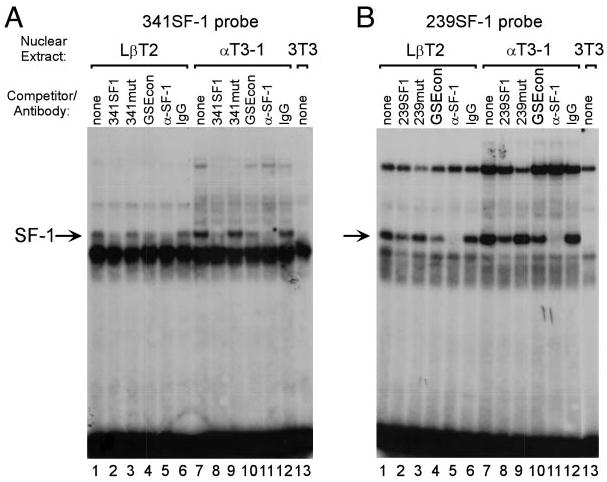
EMSA was performed using a probe spanning either the −341 SF-1 element (A) or the −239 SF-1 element (B) along with nuclear extracts from LβT2 (lanes 1–6), αT3-1 (lanes 7–12), or NIH3T3 (lane 13) cells. Competitions were performed using 100-fold excess unlabeled oligonucleotide as indicated above each lane: None (lanes 1, 7, and 13; no competitor), 341SF1 (A, lanes 2 and 8; wild-type 341SF-1 element; 5′-TTGGTTTACCTTCGCAATGGAG-3′), 341mut (A, lanes 3 and 9; mutated 341SF-1 element; 5′-TTGGTTTAAATTCGCAATGGAG -3′), 239SF1 (B, lanes 2 and 8; wild-type 239SF-1 element; 5′-TTTAATTTACAAGGTGAGGGAG-3′), 239mut (B, lanes 3 and 9; mutated 239SF-1 element; 5′-TTTAATTTAGAATTTGAGGGAG-3′), GSEcon (lanes 4 and 10, consensus GSE from the α-GSU promoter; 5′-GCTGCTGACCTTGTCACTAGCT-3′). The GSE is underlined and the mutated sequences are shown in bold. Antibodies directed against SF-1 (α-SF-1) or normal rabbit IgG (IgG) were included in the reactions as indicated. Arrow indicates the SF-1 containing complex.
All three subunits of NF-Y are required for DNA binding; however, the NF-YA subunit is responsible for most of the sequence-specific contacts (23). Furthermore, expression of NF-YA is the most regulated among the subunits; its levels are cell cycle dependent and post-transcriptionally regulated, and its subcellular localization can change during embryogenesis (23). We therefore examined the presence of NF-YA in LβT2, αT3-1, and NIH3T3 cells by immunocytochemistry and found it to be highly expressed and localized to the nucleus in each cell line (Fig. 6A). Binding of NF-Y to its putative element was examined by EMSA using a probe spanning the mouse FSHβ promoter region from −99 bp to −65 bp (FSH 99/65). An antibody directed against the NF-YA subunit supershifts the single major band present in all three cell types (Fig. 6B). Experiments using oligonucleotides containing 2-bp mutations spanning the CCAAT box and surrounding sequences, shown in Fig. 6D, were used in competitions (Fig. 6C) and as probes (not shown) and confirm that NF-Y is interacting with the specific element that was identified (lanes 7–9).
Fig. 6. NF-Y Interacts with the FSHβ Promoter.
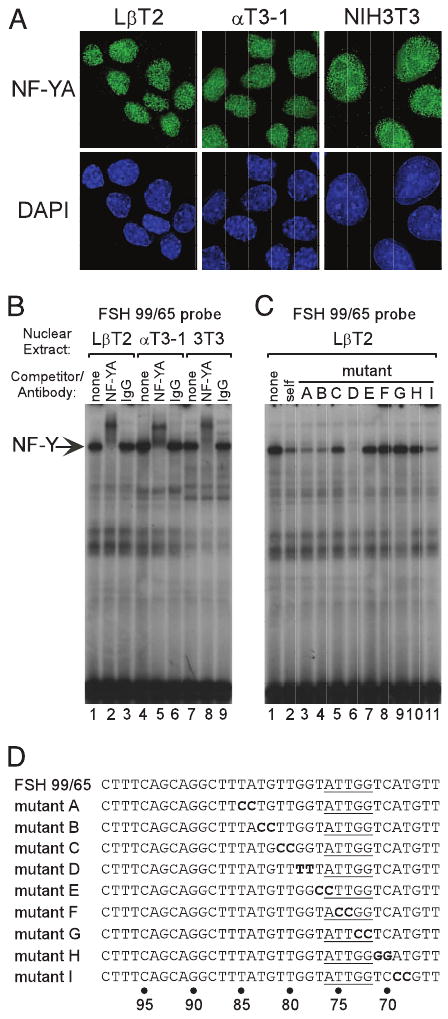
A, Immunostaining demonstrates nuclear localization of NF-YA in LβT2, αT3-1, and NIH3T3 cells. B, EMSA was performed using a probe corresponding to the mouse FSHβ promoter sequence from −99 to −65 (FSH 99/65) along with nuclear extracts from LβT2 (lanes 1–3), αT3-1 (lanes 4–6), and NIH3T3 (lanes 7–9) cells. Antibodies directed against either NF-YA (α-NF-YA; lanes 2, 5, and 8) or normal rabbit IgG (IgG; lanes 3, 6, and 9) were included in the reactions as indicated. C, Competitions using mutated FSH 99/65 oligonucleotides confirm that NF-Y binds its consensus element: none (no competitor), self (wild-type FSH 99/65). The sequences of mutations are shown in panel D. The core NF-Y consensus sequence is underlined. Identical results were observed using nuclear extracts from LβT2 (shown), αT3-1, and NIH3T3 cells (data not shown). Arrow indicates the NF-Y-containing complex.
NF-Y and SF-1 Functionally and Physically Interact and Contribute to LβT2 Cell-Specific Expression of the FSH β-Subunit Gene
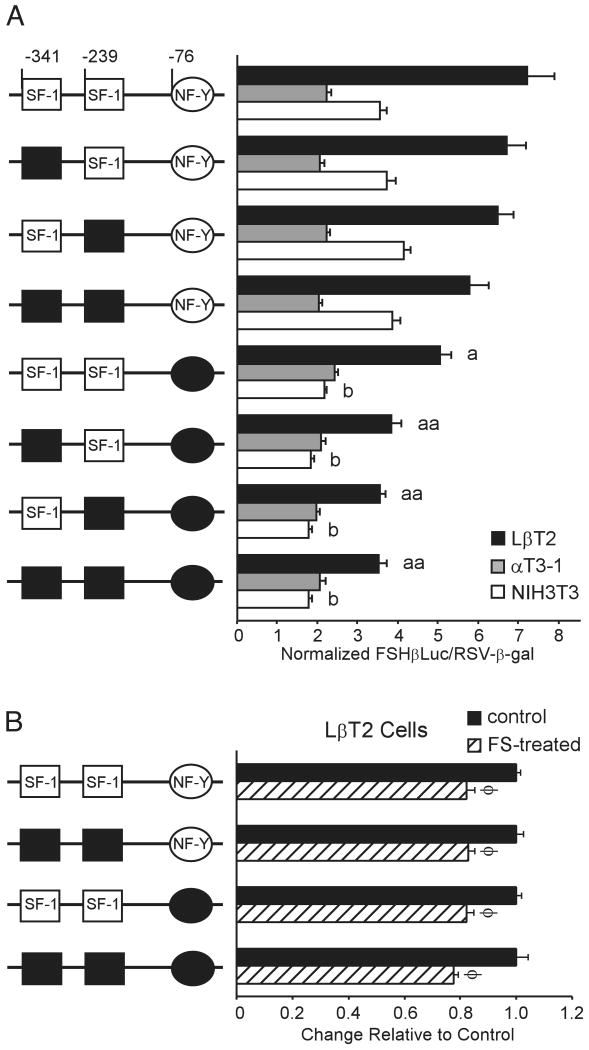
The contribution of the SF-1 and NF-Y binding sites to FSHβ promoter activity was assessed by mutating these elements alone, and in combination, within the context of −398FSHβLuc. The NF-Y mutation used corresponds to mutant F described in Fig. 6D. Interestingly, mutation of the SF-1 and NF-Y sites has a cell-dependent effect on activity of the FSHβ promoter (Fig. 7A). SF-1 is expressed in LβT2 and αT3-1 cells and, although they trend lower, neither the single SF-1 site mutants nor the double SF-1 mutant has a statistically significant effect on promoter activity in either cell line. Mutation of the NF-Y element results in an approximate 30% reduction in activity in LβT2 cells when compared with wild type and a 38% reduction in NIH3T3 cells. Surprisingly, this NF-Y mutation has no effect on promoter activity in αT3-1 cells. When the SF-1 and NF-Y mutations are combined, a greater than additive effect is observed only in LβT2 cells (50% reduction); expression is not affected by combining the mutations in either αT3-1 or NIH3T3 cells. This results in a corresponding decrease in the specificity between LβT2 and αT3-1 cells to 1.7-fold. The cooperative effect of the NF-Y and SF-1 mutations indicates that the proteins acting through these elements are functionally interacting in an LβT2 cell-specific manner.
Fig. 7. The NF-Y and SF-1 Binding Sites Contribute to LβT2 Cell Specificity but Are Not Involved in Follistatin-Regulated Expression of FSHβ.
Site-directed mutagenesis was performed to mutate the SF-1 and NF-Y binding sites in the −398FSHβLuc reporter gene. A, Transient transfections of mutant FSHβLuc plasmids were performed in LβT2, αT3-1, and NIH3T3 cells. The mutant plasmids are depicted in the diagram at left. Results represent the mean ± sem of five independent experiments, each performed in triplicate (n = 15). The letter a denotes a statistically significant difference of the NF-Y mutant plasmid as compared with wild type in LβT2 cells (P < 0.05); the double letter aa denotes a statistically significant difference between the NF-Y and SF-1 double or triple mutants and the NF-Y single mutant in LβT2 cells (P < 0.05); the letter b denotes a statistically significant difference of the mutant plasmid as compared with wild type in NIH3T3 cells (P < 0.05). B, Follistatin responsiveness of −398FSHβLuc does not require the NF-Y and SF-1 binding sites. LβT2 cells were transiently transfected with mutant FSHβLuc plasmids and treated with either vehicle (control) or 100 ng/ml follistatin for 24 h (FS-treated, hatched bars). The mutant plasmids are depicted in the diagram at left. To facilitate comparisons between promoter responses, the control samples were set at 1 and the FS-treated samples are shown relative to this control. Results represent the mean ± sem of four independent experiments, each performed in triplicate (n = 12). Bars marked with φ are statistically significant from the untreated controls (P < 0.05).
To ascertain whether the NF-Y and SF-1 binding sites are involved in activin regulation of FSHβ, the endogenous activin autocrine loop was blocked with follistatin in LβT2 cells transiently transfected with mutant FSHβLuc plasmids (Fig. 7B). If the activin system functions through the NF-Y and SF-1 binding sites, then mutation of these elements should eliminate or reduce the contribution of endogenous activin to FSHβ promoter activity, and the follistatin effect would be lost. In fact, the NF-Y mutant, SF-1 double mutant, and NF-Y/double SF-1 triple mutant promoters are all inhibited by follistatin equally as well as the wild-type promoter, indicating that these elements are not involved in activin regulation of the FSHβ gene. This is consistent with our localization of the follistatin-responsive sequences to an upstream region of the mouse FSHβ promoter, which does not encompass the NF-Y and SF-1 binding sites.
The functional interaction between the NF-Y and SF-1 binding sites on basal activity suggests that the proteins may be physically interacting with one another. To test this possibility, glutathione-S-transferase (GST) pull-down assays were performed. As seen in Fig. 8A, GST-SF-1 is capable of interacting with 35S-labeled NF-YA but not with the NF-YB or NF-YC subunits. Pituitary homeobox 1 (Ptx1) has previously been shown to interact with SF-1 and serves here as a positive control, whereas green fluorescent protein (GFP) does not interact with SF-1 and serves here as a negative control. No interactions were observed using GST alone. The converse experiment was also performed using GST-NF-YA and GST-NF-YB proteins with 35S-labeled SF-1 (Fig. 8B). Once again, SF-1 interacts with NF-YA but not with NF-YB. These experiments confirm the ability of NF-Y and SF-1 to physically interact in addition to the functional interaction observed in LβT2 cells.
Fig. 8. NF-Y and SF-1 Physically Interact.
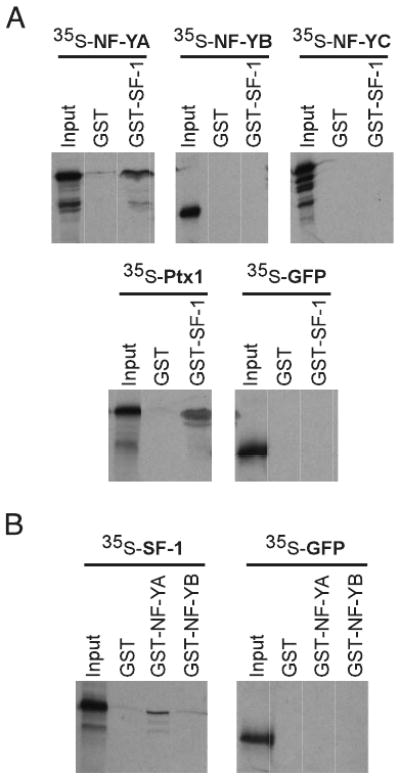
SF-1 interacts with the NF-YA subunit, but not the NF-YB or NF-YC subunits. A physical interaction between NF-Y and SF-1 was examined by a GST interaction assay. A, GST-SF-1 or GST alone was bound to glutathione sepharose beads and incubated with in vitro translated 35S-labeled NF-Y subunits, as indicated. GFP serves as a negative control and Ptx1 protein serves as a positive control for interaction with SF-1. B, GST alone, GST-NF-YA, or GST-NF-YB was incubated with 35S-labeled SF-1 or GFP, as indicated. GST alone serves as a control for the specificity of the interaction. Input shows 10% of the amount of in vitro translated protein used for the interaction.
Discussion
At the molecular level, regulation of the FSH β-subunit gene is the least well understood of the gonadotrope genes. Until recently, no appropriate cell model existed in which to study FSHβ gene regulation, and transgenic mice have been limited in their ability to help define discrete elements. Our characterization of the hormonal content of LβT2 and αT3-1 cells at the protein level provides further support for classifying these cells as relatively mature and immature gonadotrope cells, respectively. Prior work examining FSHβ gene expression in LβT2 cells demonstrated the presence of FSHβ mRNA under basal, nonstimulated conditions and showed that it is appropriately regulated by activin and follistatin (10), physiological regulators of FSH synthesis in vivo. A separate study detected low levels of FSH secretion from untreated LβT2 cells and showed that FSH secretion is dramatically increased in response to activin (11). Our current work confirms the findings suggested by the secretion studies, i.e. that LβT2 cells make FSHβ protein under nonstimulatory conditions. Although the level of FSHβ expression in LβT2 cells may be low, intracellular FSHβ protein is both present and detectable. Furthermore, both mouse and ovine transfected FSHβ-Luciferase reporter genes are expressed very well in these cells, at an activity level comparable to that of the 1800-bp rat LHβ promoter (data not shown). The expression and appropriate hormonal regulation of FSHβ in LβT2 cells (10, 11, 14) should help resolve the impasse in our progress in understanding of FSHβ gene regulation at the molecular level.
We have previously shown that an endogenous activin autocrine loop is present in LβT2 cells and contributes to cell-specific activity of the ovine FSHβ regulatory region (10). In this study, we examine the contribution of activin to cell-specific activity of the mouse FSHβ promoter, by neutralizing endogenous activin with follistatin. Whereas activity of the 5.5-kb ovine FSHβ regulatory region was reduced approximately 50% by follistatin treatment, activity of the 398-bp mouse FSHβ promoter is reduced approximately 25%. The experiments on the ovine FSHβ gene were performed in 1% serum (10), while the current experiments on the mouse FSHβ gene were performed in 10% serum to allow for direct comparison with the transfections analyzing basal and cell-specific expression. However, we find no significant differences due to serum conditions, having performed the activin and follistatin treatments of the mouse FSHβ gene under both conditions in parallel (data not shown). Another difference is the source of follistatin: whereas the previous experiments used follistatin kindly provided by Dr. Shunichi Shimasaki, the current experiments utilize commercially available follistatin. Despite this change, we now show that the current follistatin treatment is sufficient to block induction by exogenous activin treatment as well as the endogenous activin loop. Finally, this difference in magnitude of the follistatin effect could be due to species-specific differences in regulation of the ovine and mouse FSHβ genes. Whatever the cause for the difference in magnitude, the contribution of endogenous activin to both ovine and mouse FSHβ promoter activity is LβT2 cell specific; neither promoter responds to follistatin in αT3-1 or NIH3T3 cells. Taking into account the contribution of activin to mouse FSHβ promoter activity in LβT2 cells, an approximate 3- to 3.5-fold basal specificity between LβT2 and αT3-1 cells remains. In this report, we identify specific regulatory elements involved in basal expression of the FSHβ gene (binding sites for SF-1 and NF-Y) and show that follistatin regulation is acting independently of these elements.
SF-1 is an orphan nuclear receptor that, within the pituitary, is specifically expressed in the gonadotrope cell population (20). SF-1 binding sites have previously been identified in the LHβ, α-GSU, and GnRH receptor promoters (3, 4, 24); therefore, it is not surprising to find them within the FSHβ regulatory region. The importance of SF-1 to gonadotrope function in vivo has been demonstrated by the generation of SF-1-null mice (20). These animals are infertile and have markedly reduced levels of α-GSU, LHβ, FSHβ, and GnRH receptor. Pituitary-specific SF-1 knockout animals indicate that the impaired gonadotropin expression is a primary defect due to the lack of SF-1 in the pituitary and not a secondary defect due to SF-1 deficiency in other tissues (21). Although SF-1 is an important basal regulator of gonadotrope function, it is not sufficient to confer gonadotrope-specific expression since it is made in steroidogenic tissues outside of the pituitary (25), as well as in αT3-1 cells, which do not make the FSH or LH β-subunits. Moreover, its expression commences significantly earlier in pituitary development than the emergence of FSHβ gene expression (25). In the context of the LHβ promoter, SF-1 regulates gene expression, in part, by interacting with proteins acting through early growth response 1 and homeodomain-binding elements (26). The role of SF-1 in FSHβ gene regulation is less clear but appears to be dependent on other regulatory factors. Although not statistically significant, deletion of the −341 SF-1 site or mutation of both SF-1 sites reduces basal expression of FSHβLuc in LβT2 cells approximately 20% compared with the wild-type promoter. However, when combined with a mutation in the NF-Y element, the SF-1 binding site mutations do indeed have a statistically significant effect on FSHβ promoter activity, reducing expression an additional 30% from the level of the NF-Y mutant to 50% that of the wild-type promoter. This indicates that the SF-1 elements contribute to FSHβ gene expression, in part, through a functional interaction with NF-Y. The necessity for mutating the NF-Y element to observe a role for the SF-1 sites may be due to the ability of these factors to physically interact. For example, if SF-1 is capable of conferring its activity on the FSHβ promoter through either binding its own sites or through interacting with NF-Y on the NF-Y site in LβT2 cells, then the result that mutation or deletion of the SF-1 sites alone is insufficient to decrease expression to a significant level would be observed. In addition to NF-Y, SF-1 has been shown to physically interact with many transcription factors including transcription factor IIB, GCN5, c-jun, Ptx1, early growth response 1, SP1, androgen receptor, and GATA-4 (26–31). It is possible that SF-1 activates transcription of FSHβ through interactions with these and other proteins and that these interactions mask the direct role of the SF-1 sites themselves on FSHβ promoter activity unless the NF-Y site is mutated.
Consistent with most CCAAT boxes in TATA-containing promoters, the NF-Y site in the mouse FSHβ gene is located within the proximal 100 bp of the promoter (32). NF-Y is an important basal regulator of many genes and has been shown to physically interact with several key components of transcriptional machinery, including TATA binding protein (TBP), p300/CREB binding protein (CBP), GCN5, and p300/CBP-associated factor, as well as many other transcription factors, such as SP1, hepatocyte nuclear factor 4, estrogen receptor-α, and sterol regulatory element binding proteins (23, 33–38). These protein-protein interactions can occur through different mechanisms. In many instances, they involve just one of the NF-Y subunits, but in some cases, the interaction requires the NF-Y(BC) dimer or the complete NF-Y trimer. In the case of SF-1, the interaction occurs through the NF-YA subunit. Recently, NF-Y binding sites have been demonstrated to be essential for activity of the Hoxb4 regulatory region (39). Although, an NF-Y site alone is not sufficient to confer tissue-specific expression on a heterologous promoter, inclusion of flanking sequences does lead to a spatially restricted pattern of expression (39). This indicates that NF-Y cooperates with adjacent factors to regulate expression of Hoxb4. The ability of NF-Y to recruit cofactors may be critical to its role as a basal regulator of transcription. It may also represent a mechanism through which NF-Y is involved in cell-specific expression despite its relatively ubiquitous pattern of expression.
Despite the presence of both SF-1 and NF-Y in LβT2 and αT3-1 cells and our demonstration that proteins from both cell lines exhibit DNA-binding activity in vitro, neither SF-1 nor NF-Y contributes to activity of the FSHβ promoter in αT3-1 cells. This specific lack of activity in αT3-1 cells can be explained by several possible mechanisms. The activity generated by the interaction of SF-1 with NF-Y may require LβT2 cell-specific posttranslational modification of one or both factors or an LβT2 cell-specific coactivator. Conversely, αT3-1 cells may contain a factor that blocks the interaction between NF-Y and SF-1, thus preventing their cooperative activity. The specificity of the SF-1/NF-Y functional interaction to LβT2 cells suggests that additional proteins are interacting with SF-1 and NF-Y and that the components of this complex vary in each cell line. It is likely that additional regulatory elements and transcription factors are also important for FSHβ gene expression, which may or may not interact with SF-1 or NF-Y. It is also possible that FSHβ gene expression in αT3-1 cells may be repressed by a factor binding inside the −64-bp region, preventing activation of any of the promoters used. Furthermore, it is likely that the combination of factors contributing to LβT2 cell specificity when compared with immature gonadotropes (αT3-1 cells) differs from the combination of factors contributing to LβT2 cell specificity when compared with a non gonadotrope cell (NIH3T3 cells). Some of these factors may be involved in activin regulation of FSHβ, and some may be independent of the activin system, as are SF-1 and NF-Y. One possible regulator of FSHβ is the homeodomain-containing transcription factor Ptx1. Ptx1 is important for pituitary development, can physically interact and synergize with SF-1, and has been implicated in LHβ gene regulation (26, 40, 41). Recently, a Ptx1 binding site was identified within the proximal region of the rat FSHβ promoter and shown to be important for basal expression (42). This site is conserved in the mouse FSHβ promoter, located at approximately −54 bp, and may be involved in some of the remaining basal activity present in our −64-bp truncation. However, as with SF-1 and NF-Y, Ptx1 is expressed in both LβT2 and αT3-1 cells (7) and is unlikely to be solely responsible for the differential expression of FSHβ between the cell lines.
Tissue-specific gene expression is often conferred by complex control regions in which several cell-restricted, but not necessarily cell-specific, factors interact (43, 44). The spatial and temporal pattern in which regulatory factors are expressed is crucial when cell-fate decisions are occurring since it is the repertoire of factors present that ultimately determines cell types. It is likely that a combination of transcriptional activators conferring FSHβ gene expression in LβT2 cells and repressors preventing FSHβ gene expression in αT3-1 cells produces the differential gene expression in these gonadotrope-derived cell lines as well as during gonadotrope maturation. Here we demonstrate that cell specificity can involve the interaction between tissue-restricted factors, such as SF-1, and ubiquitous factors, such as NF-Y. Although NF-Y and SF-1 are not sufficient to confer FSHβ gene expression or to fully distinguish between the gonadotrope-derived cell lines, they are clearly required to provide full, basal expression in LβT2 cells. Moreover, the LβT2 cell-specific interaction between these sites contributes to the specific expression of FSHβ in these mature gonadotrope cells.
It is of particular interest that functional NF-Y, SF-1, and Ptx1 binding sites have been identified in the FSHβ promoter, since they have also been described in the LHβ promoter (4, 6, 45, 46). In transgenic mice, the elements through which Ptx1, SF-1, and NF-Y act are essential for basal activity of the LHβ promoter (5, 6, 45). This parallel mechanism of gene regulation of LHβ and FSHβ may be involved in coordinating gonadotrope-specific expression of these genes. SF-1 is clearly involved in gonadotrope determination as it is essential for expression of many gonadotrope markers. Similarly, Ptx1 is important for pituitary development and expression of the gonadotropin genes in vivo (40) and can transactivate numerous pituitary-specific promoters, including FSHβ, LHβ, α-GSU, and GnRH-receptor (41, 47). Additionally, NF-Y can regulate expression of genes involved in similar functions, such as major histocompatibility complex II genes involved in peptide presentation (48) and several genes involved in cholesterol metabolism (23, 38). In the gonadotrope, NF-Y might function to coordinate expression of gonadotrope-specific genes by interacting with SF-1. It will be interesting to determine whether NF-Y binding sites are also present in the α-GSU- and GnRH receptor-regulatory regions, whether they are important for cell-specific expression of those genes, and if they interact with the well characterized SF-1 elements and/or the Ptx1 sites. The onset of LHβ expression precedes the onset of FSHβ expression by 1 d during embryonic development (e16.5 vs. e17.5) (49), indicating that differences in gonadotropin β-subunit gene activation must also exist. The work presented here is a key step in understanding factors involved in the developmental activation of FSHβ gene expression in the maturing gonadotrope cell. Further characterization of the interactions occurring in the regulatory regions of the various gonadotropin genes will likely lead to a greater understanding of the molecular mechanisms, both common and unique, involved in their coordinated and differential expression.
Materials and Methods
Immunocytochemistry
Cells were trypsinized and plated onto glass coverslips in 24-well tissue culture dishes at a density of 150,000 cells per well (LβT2), 100,000 cells per well (αT3-1), or 20,000 cells per well (NIH3T3). Cell culture conditions have been described previously (7). Immunostaining was performed as follows: Cells were washed twice with 1× PBS containing 1 mm MgCl2 and then fixed for 20 min with 4% formaldehyde and 1 mm MgCl2 in 1× PBS. Cells were blocked and permeabilized with 20% donkey serum in wash buffer (1% BSA, 0.5% NP-40 in 1× PBS) for 1 h. The primary antibodies were diluted in wash buffer and bound for 1 h. The following dilutions were used: FSHβ (1:2000), LHβ (1:2000), α-GSU (1:2000), SF-1 (1:1000), or NF-YA (1:2000). The FSHβ, LHβ, and α-GSU antibodies were purchased from the National Hormone and Peptide Program; the SF-1 antibody was provided by Dr. Bon-Chu Chung, and the NF-YA antibody is from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The secondary antibodies (Texas Red-conjugated donkey α-rabbit IgG or fluorescein isothiocyanate-conjugated donkey α-mouse IgG from Jackson ImmunoResearch Laboratories, West Grove, PA) were diluted 1:200 in wash buffer and bound for 30 min. After three washes each with wash buffer and 1× PBS, nuclei were stained with 4,6-diamidino-2-phenylindole (1 μg/ml; Roche Biochemical, Indianapolis, IN) for 5 min. Coverslips were mounted in a solution containing 10% gelvatol (polyvinyl alcohol), 33% glycerol, and 0.1 m Tris-Cl, pH 8.5. Images (0.2-μm sections) were captured using a Delta-Vision deconvolution microscope system (Applied Precision, Inc., Issaquah, WA) and a Nikon ×100 (NA 1.4) lens. The data sets were deconvolved and analyzed using SoftWorx software (Applied Precision, Inc.). Deconvolution microscopy was performed with assistance from Steve McMullen at the UCSD Cancer Center Digital Imaging Shared Resource.
Plasmid Construction and Preparation
The 398-bp mouse FSHβ promoter was PCR amplified from a genomic clone (kindly provided by Dr. Malcolm Low) and ligated into the SmaI restriction site of the pGL3 luciferase reporter plasmid (Promega Corp., Madison, WI) to generate −398FSHβLuc. The −304FSHβLuc and −230FSHβLuc truncations were created by digesting the MluI/BglII fragment of −398FSHβLuc with XmnI and BbvI restriction enzymes, respectively. The truncated promoter fragments were then blunt ended with Klenow fragment (New England Biolabs, Beverly, MA) and ligated into the SmaI restriction site of pGL3. The −129FSHβLuc and −64FSHβLuc reporter genes were created by digesting the MluI/BglII fragment of −304FSHβLuc with DpnI and HpaI restriction enzymes, respectively. The fragments were blunt ended with Klenow and ligated into the SmaI restriction site of pGL3. The −194FSHβLuc plasmid was generated by digesting the MluI/BglII fragment of −230FSHβLuc with the RsaI restriction enzyme and cloning the appropriate promoter fragment into the SmaI and BglII sites of pGL3. The −95FSHβLuc plasmid was created by PCR amplifying the promoter from −398FSHβLuc using a forward primer corresponding to the sequence of the mouse FSHβ promoter from −95 bp to −77 bp and containing a KpnI linker and a reverse primer spanning the HindIII restriction site from the pGL3 vector. The PCR product was digested with KpnI and BglII restriction enzymes and ligated into the corresponding sites in pGL3. The sequences of all promoter fragments were confirmed by dideoxynucleotide sequencing (50).
Plasmid DNA was prepared from overnight bacterial cultures using a cesium chloride protocol adapted from Sambrook et al. (51).
Transient Transfections, Normalizing Transfection Data, and Statistics
Transient transfections were performed as described previously (7) using 2.5 μg of the reporter plasmids. For the activin and follistatin experiments, cells were treated with vehicle (1× PBS), 100 ng/ml recombinant mouse follistatin 288 (R & D Systems, Minneapolis, MN), 10 ng/ml human recombinant activin A (Calbiochem, San Diego, CA), or a combination of both follistatin and activin 24 h after transfection in DMEM with 10% fetal bovine serum. Twenty-four hours after treatment, cells were harvested and assayed for luciferase and β-galactosidase activity. All transfection experiments were performed at least three times in triplicate. To control for differences in expression between the different cell types, each experiment was normalized. The RSV enhancer fused to the RSV promoter driving luciferase (RSV-Luc) was transfected in parallel with FSHβLuc in each experiment. The RSV enhancer and promoter fused to β-galactosidase (RSV-β-gal, 0.5 μg) was used as an internal control for each transfected plate of cells. The RSV-Luc luciferase values were divided by the RSV-β-gal β-galactosidase values, averaged, and set equal to 100. The FSHβLuc/RSV-β-gal values were normalized to this value in each cell type; thus, the values from the individual cell types can be directly compared. The error bars in all bar graphs represent sem. Normal or Box Cox Transformed ratios for each promoter construct in each cell type were compared by the ANOVA Factorial test, followed by the Tukey-Kramer HSD post hoc test. In all analyses, P ≤ 0.05 was considered significant.
Nuclear Extracts and EMSAs
Preparation of nuclear extracts and binding conditions for the EMSAs have been described previously (7). Binding reactions were incubated for 5 min at room temperature after addition of probe, loaded onto a 5% nondenaturing polyacrylamide gel, and electrophoresed. For competition and antibody experiments, 100-fold excess unlabeled competitor oligonucleotide or 1 μl antibody was added 20 min before addition of probe. The SF-1 antibody used in EMSA was purchased from Upstate Biotechnology (Lake Placid, NY), and the NF-YA antibody was purchased from Chemicon International (Temecula, CA).
Mutagenesis
Mutagenesis of FSHβLuc was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Oligonucleotides used for mutagenesis were the following: 341SF-1mut [5′-GGATCAATTAAGACATATTTTGGTTTAAATTCGCAATGGAGCCAAAGCAATGTTCAG-3′ (top strand)], 239SF-1mut [5′-GTCTAGACTCTAGAGTCACATTTAATTTAGAATTTGAGGGAGTGGGTGTGCTGCC-3′ (top strand)], and NF-Ymut [5′-CAGCAGGCTTTATGTTGGTACCGGTCATGTTAACACCC-3′ (top strand)]. Mutations (underlined) were confirmed by dideoxynucleotide sequencing performed by the DNA Sequencing Shared Resource, UCSD Cancer Center.
GST-Interaction Assay
The GST-SF1 fusion protein was created by cloning the SF-1 cDNA into the pGEX-4T1 vector. Sequencing confirmed that the SF-1 reading frame was cloned in-frame with the GST protein. The GST-NF-YA and GST-NF-YB vectors were provided by Dr. Sankar Maity and Dr. David Gardner, respectively (37, 52). The SF-1 expression vector was made by cloning the SF-1 cDNA into pcDNA3. The NF-Y and GFP expression vectors were provided by Dr. Roberto Mantovani and Dr. Douglass Forbes, respectively. The Ptx1 vector has been described previously (7). 35S-labeled proteins were produced using the TnT T7 Coupled Reticulocyte Lysate System (Promega Corp.). Bacteria transformed with the pGEX vectors were grown to an OD of 0.5 and then induced with 0.2 mm isopropyl-β-d-thiogalactoside overnight. Bacterial pellets were sonicated in 0.1% Triton X-100, 5 mm EDTA in 1× PBS and centrifuged, and the supernatant was bound to glutathione sepharose 4B resin (Amersham Pharmacia, Piscataway, NJ). The interaction assay was performed as described previously (53).
Acknowledgments
We thank Malcolm Low for generously providing the mouse FSHβ genomic clone; Bon-Chu Chung for the SF-1 antibody and expression vector; David Gardner for the GST-NF-YB plasmid; Sankar Maity for the GST-NF-YA plasmid; and Roberto Mantovani for the NF-Y expression vectors. We are grateful to Stephen McMullen and James Feramisco at The UCSD Digital Imaging Core for technical assistance; and to Naama Rave-Harel, Marjory Givens, Mark A. Lawson, and Suzie Bailey for helpful discussions.
This work was supported by NIH Grant R37 HD-20377 (to P.L.M.). This work was also supported by NICHD/NIH through cooperative agreement (U54 HD12303) as part of the Specialized Cooperative Centers Program in Reproduction Research (P.L.M.). S.B.R.J. and S.M.M. were partially supported by NIH Training Grant T32 DK-07451. D.C. was supported by NIH Grants NRSA HD-41301 and T32 DK-07044. The UCSD Cancer Center Digital Imaging and DNA Shared Resources are supported in part by NCI Cancer Center Support Grant P30 CA23100.
Abbreviations
- GFP
Green fluorescent protein
- GSE
gonadotrope-specific element
- GST
glutathione-S-transferase
- α-GSU
α-glycoprotein subunit
- NF-Y
nuclear factor 1
- Ptx1
pituitary homeobox 1
- RSV
Rous sarcoma virus
- RSV-β-gal
RSV-β-galactosidase
- RSV-Luc
RSV-luciferase
- SF-1
steroidogenic factor 1
References
- 1.Pierce JG, Parsons TF. Glycoprotein hormones: structure and function. Annu Rev Biochem. 1981;50:465–495. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- 2.Schoderbek WE, Kim KE, Ridgway EC, Mellon PL, Maurer RA. Analysis of DNA sequences required for pituitary-specific expression of the glycoprotein hormone α-subunit gene. Mol Endocrinol. 1992;6:893–903. doi: 10.1210/mend.6.6.1379672. [DOI] [PubMed] [Google Scholar]
- 3.Barnhart KM, Mellon PL. The orphan nuclear receptor, steroidogenic factor-1, regulates the glycoprotein hormone α-subunit gene in pituitary gonadotropes. Mol Endocrinol. 1994;8:878–885. doi: 10.1210/mend.8.7.7527122. [DOI] [PubMed] [Google Scholar]
- 4.Halvorson LM, Ito M, Jameson JL, Chin WW. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone β-subunit gene expression. J Biol Chem. 1998;273:14712–14720. doi: 10.1074/jbc.273.24.14712. [DOI] [PubMed] [Google Scholar]
- 5.Quirk CC, Lozada KL, Keri RA, Nilson JH. A single Pitx1 binding site is essential for activity of the LHβ promoter in transgenic mice. Mol Endocrinol. 2001;15:734–746. doi: 10.1210/mend.15.5.0628. [DOI] [PubMed] [Google Scholar]
- 6.Keri RA, Bachmann DJ, Behrooz A, Herr BD, Ameduri RK, Quirk CC, Nilson JH. An NF-Y binding site is important for basal, but not gonadotropin-releasing hormone-stimulated, expression of the luteinizing hormone β subunit gene. J Biol Chem. 2000;275:13082–13088. doi: 10.1074/jbc.275.17.13082. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg SB, Mellon PL. An Otx-related homeodomain protein binds an LHβ promoter element important for activation during gonadotrope maturation. Mol Endocrinol. 2002;16:1280–1298. doi: 10.1210/mend.16.6.0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar TR, Fairchild-Huntress V, Low MJ. Gonadotrope-specific expression of the human follicle-stimulating hormone β-subunit gene in pituitaries of transgenic mice. Mol Endocrinol. 1992;6:81–90. doi: 10.1210/mend.6.1.1738375. [DOI] [PubMed] [Google Scholar]
- 9.Huang HJ, Sebastian J, Strahl BD, Wu JC, Miller WL. The promoter for the ovine follicle-stimulating hormone-β gene (FSHβ) confers FSHβ-like expression on luciferase in transgenic mice: regulatory studies in vivo and in vitro. Endocrinology. 2001;142:2260–2266. doi: 10.1210/endo.142.6.8202. [DOI] [PubMed] [Google Scholar]
- 10.Pernasetti F, Vasilyev VV, Rosenberg SB, Bailey JS, Huang HJ, Miller WL, Mellon PL. Cell-specific transcriptional regulation of follicle-stimulating hormone-β by activin and gonadotropin-releasing hormone in the LβT2 pituitary gonadotrope cell model. Endocrinology. 2001;142:2284–2295. doi: 10.1210/endo.142.6.8185. [DOI] [PubMed] [Google Scholar]
- 11.Graham KE, Nusser KD, Low MJ. LβT2 gonadotroph cells secrete follicle stimulating hormone (FSH) in response to activin A. J Endocrinol. 1999;162:R1–R5. doi: 10.1677/joe.0.162r001. [DOI] [PubMed] [Google Scholar]
- 12.Ling N, Ying SY, Ueno N, Shimasaki S, Esch F, Hotta M, Guillemin R. Pituitary FSH is released by a heterodimer of the β-subunits from the two forms of inhibin. Nature. 1986;321:779–782. doi: 10.1038/321779a0. [DOI] [PubMed] [Google Scholar]
- 13.Carroll RS, Kowash PM, Lofgren JA, Schwall RH, Chin WW. In vivo regulation of FSH synthesis by inhibin and activin. Endocrinology. 1991;129:3299–3304. doi: 10.1210/endo-129-6-3299. [DOI] [PubMed] [Google Scholar]
- 14.Vasilyev VV, Pernasetti F, Rosenberg SB, Barsoum MJ, Austin DA, Webster NJ, Mellon PL. Transcriptional activation of the ovine follicle-stimulating hormone-β gene by gonadotropin-releasing hormone involves multiple signal transduction pathways. Endocrinology. 2002;143:1651–1659. doi: 10.1210/endo.143.5.8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alarid ET, Windle JJ, Whyte DB, Mellon PL. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329. doi: 10.1242/dev.122.10.3319. [DOI] [PubMed] [Google Scholar]
- 16.Sosnowski R, Mellon PL, Lawson MA. Activation of translation in pituitary gonadotrope cells by gonadotropin-releasing hormone. Mol Endocrinol. 2000;14:1811–1819. doi: 10.1210/mend.14.11.0550. [DOI] [PubMed] [Google Scholar]
- 17.Ying SY. Inhibins, activins, and follistatins: gonadal proteins modulating the secretion of follicle-stimulating hormone. Endocr Rev. 1988;9:267–293. doi: 10.1210/edrv-9-2-267. [DOI] [PubMed] [Google Scholar]
- 18.Shimasaki S, Koga M, Esch F, Cooksey K, Mercado M, Koba A, Ueno N, Ying SY, Ling N, Guillemin R. Primary structure of the human follistatin precursor and its genomic organization. Proc Natl Acad Sci USA. 1988;85:4218–4222. doi: 10.1073/pnas.85.12.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimonaka M, Inouye S, Shimasaki S, Ling N. Follistatin binds to both activin and inhibin through the common subunit. Endocrinology. 1991;128:3313–3315. doi: 10.1210/endo-128-6-3313. [DOI] [PubMed] [Google Scholar]
- 20.Ingraham HA, Lala DS, Ikeda Y, Luo X, Shen WH, Nachtigal MW, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2302–2312. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Bakke M, Krimkevich Y, Cushman LJ, Parlow AF, Camper SA, Parker KL. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development. 2001;128:147–154. doi: 10.1242/dev.128.2.147. [DOI] [PubMed] [Google Scholar]
- 22.Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 24.Ngan ES, Cheng PK, Leung PC, Chow BK. Steroidogenic factor-1 interacts with a gonadotrope-specific element within the first exon of the human gonadotropin-releasing hormone receptor gene to mediate gonadotrope-specific expression. Endocrinology. 1999;140:2452–2462. doi: 10.1210/endo.140.6.6759. [DOI] [PubMed] [Google Scholar]
- 25.Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 26.Tremblay JJ, Drouin J. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone β gene transcription. Mol Cell Biol. 1999;19:2567–2576. doi: 10.1128/mcb.19.4.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugawara T, Saito M, Fujimoto S. Sp1 and SF-1 interact and cooperate in the regulation of human steroidogenic acute regulatory protein gene expression. Endocrinology. 2000;141:2895–2903. doi: 10.1210/endo.141.8.7602. [DOI] [PubMed] [Google Scholar]
- 28.Li LA, Chiang EF, Chen JC, Hsu NC, Chen YJ, Chung BC. Function of steroidogenic factor 1 domains in nuclear localization, transactivation, and interaction with transcription factor TFIIB and c-Jun. Mol Endocrinol. 1999;13:1588–1598. doi: 10.1210/mend.13.9.0349. [DOI] [PubMed] [Google Scholar]
- 29.Jacob AL, Lund J, Martinez P, Hedin L. Acetylation of steroidogenic factor 1 protein regulates its transcriptional activity and recruits the coactivator GCN5. J Biol Chem. 2001;276:37659–37664. doi: 10.1074/jbc.M104427200. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen JS, Nilson JH. AR suppresses transcription of the LHβ subunit by interacting with steroidogenic factor-1. Mol Endocrinol. 2001;15:1505–1516. doi: 10.1210/mend.15.9.0691. [DOI] [PubMed] [Google Scholar]
- 31.Tremblay JJ, Viger RS. Transcription factor GATA-4 enhances Mullerian inhibiting substance gene transcription through a direct interaction with the nuclear receptor SF-1. Mol Endocrinol. 1999;13:1388–1401. doi: 10.1210/mend.13.8.0330. [DOI] [PubMed] [Google Scholar]
- 32.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Currie RA. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 34.Faniello MC, Bevilacqua MA, Condorelli G, de Crombrugghe B, Maity SN, Avvedimento VE, Cimino F, Costanzo F. The B subunit of the CAAT-binding factor NFY binds the central segment of the co-activator p300. J Biol Chem. 1999;274:7623–7626. doi: 10.1074/jbc.274.12.7623. [DOI] [PubMed] [Google Scholar]
- 35.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, Wolffe AP. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farsetti A, Narducci M, Moretti F, Nanni S, Mantovani R, Sacchi A, Pontecorvi A. Inhibition of ERα-mediated trans-activation of human coagulation factor XII gene by heteromeric transcription factor NF-Y. Endocrinology. 2001;142:3380–3388. doi: 10.1210/endo.142.8.8345. [DOI] [PubMed] [Google Scholar]
- 37.Liang F, Schaufele F, Gardner DG. Functional interaction of NF-Y and Sp1 is required for type a natriuretic peptide receptor gene transcription. J Biol Chem. 2001;276:1516–1522. doi: 10.1074/jbc.M006350200. [DOI] [PubMed] [Google Scholar]
- 38.Dooley KA, Millinder S, Osborne TF. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J Biol Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 39.Gilthorpe J, Vandromme M, Brend T, Gutman A, Summerbell D, Totty N, Rigby PW. Spatially specific expression of Hoxb4 is dependent on the ubiquitous transcription factor NFY. Development. 2002;129:3887–3899. doi: 10.1242/dev.129.16.3887. [DOI] [PubMed] [Google Scholar]
- 40.Szeto DP, Rodriguez-Esteban C, Ryan AK, O'Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisúa-Belmonte JC, Rosenfeld MG. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tremblay JJ, Lanctôt C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12:428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 42.Zakaria MM, Jeong KH, Lacza C, Kaiser UB. Pituitary homeobox 1 activates the rat FSHβ (rFSHβ) gene through both direct and indirect interactions with the rFSHβ gene promoter. Mol Endocrinol. 2002;16:1840–1852. doi: 10.1210/me.2002-0088. [DOI] [PubMed] [Google Scholar]
- 43.Struhl K. Mechanisms for diversity in gene expression patterns. Neuron. 1991;7:177–181. doi: 10.1016/0896-6273(91)90256-y. [DOI] [PubMed] [Google Scholar]
- 44.He X, Rosenfeld MG. Mechanisms of complex transcriptional regulation: implications for brain development. Neuron. 1991;7:183–196. doi: 10.1016/0896-6273(91)90257-z. [DOI] [PubMed] [Google Scholar]
- 45.Keri RA, Nilson JH. A steroidogenic factor-1 binding site is required for activity of the luteinizing hormone β subunit promoter in gonadotropes of transgenic mice. J Biol Chem. 1996;271:10782–10785. doi: 10.1074/jbc.271.18.10782. [DOI] [PubMed] [Google Scholar]
- 46.Tremblay JJ, Marcil A, Gauthier Y, Drouin J. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J. 1999;18:3431–3441. doi: 10.1093/emboj/18.12.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tremblay JJ, Goodyer CG, Drouin J. Transcriptional properties of Ptx1 and Ptx2 isoforms. Neuroendocrinology. 2000;71:277–286. doi: 10.1159/000054547. [DOI] [PubMed] [Google Scholar]
- 48.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 49.Japon MA, Rubinstein M, Low MJ. In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J Histochem Cytochem. 1994;42:1117–1125. doi: 10.1177/42.8.8027530. [DOI] [PubMed] [Google Scholar]
- 50.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Sinha S, Maity SN, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zappavigna V, Sartori D, Mavilio F. Specificity of HOX protein function depends on DNA-protein and protein-protein interactions, both mediated by the homeo domain. Genes Dev. 1994;8:732–744. doi: 10.1101/gad.8.6.732. [DOI] [PubMed] [Google Scholar]