Abstract
Background
Recent discoveries of risk alleles have made it possible to define genetic risk profiles for patients with rheumatoid arthritis (RA). We examined whether a cumulative score based on 22 validated genetic risk alleles for seropositive RA would identify high-risk, asymptomatic individuals who might benefit from preventive interventions.
Methods
We genotyped 14 single nucleotide polymorphisms (SNPs) at 13 validated RA risk loci and 8 HLA alleles among (1) 289 Caucasian seropositive cases and 481 controls from the US Nurses' Health Studies (NHS), and (2) 629 Caucasian CCP antibody positive cases and 623 controls from the Swedish Epidemiologic Investigation of RA (EIRA). We created a weighted genetic risk score (GRS), where the weight for each risk allele is the log of the published odds ratio. We used logistic regression to study associations with incident RA. We compared AUCs from a clinical-only model and clinical + genetic model in each cohort.
Results
Patients with GRS > 1.25 standard deviations of the mean had a significantly higher OR of seropositive RA in both NHS (OR=2.9, 95%CI 1.8–4.6) and EIRA (OR=3.4, 95% CI 2.3–5.0) referent to the population average. In NHS, the AUC for a clinical model was 0.57 and for a clinical + genetic model was 0.66, and in EIRA was 0.63 and 0.75, respectively.
Conclusion
The combination of 22 risk alleles into a weighted genetic risk score significantly stratifies individuals for RA risk beyond clinical risk factors alone. However, given the low incidence of RA, the clinical utility of a weighted genetic risk score is limited in the general population.
Keywords: rheumatoid arthritis, polymorphism, autoantibodies, anti-CCP, smoking
RA is a complex autoimmune disease thought to develop in genetically predisposed individuals when exposed to certain environmental factors. Early diagnosis and treatment strategies are critical to minimize disability from joint destruction1. Although epidemiologic research has produced convincing data linking cigarette smoking to RA risk2–4, and identified genetic variants associated with RA risk in the major histocompatibility complex (MHC) region were discovered over 30 years ago5, these risk factors are not used clinically for behavior modification, preventive therapy, or in establishing a diagnosis of RA. Similarly, the presence of RA-specific autoantibodies and inflammatory biomarkers appear years before disease onset and predict more severe disease, but are not used in clinical medicine prior to the onset of symptoms6–9.
Advances in human genetics have led to a dramatic increase in the number of validated disease risk alleles in RA. There are now up to 22 risk alleles that explain approximately one-third of the genetic burden of seropositive RA risk5, 10–20. Much of the risk is derived from 8 alleles that reside within the MHC region5, with up to 5% of risk explained by the 14 alleles outside of the MHC20. The discoveries of these alleles for RA, and similar discoveries for risk alleles in other diseases, has spurned much discussion about the clinical validity of using genetic results in personalized medicine21–24.
Despite these advances, it is not clear how to utilize genetic information for prediction of RA risk in clinical practice. A critical first step is to understand the role of aggregate genetic risk factors, rather than associations of individual alleles with RA. Towards this end, we used 22 validated RA risk alleles to derive an aggregate genetic risk score (GRS) in seropositive RA patients derived from over 238,000 prospectively followed subjects from the United States (Nurses' Health Study) and seropositive RA patients derived from a large case control study of > 3600 subjects from Sweden (Epidemiologic Investigation of Rheumatoid Arthritis). We calculated odds ratios for seropositive RA relative to the median risk in these datasets and estimated genotype-specific incidence, which is a more useful measure of risk in a clinical setting. We compared predicted multi-locus odds ratios—formed by taking the product of individual-locus odds ratios estimated in a previous meta-analysis20 — to multi-locus odds ratios estimated in this data set. We included the strongest epidemiologic risk factors for RA in the general population in the models (age, sex, and smoking) as “clinical” risk factors. Although the GRS is strongly associated with seropositive RA and adds significantly to the discrimination of a clinical model, the genotype-specific incidence remains low, suggesting that genetic information is not yet clinically useful in an asymptomatic individual patient.
Methods
STUDY SAMPLE
The Nurses' Health Study (NHSI) is a prospective cohort of 121,700 female nurses, aged 30–55 years in 1976 in which 32,826 (27%) NHSI participants aged 43 to 70 years provided blood samples for future studies and an additional 33,040 (27%) provided buccal cell samples, a total of 65,866 (54% of the cohort). The Nurses' Health Study II (NHSII) is a similar prospective cohort, established in 1989, with 116,609 female nurses aged 25–42 years in which 29,611 (25%) provided blood samples for future studies. In the current study, we combine both NHSI and NHSII, herein referred to simply as `NHS'. All women in both cohorts completed an initial questionnaire and have been followed biennially by questionnaire to update exposures and disease diagnoses. The specificity of CTD detection using a staged series design is very high, reducing misclassification of healthy subjects25. RA cases were validated, using previously described methods4, in which two board-certified rheumatologists trained in chart abstraction independently conducted a medical record review blinded to the second reviewer's result, examining the charts for the American College of Rheumatology (ACR) classification criteria for RA26, date of first RA symptom, evidence of RA-specific medication treatment, and the treating physician's diagnosis. Definite RA included subjects with four of the seven ACR criteria documented in the medical record or agreement by 2 rheumatologists on the diagnosis of RA with 3 documented ACR criteria for RA and a diagnosis of RA by their physician. Seropositive status was determined by chart review, and in some cases by direct assay, as previously described9. Each NHS participant with confirmed incident or prevalent RA was matched by year of birth, race/ethnicity, menopausal status, and postmenopausal hormone use to a single healthy woman in the same cohort without RA.
This initial NHS nested case-control dataset consists of 585 RA cases and 585 matched controls. To minimize potential population stratification, we excluded non-Caucasian women (based on self-report), resulting in 564 total RA cases and 571 controls. We restricted our analysis to only seropositive RA, resulting in a sample of 327 seropositive RA cases and 571 controls. Covariate information was collected from the subjects in both cohorts via prospective biennial questionnaires regarding diseases, lifestyle, and health practices. All aspects of this study were approved by the Partners' HealthCare Institutional Review Board.
The Epidemiological Investigation of Rheumatoid Arthritis (EIRA) is a population based case-control study on incident RA in Sweden. Data on > 3,600 cases and controls was collected between May 1996 and December 2006. As described previously3, 27, a case is defined as an individual who fulfills the American College of Rheumatology (ACR) 1987 criteria for the classification of RA, and had symptoms for less than 1 year. For each potential case, a control subject was randomly selected from the study base, taking into consideration the subject's age, sex and geographic location. In total, 659 confirmed CCP positive RA cases and 650 controls were included. All aspects of the EIRA study were approved by the Karolinska Institutet Institutional Review Board.
SELECTION OF GENETIC RISK FACTORS AND GENOTYPING
We selected all validated seropositive RA susceptibility SNPs established prior to September 2008. We define validated as those alleles demonstrating p < 5×10−7 with evidence of replication at p < 0.05 in at least one independent study10–17, 20. One locus, CDK6, has a strong but not unequivocal evidence of association based on these criteria. In NHS, low resolution HLA-DRB1 genotyping was performed using polymerase chain reaction with sequence specific primers (PCR-SSP) using OLERUP SSP kits (QIAGEN, West Chester, PA), as previously described.28 For samples with positive 2-digit HLA signals, sequence specific primers were used for high-resolution 4-digit allele detection of DRB1*0401, *0404, *0405, *0408, *0101, *0102, *09 and *1001. In EIRA, low-resolution HLA typing was performed using Olerup PCR-SSP (DR low resolution and DR4 kits, Olerup SSP AB, Saltsjöbaden, Sweden). High resolution typing was performed for positive *04 samples. Thus 4 digit HLA subtypes were available from EIRA for *0401, *0404, *0405, *0408 and 2 digit subtypes were available for other alleles. All non-MHC risk alleles for both NHS and EIRA were genotyped using iPlex (Sequenom) at the Broad Institute, as previously described20. All SNPs had call rates >95% and Hardy-Weinberg equilibrium p-values > 0.01.
We filtered our data to account for missing genotype information, dropping individuals with >10% missing SNP data and dropping individuals missing any HLA data. In NHS, among 327 seropositive RA cases, 6 (2%) were missing HLA data and 32 (10%) were missing >10% SNP information, leaving 289 seropositive RA cases in the analysis. Among 571 controls, 20 (4%) were missing HLA and 70 (12%) were missing >10% SNP information data, leaving us with 481controls in the analysis. In EIRA, among 659 cases, 3 (0.5%) were missing HLA data and 27 (4%) were missing > 10% SNP information leaving 629 cases in the analysis. Among 650 controls 1 (0.1%) was missing HLA results and 25 (4%) were missing > 10% SNP information, leaving 623 controls in the analysis. in a sample of 656 cases and 648 controls. The higher rates of genotyping failure in NHS were due primarily to poor quality cheek cell DNA samples. We are confident that this missingness is completely at random, and therefore does not bias our results, since the case and control samples were randomly interspersed on the genotyping plate and our resulting odds ratios are consistent with previously published results (see Table 2).
Table 2.
Allele frequencies and association with seropositive RA in NHS and CCP positive RA in EIRA for 22 alleles
| NHS | EIRA | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Loci | SNP | Risk Allele | Published OR1 | Cases RAF | Controls RAF | OR (95% CI) | Cases RAF | Controls RAF | OR (95% CI) |
| DRB*0401 | SE | SE | 3.30 | 0.15 | 0.09 | 1.70 (1.25–2.29) | 0.30 | 0.13 | 3.04 (2.45–3.77) |
| DRB*0404 | SE | SE | 1.85 | 0.07 | 0.04 | 1.81 (1.13–2.90) | 0.10 | 0.04 | 2.94 (2.07–4.18) |
| DRB*0405 | SE | SE | 3.84 | 0.02 | 0.003 | 6.31 (1.75–22.81) | 0.008 | 0.004 | 1.98 (0.67–5.83) |
| DRB*0408 | SE | SE | 1.04 | 0.003 | 0.006 | 0.55 (0.11–2.76) | 0.02 | 0.004 | 5.07 (1.923–13.34) |
| DRB*0101 | SE | SE | 1.60 | 0.11 | 0.06 | 1.83 (1.27–2.63) | 0.142 | 0.10 | 1.47 (1.15–1.89) |
| DRB*0102 | SE | SE | 1.10 | 0.01 | 0.01 | 1.69 (0.63–4.54) | --- | --- | --- |
| DRB*1001 | SE | SE | 2.35 | 0.01 | 0.01 | 2.26 (0.78–6.58) | 0.02 | 0.01 | 1.53 (0.79–2.97) |
| DRB*09 | SE | SE | 1.48 | 0.01 | 0.01 | 1.34 (0.36–5.03) | 0.02 | 0.02 | 0.94 (0.53–1.67) |
| PTPN22 | rs2476601 | T | 1.75 | 0.14 | 0.09 | 1.75 (1.27–2.41) | 0.17 | 0.12 | 1.49 (1.19–1.86) |
| TRAF1-C5 | rs3761847 | G | 1.32 | 0.44 | 0.40 | 1.18 (0.95–1.45) | 0.51 | 0.45 | 1.29 (1.1–1.51) |
| STAT4 | rs7574865 | T | 1.27 | 0.24 | 0.21 | 1.18 (0.93–1.51) | 0.24 | 0.22 | 1.07 (0.89–1.29) |
| TNFAIP3 | rs17066662 | C* | 1.33 | 0.73 | 0.73 | 1.00 (0.80–1.25) | 0.80 | 0.79 | 1.04 (0.86–1.26) |
| TNFAIP3 | rs6920220 | A | 1.22 | 0.22 | 0.21 | 1.05 (0.82–1.35) | 0.26 | 0.22 | 1.23 (1.02–1.48) |
| CD40 | rs4810485 | G* | 1.15 | 0.76 | 0.75 | 1.05 (0.83–1.35) | 0.78 | 0.72 | 1.36 (1.13–1.64) |
| CCL21 | rs2812378 | C | 1.12 | 0.35 | 0.34 | 1.07 (0.87–1.33) | 0.36 | 0.34 | 1.11 (0.94–1.31) |
| CTLA4 | rs3087243 | G* | 1.11 | 0.54 | 0.52 | 1.06 (0.87–1.30) | 0.63 | 0.59 | 1.20 (1.02–1.4) |
| PADI4 | rs2240340 | A | 1.02 | 0.43 | 0.40 | 1.10 (0.89–1.35) | 0.42 | 0.39 | 1.14 (0.97–1.33) |
| CDK6 | rs42041 | G | 1.11 | 0.27 | 0.24 | 1.21 (0.96–1.53) | 0.25 | 0.24 | 1.02 (0.85–1.23) |
| TNFRSF14 | rs3890745 | T* | 1.12 | 0.67 | 0.68 | 0.93 (0.75–1.16) | 0.70 | 0.70 | 1.02 (0.86–1.21) |
| PRKCQ | rs4750316 | C* | 1.14 | 0.85 | 0.82 | 1.25 (0.95–1.65) | 0.82 | 0.80 | 1.09 (0.89–1.34) |
| KIF5A | rs1678542 | G* | 1.12 | 0.70 | 0.64 | 1.28 (1.02–1.60) | 0.59 | 0.58 | 1.06 (0.91–1.24) |
| IL2/IL21 | rs6822844 | G* | 1.09 | 0.84 | 0.84 | 0.99 (0.74–1.31) | 0.85 | 0.82 | 1.29 (1.04–1.62) |
Major Allele
Published OR with respect to the risk allele
Only 2 digit DR1 data available. NHS: Nurses' Health Studies, EIRA: Epidemiologic Investigation of Rheumatoid Arthritis, RAF: risk allele frequency, OR: odds ratio, SE: HLA-DRB1 shared epitope.
STATISTICAL METHODS
Characteristics of RA cases and controls were summarized by means and standard deviations for continuous variables and frequency and percent for categorical variables. Data for NHS was presented separately from data from EIRA. All analyses were performed using SAS version 9.1 or version 9.2 (SAS Institute, Cary, NC).
SELECTION OF EPIDEMIOLOGIC COVARIATES
In NHS and EIRA, lifetime history of smoking was collected at baseline. In the NHS cohorts, data concerning current smoking, and number of cigarettes smoked per day were updated in two year questionnaire cycles and data on pack years of smoking (number of packs per day × number of years smoking) was selected from the questionnaire cycle prior to the date of RA diagnosis (or index date in controls). In EIRA, pack-years of smoking was calculated prior to RA onset for cases or index date for controls. We included age, sex, and pack-years of smoking as “clinical” risk factors in the models.
ASSOCIATION BETWEEN GENETIC RISK ALLELES AND RA
We used logistic regression to study the association of each allele with risk of seropositive RA according to an additive log-odds model in NHS and in EIRA (Table 2).
WEIGHTED GENETIC RISK SCORE
We developed a “weighted-GRS” (wGRS) that utilized the allelic odds ratios (OR) from published studies to account for the strength of the genetic association within each allele. We calculated a wGRS22 that included 8 HLA-DRB1 “shared epitope” (HLA-SE) alleles and 14 non-MHC risk alleles, and a wGRS14 (no HLA) that included only the 14 non-MHC risk alleles. This is preferred over a simple count GRS, equal to the sum the number of risk alleles carried, since PTPN22 and HLA-SE have substantially higher odds ratios for RA than do the more recently discovered SNPs. The weights used in the wGRS were calculated as the natural log of the published OR with respect to the risk allele as presented in Table 2. The odds ratios for HLA-SE alleles were derived from a recent meta-analysis of all published studies29. The ORs for the 14 non-MHC alleles were derived from published studies in which results have been extensively replicated, including the following alleles: PTPN22 (rs2476601)10, TRAF1-C5 (rs3761847)13, STAT4 (rs7574865)12, TNFAIP3 (rs17066662, in LD with 10499194, R2 = 1.0)14), TNFAIP3 (rs6920220)14. We also included 9 alleles from a meta-analysis of GWAS data for 3,393 cases and 12,462 controls with replication in 3,929 seropositive RA cases and 5,807 matched controls by Raychaudhuri et al.20: CD40 (rs4810485), CCL21 (rs2812378), CTLA4 (rs3087243), PADI4 (rs2240340), CDK6 (rs42041), TNFRSF14 (rs3890745), PRKCQ (rs4750316), KIF5A (rs1678542), and 4q27 (rs6822844). For each non-MHC allele, we chose the OR in replication samples to avoid over-estimation of the true effect size30. In EIRA, we used a proxy SNP for STAT4 (rs11889341, r2=1.0 with rs7574865) and a proxy SNP for KIF5A rs775322, r2=1.0 with rs 1678542). For any individual with missing genotype data for a particular SNP, we assigned the expected allele count (twice the risk allele frequency) to that individual. We tested for epistasis and did not find any significant gene-gene interaction, in agreement with our previous studies13, 14, 20. Our results are consistent with a multiplicative genetic model. We did not consider more complex HLA associations, including analysis of compound heterozygotes that have substantially higher risk such as HLA 0401/0404 (9 cases and 3 controls (n=12 total) in NHS and 52 cases and 4 controls (n=56 total) in EIRA).
To determine the cumulative effect of the 14 or 22 alleles on risk of RA we first divided wGRS scores into 7 categories based on the mean and standard deviation (SD) of the wGRS distribution in the controls. Dividing our score into 7 categories provided the most robust distribution, allowing us to parse out the highest and lowest risk groups while assuring that there were sufficient numbers of cases and controls in these extreme categories of interest. Additional details on determination of the groupings are available in the Supplementary Methods. We used logistic regression models adjusting for year of birth, sex and total pack-years of smoking to study the association of wGRS22 with seropositive RA and wGRS14 (no HLA) with seropositive RA (Table 3), comparing each group to a referent median group. An ordinal wGRS variable based on our groupings was used to calculate a p-value for trend. Finally, we calculated the odds of RA for the top group (group 7) as compared to the bottom group (group 1) in two ways. First, by using group 1 as the referent group, the method used in other GRS analyses of complex diseases (eg. macular degeneration31, prostate cancer32, 33, lipid levels and heart disease34–37, and diabetes38–40). Second, because group 1 has few cases and the first method only considers subjects in group 7 and 1, we also compared the median wGRS score in group 7 to the median wGRS score in group 1 using a model derived from an ordinal wGRS variable in which each group was given its median wGRS value as a score.
Table 3.
Weighted GRS scores and odd ratios of seropositive RA in NHS and CCP positive RA in EIRA
| NHS | EIRA | |||||
|---|---|---|---|---|---|---|
| Sero+ Cases | Controls | OR (95%CI)1 | CCP+ Cases | Controls | OR (95% CI)2 | |
| wGRS22 | ||||||
| Groups | ||||||
| 1 | 8 | 32 (7%) | 0.51 (0.22–1.19) | 20 | 54 (9%) | 0.38 (0.20–0.71) |
| 2 | 27 | 79 (16%) | 0.67 (0.38–1.19) | 32 | 101 (16%) | 0.32 (0.19–0.53) |
| 3 | 37 | 113 (23%) | 0.67 (0.40–1.13) | 54 | 133 (21%) | 0.44 (0.28–0.70) |
| 4 | 49 | 96 (20%) | 1.00 (ref) | 96 | 107 (17%) | 1.00 (ref) |
| 5 | 51 | 64 (13%) | 1.69 (1.01–2.82) | 78 | 85 (14%) | 1.01 (0.65–1.57) |
| 6 | 32 | 38 (8%) | 1.77 (0.98–3.20) | 96 | 65 (10%) | 1.52 (0.98–2.36) |
| 7 | 85 | 59 (12%) | 2.85 (1.75–4.64) | 253 | 78 (13%) | 3.36 (2.27–4.97) |
| 7 vs. 13 | 5.61 (2.41–13.07) | 8.83 (4.77–6.32) | ||||
| wGRS22 | ||||||
| Ordinal Model | ||||||
| 7 vs. 14 | 6.30 (3.78–10.48) | 12.31 (8.12–18.67) | ||||
| wGRS14(noHLA) | ||||||
| Groups | ||||||
| 1 | 17 | 47 (10%) | 0.74 (0.38–1.43) | 42 | 60 (10%) | 0.86 (0.53–1.42) |
| 2 | 38 | 68 (14%) | 1.13 (0.66–1.94) | 61 | 79 (13%) | 1.01 (0.65–1.58) |
| 3 | 46 | 82 (17%) | 1.09 (0.65–1.83) | 88 | 116 (19%) | 0.95 (0.64–1.41) |
| 4 | 45 | 90 (19%) | 1.00 (ref) | 114 | 141 (23%) | 1.00 (ref) |
| 5 | 55 | 89 (19%) | 1.20 (0.73–1.98) | 107 | 94 (15%) | 1.62 (1.09–2.41) |
| 6 | 28 | 57 (12%) | 1.00 (0.55–1.80) | 85 | 60 (10%) | 1.89 (1.22–2.94) |
| 7 | 60 | 48 (10%) | 2.52 (1.49–4.28) | 132 | 73 (12%) | 2.43 (1.62–3.63) |
| 7 vs. 13 | 3.43 (1.74–6.74) | 2.81 (1.66–4.73) | ||||
| wGRS14 (no HLA) | ||||||
| Ordinal Model | ||||||
| 7 vs. 14 | 2.39 (1.44–3.98) | 3.22 (2.14–4.86) | ||||
Odds ratio adjusted for age and pack-years smoking
Odds ratio adjusted for age, sex, geographic region and pack-years smoking
Odds ratio adjusted for age, sex, geographic region and pack-years smoking using group 1 as referent group
Model based estimate of group 7 vs. group 1 using an ordinal model and takes into account all data in the model. NHS: Nurses' Health Studies, EIRA: Epidemiologic Investigation of Rheumatoid Arthritis, sero+: seropositive, CCP: cyclic-citrullinated peptide antibody, OR: odds ratio, CI: confidence interval; wGRS22: weighted genetic risk score with 22 alleles; wGRS14 (no HLA): weighted genetic risk score with 14 alleles, without HLA alleles
ADDITIONAL STATISTICAL ANALYSIS
To determine how well our wGRS predictors discriminate between cases and controls, we generated ROC curves by plotting sensitivity of the wGRS22 score (continuous) against 1-specificity and calculated the area-under-the-curve (AUC) for both NHS and EIRA. Because there are few established epidemiologic predictors other than age, sex and smoking in the asymptomatic general population, any improvement in the ROC curve contributed by the wGRS may have value in a clinical setting. ROC curves were plotted for a “clinical” model that included year of birth and pack-years of smoking in NHS and age, sex, pack-years of smoking, and geographic region in EIRA, for a “clinical + genetic” model based on adding wGRS14 (no HLA) and a full “clinical + genetic” model that included age, smoking, (residential area in EIRA only) and wGRS22. The AUCs were compared using a non-parametric approach with each “clinical + genetic” model compared to the “clinical” model as described by DeLong et.al.41
To judge how well previously-reported association results could be used to distinguish cases and controls in this data set, using a likelihood ratio test we studied the calibration of a model for the multilocus odds ratio, formed by multiplying the individual-locus odds ratios, from the published odds ratios in Table 2 (i.e. exponentiating the continuous wGRS) (see Supplementary Methods).
To determine whether wGRS22 is clinically useful on an individual patient basis, we estimated risk-score specific incidence among US women. We used the average annual incidence estimated from the full NHS cohort: λ=33/100,000; the risk-score specific odds ratios ORG; and one minus the population attributable risk 1-PAR = 1/(ΣG ORG πG), where πG is the prevalence of genotype G in the controls. The risk score specific incidence is then: λ (1-PAR) ORGπG42. To estimate risk-score specific absolute risks among Swedish men and women we used data on RA incidence rates in Northern Europe from Alamanos et al, and estimated Swedish annual incidence rates λ=40/100,000 for women, and λ=20/100,000 for men43.
RESULTS
PATIENTS
Characteristics of RA cases and controls for NHS and EIRA are presented in Table 1. The demographics of both groups are similar although (a) seropositive status in NHS was defined as either rheumatoid factor (RF) or CCP positive and in EIRA as those who were CCP positive, (b) NHS includes patients with new-onset and long-standing disease, whereas EIRA patients are of new-onset only, and (c) NHS is all female, whereas EIRA is both female and male (at the expected ratio of approximately 3:1).
Table 1.
Characteristics of Seropositive RA cases and matched controls in the Nurses' Health Studies and CCP Positive RA in Epidemiologic Investigation of RA (NHS)
| NHS | EIRA | |||
|---|---|---|---|---|
| RA cases (n=289) | Controls (n=481) | RA cases1 (n=629) | Controls2 (n=623) | |
| Age, mean (SD)3 | 55.3 (±8.1) | 55.7 (±7.9) | 51.2 (±11.7)5 | 52.1 (±11.8)6 |
| Pack-years among smokers, | 25.2 (±17.5) | 23.0 (±21.4) | 18.3 (±15.3) | 14.7 (±13.8) |
| mean (SD) | ||||
| RA features | ||||
| Mean age at diagnosis, (SD) | 56.2 (±9.9) | - | 51.2 (±11.7)5 | - |
| Rheumatoid factor, positive (%) | 270 (93.4%) | - | 523 (87.3%)5 | - |
| Anti-CCP4 positive (%) | 106 (55.5%) | - | 629 (100%) | - |
| Seropositive (%) | 289 (100.0%) | - | 629 (100%) | - |
| Rheumatoid nodules, (%) | 45 (15.6%) | - | ||
| Radiographic changes, (%) | 95 (32.9%) | - | ||
EIRA: 184 male cases; 446 female cases
EIRA: 163 male controls, 460 female controls
Age at blood draw
Citric citrullinated protein antibodies assayed in subset of NHS cases (n=191) with stored blood samples at collected at different points with respect to RA onset, up to 12 years prior to onset or after diagnosis
31 EIRA cases missing covariate data
5 EIRA controls missing covariate data. NHS: Nurses' Health Studies, EIRA: Epidemiologic Investigation of Rheumatoid Arthritis, anti-CCP: cyclic-citrullinated peptide antibody
ASSOCIATION BETWEEN GENETIC RISK ALLELES AND RA
The results for each of the 22 risk alleles with risk of RA are presented in Table 2. The majority of the odds ratios are in the same direction for the risk allele and of the same magnitude as from published discovery studies. Not surprisingly, many of the 95% confidence intervals cross 1.0, as might be expected given the modest odds ratios of the non-MHC alleles and the sample size of the two cohorts.
OBSERVED RELATIVE RISK WITH GENETIC RISK SCORE
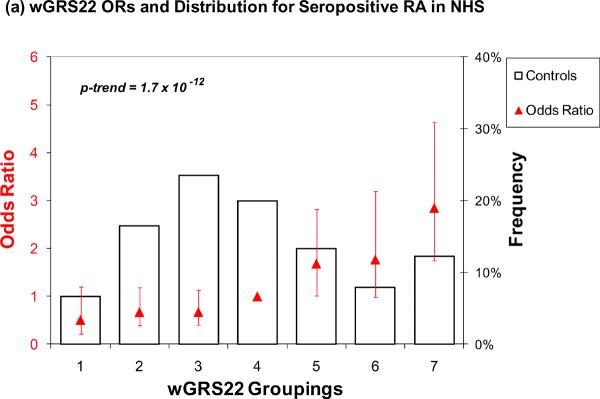
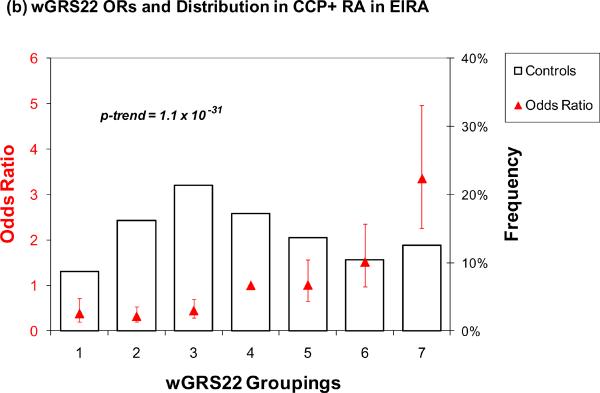
The results for wGRS22 as a predictor of seropositive RA are presented in Table 3 and Figure 1. For wGRS22, the median level of risk (group 4, containing 20% of controls) was used as the referent group. Those with the highest risk (group 7) had a significantly higher odds of RA as compared to group 4 in both NHS (OR = 2.85, 95% CI 1.75 – 4.64) and in EIRA (OR = 3.36, 95% CI 2.27 – 4.97) (Table 3, Figure 1a and 1b). Using group 1 (lowest level of risk) as a reference group, group 7 had a higher odds of RA, 5.61 (95% CI 2.41 – 13.07) in NHS and 8.83 (95% CI 4.77 – 16.32) in EIRA. In the ordinal model that takes into account all data in the model, group 7 had even higher odds of RA, 6.30 (95% CI 3.78 – 10.48) for NHS and 12.31 (95% CI 8.12 – 18.67) for EIRA. The trends across all 7 categories of risk were highly significant, with p < 0.0001 for both NHS and EIRA.
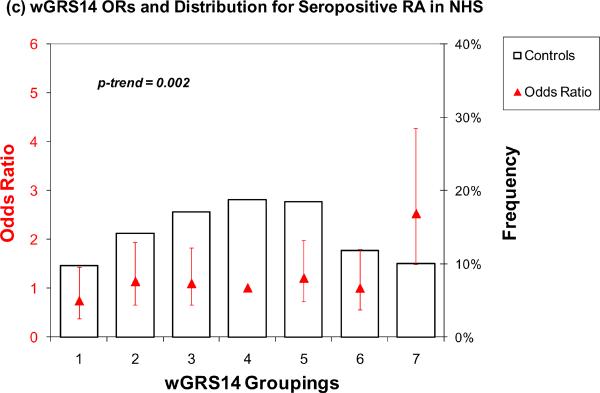
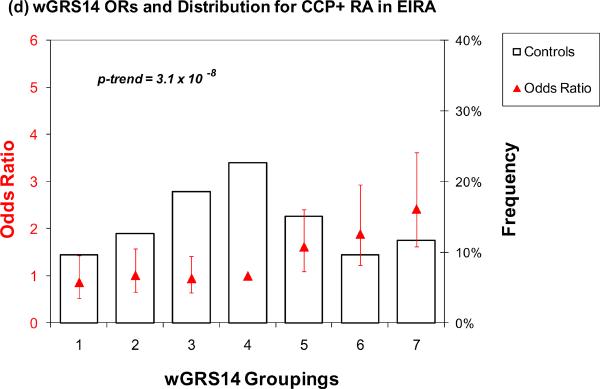
Figure 1.
Odds ratios for wGRS22 and wGRS14 (no HLA) in NHS and EIRA. Weighted GRS distribution among controls shown in bars, odds ratios shown in red triangles. (a) Odds ratios for wGRS22 and seropositive RA in NHS; b) Odds ratios for wGRS22 and CCP+ RA in EIRA; (c) Odds ratios for wGRS14 (no HLA) and Sero+ RA in NHS; (d) Odds ratios for wGRS14(no HLA) and CCP+ RA in EIRA. NHS: Nurses' Health Studies, EIRA: Epidemiologic Investigation of Rheumatoid Arthritis, CCP: cyclic-citrullinated peptide antibody, wGRS22: weighted genetic risk score with 22 alleles; wGRS14 (no HLA): weighted genetic risk score with 14 alleles, without HLA alleles.
A similar analysis was performed using only the 14 non-HLA risk alleles (Table 3, Figure 1c and 1d). For wGRS14 (no HLA), those in group 7 (highest risk) relative to group 4 (median) had an elevated OR of 2.52 (95% CI 1.49 – 4.28) and 2.43 (95% CI 1.62 – 3.63) in both NHS and EIRA, respectively. Using group 1 as the reference, group 7 had a higher odds of RA 3.43 (95% CI 1.74 – 6.74) and 2.81 (95% CI 1.66 – 4.73) in NHS and EIRA respectively. The OR from an ordinal model for group 7 was 2.39 (95% CI 1.44 – 3.98) in NHS and 3.22 (95% CI 2.14 – 4.86) in EIRA. The trends across all 7 categories were highly significant (p = 0.002 for NHS, p < 0.0001 for EIRA).
DISCRIMINATION OF CASES AND CONTROLS BY GRS SCORES
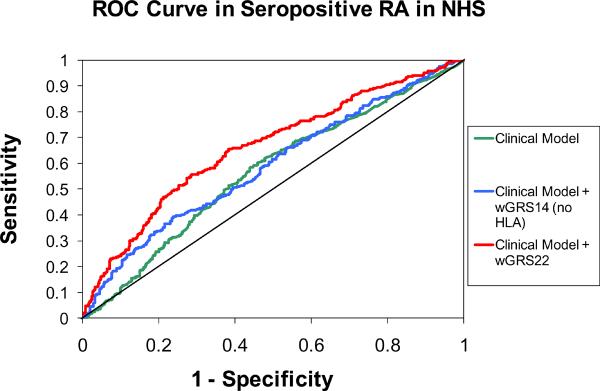
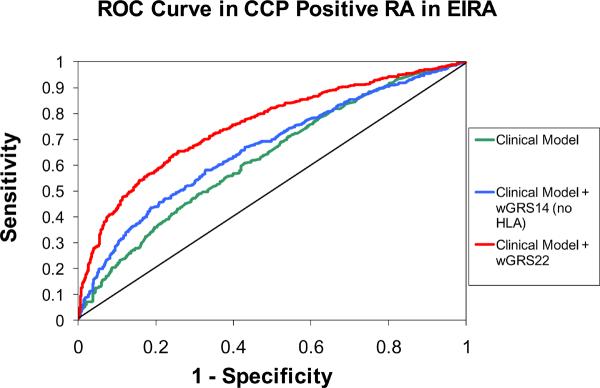
The statistics used during the discovery phase of research (such as odds ratios or P-values for association) are not the most appropriate measures for evaluating the predictive value of genetic profiles in clinical practice. Other measures - sensitivity, specificity, and risk classification - are more useful when proposing a genetic profile for risk prediction23, 24, 44. ROC curves that plot sensitivity of the GRS score (continuous) against 1-specificity, and calculated the area-under-the-curve (AUC), also known as the c-statistic, for both NHS and EIRA are shown in Figure 2. In the NHS, the AUC for the clinical model including age and pack-years of smoking was 0.566. Adding wGRS14 (no HLA) to this model did not significantly improve discrimination (AUC = 0.589; p=0.31). Adding HLA subtypes to the clinical + genetic model significantly improved discrimination relative to both the clinical model and the clinical + wGRS14 model (AUC = 0.660; p < 0.001 for both comparisons). In EIRA, ROC curves for the clinical model adjusted for age, sex, geographic region, and pack-years of smoking demonstrate significant improvements in discrimination with the addition of wGRS14 (no HLA) or wGRS18 scores, with AUCs of 0.627 0.662, and 0.752 (clinical + wGRS22 vs. wGRS14 (no HLA), p< 0.0001; clinical + wGRS22 vs. clinical, p< 0.0001; clinical + wGRS14 vs. clinical p=0.002).
Figure 2.
Receiver Operator-Characteristic (ROC) curves for predicting seropositive RA in NHS and CCP+ RA in EIRA. NHS clinical model is adjusted for year of birth and pack-years of smoking. EIRA clinical model is adjusted for age, sex, geographic region and pack-years of smoking. NHS AUCs: clinical model: AUC=0.566; clinical + wGRS14 (no HLA): AUC=0.589; clinical + wGRS22: AUC=0.660. NHS AUC comparisons: clinical + wGRS22 vs. clinical + wGRS14 (no HLA), p< 0.001; clinical + wGRS22 vs. clinical, p< 0.001; clinical + wGRS14 vs. clinical p = 0.31. EIRA AUCs: clinical model: AUC=0.626; clinical + wGRS14 (no HLA): AUC=0.662; clinical + wGRS22: AUC=0.752. EIRA AUC comparisons: clinical + wGRS22 vs. clinical + wGRS14 (no HLA), p< 0.0001; clinical + wGRS22 vs. clinical, p< 0.0001; clinical + wGRS14 vs. clinical p=0.002. NHS: Nurses' Health Studies, EIRA: Epidemiologic Investigation of Rheumatoid Arthritis, CCP: cyclic-citrullinated peptide antibody, wGRS22: weighted genetic risk score with 22 alleles; wGRS14 (no HLA): weighted genetic risk score with 14 alleles, without HLA alleles
GENOTYPE-SPECIFIC RISK AND COMPARISON BETWEEN PREDICTED AND OBSERVED ODDS RATIOS
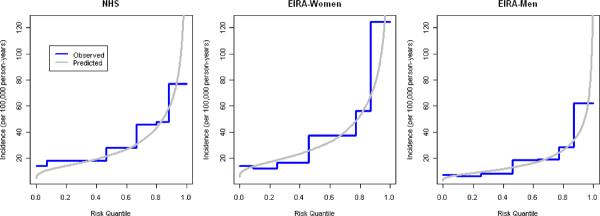
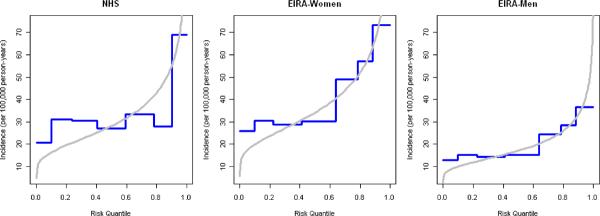
Figure 3 plots the distribution of genotype (or genotype-category) annual incidence for predicted models based on previous locus-specific odds ratio estimates and the observed categorized wGRS models fit to these data sets. For NHS and women in EIRA, the observed risks from our groupings approximate the predicted risk from a continuous wGRS, except for the lowest risk group (group 1) where observed risk exceeds predicted risk. For men in EIRA the observed risks from our groupings approximate the predicted risk from a continuous wGRS except for the highest risk group (group 7) where predicted risk exceeds observed risk, suggesting that in the highest risk group the risk based on grouping the wGRS is biased toward the null or the predicted risk is an overestimate. Figure 3 also shows that despite the statistically significant improvement in the AUC after incorporating the wGRS22, the predicted risks of RA were still small (<1% annual risk) for all of the observed genotypes.
Figure 3.
a) predicted vs. observed incidence rates for wGRS22 in NHS women, EIRA women and EIRA men; b) predicted vs. observed incidence rates for wGRS14 in NHS women, EIRA women and EIRA men; NHS: Nurses' Health Studies, EIRA: Epidemiologic Investigation of Rheumatoid Arthritis, CCP: cyclic-citrullinated peptide antibody, wGRS22: weighted genetic risk score with 22 alleles; wGRS14 (no HLA): weighted genetic risk score with 14 alleles, without HLA alleles
Discussion
Until 2004, only two genetic loci had been unequivocally associated with risk of RA susceptibility: HLA-DRB1 and PTPN225, 10. Recent large studies using genome-wide scans or related methodologies have discovered and replicated 12 additional non-MHC risk loci12–15, 20. In the current study, we develop a weighted genetic risk score including established 14 risk alleles from 13 non-MHC RA loci and 8 HLA subtypes based on high resolution genotyping. We demonstrate that a composite genetic risk score improves significantly the discrimination ability of the model for seropositive RA compared to no RA when compared to a risk model with epidemiologic variables alone when applied in the general population.
We found that in our top wGRS group with 22 alleles there was a 2.9 fold increase in the odds of seropositive RA compared to the most common wGRS group, and a 5.6 fold increase in odds of RA compared to the wGRS group with the lowest score in the NHS. In EIRA, the top wGRS group with 22 alleles had a higher increase in the odds of RA than in the US cohort, with a 3.4 fold compared to the most common wGRS group and 8.8 fold compared to the lowest wGRS group. However, comparing results from the cumulative score with 14 alleles, without the HLA-SE alleles, there were similar increased odds ratios for RA in both cohorts (2.5 fold in NHS and 2.4 fold in EIRA). This suggests that the increased risk in the Swedish cohort is primarily due to the higher frequency of HLA-SE alleles in that population, which may reflect the higher percentage of patients seropositive for CCP autoantibodies (Table 1).
Publications on genetic risks for other complex human diseases and quantitative traits such as macular degeneration31, prostate cancer32, 33, lipid levels and heart disease34–37, height45, 46 and diabetes38–40. These studies have combined risk alleles into a single risk score simply by summing the number of risk alleles carried. Our study extends the methodology by weighting the risk score by the published allelic odds ratios, thus accounting for the different strengths of association for genes such as the HLA-SE and PTPN22. Although models have been developed to identify which patients presenting with early inflammatory arthritis will progress to RA47, this is the first demonstration of risk models that include all known genetic risk factors and the two strongest epidemiologic factors, age and smoking, in prediction of incident RA among healthy subjects without symptoms.
Our wGRS is a first step towards development of RA risk prediction models that incorporate aggregate genetic factors. In contrast to other complex diseases such as diabetes38, 39 and heart disease34–37, where adding genetic markers to clinical risk factors does not add to discrimination, the addition of genetic factors to a clinical model that includes epidemiologic risk factors improves discrimination significantly for RA, which supports the clinical validity of this approach. The AUCs of 0.566 and 0.627 in NHS and EIRA, respectively, suggest that clinical risk factors alone – in subjects without symptoms – do not provide much discrimination between RA cases and controls. Adding genetic alleles to the aggregate score significantly improves the model AUCs to 0.660 in NHS and 0.752 in EIRA. However, there is variance in risk that remains unexplained, suggesting that further work is needed to incorporate environmental exposure data and gene-environment interactions into risk models and to discover additional genetic variants. We note that in patients with early symptoms consistent with an inflammatory arthritis, clinical prediction models that include sex, age, localization of symptoms, morning stiffness, the tender joint count, the swollen joint count, the C-reactive protein level, rheumatoid factor positivity, and the presence of anti-cyclic citrullinated peptide antibodies accurately predict who will go on to develop RA47, 48. Under this clinical scenario, it will be important to test whether genetic factors helps discriminate which patients will develop RA.
Odds ratios alone are difficult to interpret for patients and physicians in a clinical setting24. However, as suggested by Kraft et. al.24, measures of absolute risk (i.e. risk that a disease-free subject will develop disease) such as the results shown in Figure 3, provide a more intuitive context of RA risk at the individual level. A strength of our study is that we have data on the entire prospective NHS cohort from which our nested samples were taken and thus we have an accurate estimate of the population annual incidence. Using data from the full NHS cohort, we see an absolute risk of RA among US women aged 25–50 of 0.3%, thus a wGRS22 in group 7 increases the absolute risk to 0.7%. In EIRA women, the wGRS22 score in group 7 increases the absolute risk from 0.4% to 1.3%. In EIRA men, the wGRS22 score in group 7 increases the absolute risk from 0.2% to 0.7%. These predictive models demonstrate that there is a small portion of the general population at very high risk
Although the hope is that we will soon be able to apply genetic information to individual patients, the wGRS for RA is unlikely to be useful in routine clinical practice for assessing risk among the healthy asymptomatic patients. Even the highest risk category - group 7 - has a modest absolute risk of RA. It is possible genetic results might eventually help us to identify subsets of patients who are at substantially elevated absolute risk, and would be willing to undergo potentially toxic therapies, to prevent RA. It will be important to perform studies in among subsets of patients at higher risk for RA, eg. patients with early undifferentiated arthritis, patients with anti-CCP + arthralgia, and first degree relatives of RA patients49. We propose that wGRS22 may be clinically useful as part of an overall risk assessment tool among high risk groups.
We recognize that the ideal setting to perform prognostic modelling analyses is a prospective cohort study, such as the Framingham Heart Study or the full Nurses' Health Study cohorts. However, no such large study has blood samples available on the full dataset and validated RA cases. Instead, we approximated risk by use of the odds, which in a population based case-control study with a proper sampling of controls approximates relative risk well. We calculated risk-score specific absolute risks using these odds ratios and the average population risk estimated from the full NHS cohort, and from the literature for Northern Europe. The estimated incidence in NHS is consistent with RA incidence rates observed in other studies in women of Northern European ancestry43, except for a single study from North America50. The NHS dataset is limited by the absence of CCP antibody information on cases that were diagnosed prior to the widespread use of the test. Thus the phenotype used in NHS analyses is seropositive RA, while the phenotype used in EIRA analyses is CCP positive RA, which is more strongly associated with genetic factors such as the HLA-SE. Although stronger associations are demonstrated in EIRA, the results from NHS are very consistent, suggesting that the general category of seropositive RA is associated with these genetic factors.
Despite the rapid advances in our understanding of the genetic basis of complex human diseases such as RA, it is not clear how to utilize this information for clinical care, prediction, or prevention. Although a combination of known genetic factors for RA aggregated into a weighted score has a 3-fold increased odds for the development of RA, the absolute risk of this disease remains low, suggesting that genetic risk scores, calculated as in this paper, have little clinical utility in predicting RA risk in asymptomatic individuals. More research to identify genetic and environmental risk factors, as well as gene-environment interactions, is critical to understanding the determinants of RA risk before this information can be used in patient counseling or preventive trials.
Supplementary Material
Acknowledgements
The authors wish to thank the participants, investigators and study staff of the Nurses' Health Studies in the United States and Epidemiologic Investigation of Rheumatoid Arthritis in Sweden for their contributions.
Sponsors. The NHS is supported by NIH grants R01 AR49880, CA87969, CA49449, CA67262, CA50385, AR047782, AR0524-01. RMP is supported by grants from NIAMS-NIH (AR056768 and AR057108) and the William Randolph Hearst Fund of Harvard University, and also holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. The EIRA study was supported by grants from the Swedish Medical Research Council, from Swedish Council for Working life and Social Research, from King Gustaf V:s 80-year foundation, from the Swedish Rheumatism Foundation, from Stockholm County Council and from the insurance company AFA.
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Annals of the Rheumatic Diseases and any other BMJPGL products to exploit all subsidiary rights, as set out in our licence. (http://ard.bmj.com/ifora/licence.pdf).
Footnotes
Competing Interests. None declared.
References
- 1.Finckh A, Liang MH, van Herckenrode CM, de Pablo P. Long-term impact of early treatment on radiographic progression in rheumatoid arthritis: A meta-analysis. Arthritis Rheum. 2006;55(6):864–72. doi: 10.1002/art.22353. [DOI] [PubMed] [Google Scholar]
- 2.Criswell LA, Merlino LA, Cerhan JR, et al. Cigarette smoking and the risk of rheumatoid arthritis among postmenopausal women: results from the Iowa Women's Health Study. Am J Med. 2002;112(6):465–71. doi: 10.1016/s0002-9343(02)01051-3. [DOI] [PubMed] [Google Scholar]
- 3.Stolt P, Bengtsson C, Nordmark B, et al. Quantification of the influence of cigarette smoking on rheumatoid arthritis: results from a population based case-control study, using incident cases. Ann Rheum Dis. 2003;62(9):835–41. doi: 10.1136/ard.62.9.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costenbader KH, Feskanich D, Mandl LA, Karlson EW. Smoking intensity, duration, and cessation, and the risk of rheumatoid arthritis in women. Am J Med. 2006;119(6):503–11. doi: 10.1016/j.amjmed.2005.09.053. [DOI] [PubMed] [Google Scholar]
- 5.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30(11):1205–13. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 6.Rantapaa-Dahlqvist S, de Jong BA, Berglin E, et al. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum. 2003;48(10):2741–9. doi: 10.1002/art.11223. [DOI] [PubMed] [Google Scholar]
- 7.Nielen MM, van Schaardenburg D, Reesink HW, et al. Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum. 2004;50(2):380–6. doi: 10.1002/art.20018. [DOI] [PubMed] [Google Scholar]
- 8.Chibnik LB, Mandl LA, Costenbader KH, Schur PH, Karlson EW. Comparison of threshold cutpoints and continuous measures of anti-cyclic citrullinated peptide antibodies in predicting future rheumatoid arthritis. J Rheumatol. 2009;36(4):706–11. doi: 10.3899/jrheum.080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlson EW, Chibnik LB, Tworoger SS, et al. Biomarkers of inflammation and development of rheumatoid arthritis in women from two prospective cohort studies. Arthritis Rheum. 2009;60(3):641–52. doi: 10.1002/art.24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begovich AB, Carlton VE, Honigberg LA, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–7. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plenge RM, Padyukov L, Remmers EF, et al. Replication of Putative Candidate-Gene Associations with Rheumatoid Arthritis in >4,000 Samples from North America and Sweden: Association of Susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77(6):1044–60. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Remmers EF, Plenge RM, Lee AT, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357(10):977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357(12):1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plenge RM, Cotsapas C, Davies L, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39(12):1477–82. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium WTCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomson W, Barton A, Ke X, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39(12):1431–3. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurreeman FA, Padyukov L, Marques RB, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS medicine. 2007;4(9):e278. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barton A, Thomson W, Ke X, et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008;40(10):1156–9. doi: 10.1038/ng.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki A, Yamada R, Kochi Y, et al. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nat Genet. 2008;40(10):1224–9. doi: 10.1038/ng.205. [DOI] [PubMed] [Google Scholar]
- 20.Raychaudhuri S, Remmers EF, Lee AT, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40(10):1216–23. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burke W, Psaty BM. Personalized medicine in the era of genomics. Jama. 2007;298(14):1682–4. doi: 10.1001/jama.298.14.1682. [DOI] [PubMed] [Google Scholar]
- 22.Ioannidis JP. Personalized genetic prediction: too limited, too expensive, or too soon? Annals of internal medicine. 2009;150(2):139–41. doi: 10.7326/0003-4819-150-2-200901200-00012. [DOI] [PubMed] [Google Scholar]
- 23.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS genetics. 2009;5(2):e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraft P, Wacholder S, Cornelis MC, et al. Beyond odds ratios-communicating disease risk based on genetic profiles. Nat Rev Genet [epub] 2009 doi: 10.1038/nrg2516. [DOI] [PubMed] [Google Scholar]
- 25.Liang MH, Meenan RF, Cathcart ES, Schur PH. A screening strategy for population studies in systemic lupus erythematosus. Series design. Arthritis Rheum. 1980;23(2):153–7. doi: 10.1002/art.1780230204. [DOI] [PubMed] [Google Scholar]
- 26.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 27.Padyukov L, Silva C, Stolt P, Alfredsson L, Klareskog L. A gene-environment interaction between smoking and shared epitope genes in HLA-DR provides a high risk of seropositive rheumatoid arthritis. Arthritis Rheum. 2004;50(10):3085–92. doi: 10.1002/art.20553. [DOI] [PubMed] [Google Scholar]
- 28.Costenbader KH, Chang SC, De Vivo I, Plenge R, Karlson EW. Genetic polymorphisms in PTPN22, PADI-4, and CTLA-4 and risk for rheumatoid arthritis in two longitudinal cohort studies: evidence of gene-environment interactions with heavy cigarette smoking. Arthritis Res Ther. 2008;10(3):R52. doi: 10.1186/ar2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernando MM, Stevens CR, Walsh EC, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS genetics. 2008;4(4):e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33(2):177–82. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 31.Maller J, George S, Purcell S, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38(9):1055–9. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 32.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358(9):910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 33.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 34.Kathiresan S, Melander O, Anevski D, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358(12):1240–9. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 35.Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aulchenko YS, Ripatti S, Lindqvist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41(1):47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41(3):334–41. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359(21):2220–32. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 39.Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359(21):2208–19. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornelis MC, Qi L, Zhang C, et al. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Annals of internal medicine. 2009;150(8):541–50. doi: 10.7326/0003-4819-150-8-200904210-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–45. [PubMed] [Google Scholar]
- 42.Rebbeck TR, Ambrosone CB, Shields PG, editors. Molecular epidemiology: applications in cancer and other human diseases. Informa Healthcare; New York: 2008. [Google Scholar]
- 43.Alamanos Y, Voulgari PV, Drosos AA. Incidence and prevalence of rheumatoid arthritis, based on the 1987 American College of Rheumatology criteria: a systematic review. Semin Arthritis Rheum. 2006;36(3):182–8. doi: 10.1016/j.semarthrit.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Kraft P, Hunter DJ. Genetic risk prediction--are we there yet? N Engl J Med. 2009;360(17):1701–3. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 45.Weedon MN, Lango H, Lindgren CM, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40(5):575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lettre G, Jackson AU, Gieger C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40(5):584–91. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Helm-van Mil AH, le Cessie S, van Dongen H, Breedveld FC, Toes RE, Huizinga TW. A prediction rule for disease outcome in patients with recent-onset undifferentiated arthritis: how to guide individual treatment decisions. Arthritis Rheum. 2007;56(2):433–40. doi: 10.1002/art.22380. [DOI] [PubMed] [Google Scholar]
- 48.van der Helm-van Mil AH, Detert J, le Cessie S, et al. Validation of a prediction rule for disease outcome in patients with recent-onset undifferentiated arthritis: moving toward individualized treatment decision-making. Arthritis Rheum. 2008;58(8):2241–7. doi: 10.1002/art.23681. [DOI] [PubMed] [Google Scholar]
- 49.Hemminki K, Li X, Sundquist J, Sundquist K. Familial associations of rheumatoid arthritis with autoimmune diseases and related conditions. Arthritis Rheum. 2009;60(3):661–8. doi: 10.1002/art.24328. [DOI] [PubMed] [Google Scholar]
- 50.Gabriel SE, Crowson CS, O'Fallon WM. The epidemiology of rheumatoid arthritis in Rochester, Minnesota, 1955–1985. Arthritis Rheum. 1999;42(3):415–20. doi: 10.1002/1529-0131(199904)42:3<415::AID-ANR4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.