SUMMARY
The generation of coordinated body movements relies on sensory feedback from mechanosensitive proprioceptors. We have found that the proper function of NompC, a putative mechanosensitive TRP channel, is not only required for fly locomotion, but also crucial for larval crawling. Calcium imaging revealed that NompC is required for the activation of two subtypes of sensory neurons during peristaltic muscle contractions. Having isolated a full-length nompC cDNA with a protein coding sequence larger than previously predicted, we demonstrate its function by rescuing locomotion defects in nompC mutants, and further show that antibodies against the novel C-terminus recognize NompC in chordotonal ciliary tips. Moreover, we show that the ankyrin repeats in NompC are required for proper localization and function of NompC in vivo and are required for association of NompC with microtubules. Taken together, our findings suggest that NompC mediates proprioception in locomotion and support its role as a mechanosensitive channel.
INTRODUCTION
Mechanosensation is a sensory modality of importance to both prokaryotes and eukaryotes. Most unicellular organisms are capable of detecting membrane tension and distortion caused by mechanical stimuli (Martinac, 2001). In higher organisms, specialized mechanosensitive cells and organs mediate the detection of touch, nociception, hearing, and proprioception (Ernstrom and Chalfie, 2002; Lumpkin and Caterina, 2007). Despite the importance of these modalities, in many instances, especially in the case of proprioception, the identity of the mechanosensitive cells and the molecules required for mechanosensation in these cells are largely unknown.
Proprioception refers to the sensory input and feedback by which animals keep track of and control the different parts of their body for balance and for locomotion. In humans, selective loss of proprioception results in a “rag doll” state – a failure to make any coordinated body movement (Smetacek and Mechsner, 2004). Proprioception is likely mediated by mechanosensitive stretch receptors located within the muscles, joints and ligaments (Windhorst, 2007). Ion channels and neurons important for proprioception have been identified in genetic studies of organisms with stereotypical patterns of locomotion. In C. elegans, mutations in trp-4, which encodes a transient receptor potential (TRP) channel, and unc-8, which encodes an epithelial sodium channel (ENaC), lead to abnormal body movement, suggesting that TRP-4 and UNC-8 mediate proprioception in C. elegans (Li et al., 2006; Tavernarakis et al., 1997). These studies also identified neurons that contribute to the regulation of proprioception. Two TRP-4-expressing neurons are located in the body wall with extended axons that span nearly the whole length of the body and may function as proprioceptor neurons (Li et al., 2006). Several UNC-8-expressing sensory neurons, interneurons, and motor neurons may also contribute to proprioception in C. elegans (Tavernarakis et al., 1997).
The Drosophila larval peripheral nervous system (PNS) provides a model for systematic analysis of the physiological function of morphologically distinct sensory neurons. The Drosophila PNS is composed of segmentally repeated sensory neurons which are classified as either type I or type II neurons. Type I neurons, which have ciliated monopolar dendrites, are located in external sensory organs and chordotonal organs. The primary function of type I neurons is mechanosensation (Kernan, 2007). Type II neurons, also known as multi-dendritic (MD) neurons, are further divided into tracheal dendrite (td) neurons, bipolar dendrite (bd) neurons, and dendritic arborization (da) neurons (Bodmer and Jan, 1987). Each subtype of MD neuron has characteristic dendrite arborization and axonal targeting patterns (Grueber et al., 2002; Grueber et al., 2007), suggesting that different subtypes of MD neurons may be functionally distinct (Ainsley et al., 2003; Hwang et al., 2007). Previously, we have shown that silencing all MD neurons results in a cessation of larval locomotion, demonstrating that the function of MD neurons is critical for larval locomotion (Song et al., 2007). Further, simultaneously silencing two specific subtypes of MD neurons, bd and class I da neurons, disrupts larval crawling ability (Hughes and Thomas, 2007), suggesting that bd and class I da neurons play an essential role in larval locomotion and could function as proprioceptor neurons. However, the molecules required for proprioception in these neurons have not been identified.
The Drosophila TRP channel TRPN1/NompC is a putative mechanosensitive channel that affects fly locomotion. Loss-of-function mutations of nompC abolish mechanoreceptor potentials in fly bristles and a missense mutation of nompC alters adaptation of mechanoreceptor potentials (Walker et al., 2000). NompC is also required for hearing in Drosophila (Gopfert et al., 2006; Kamikouchi et al., 2009; Sun et al., 2009). In addition, adult nompC mutant flies are severely uncoordinated (Kernan et al., 1994; Walker et al., 2000). To substantiate the physiological role of NompC in locomotion, it is important to identify the neurons that require NompC for locomotion, to characterize the subcellular localization of NompC, and to study how NompC function is regulated in vivo.
Here we report the molecular characterization of NompC that establishes its functional role in Drosophila locomotion. NompC is expressed in the dendrites of bd/class I da neurons and is required for activating bd/class I da neurons during larval peristaltic muscle contractions. Proper function of NompC controls the pace of larval crawling. In addition, the ankyrin repeats in NompC are required not only for microtubule association, but also for the proper localization and function of NompC in vivo, suggesting that the ankyrin repeats directly or indirectly connect NompC to the cytoskeleton to enable mechanosensitivity. Taken together, these findings reveal a critical role of NompC in locomotion and support its function as a mechanosensitive channel.
RESULTS
NompC is expressed in neurons required for larval locomotion
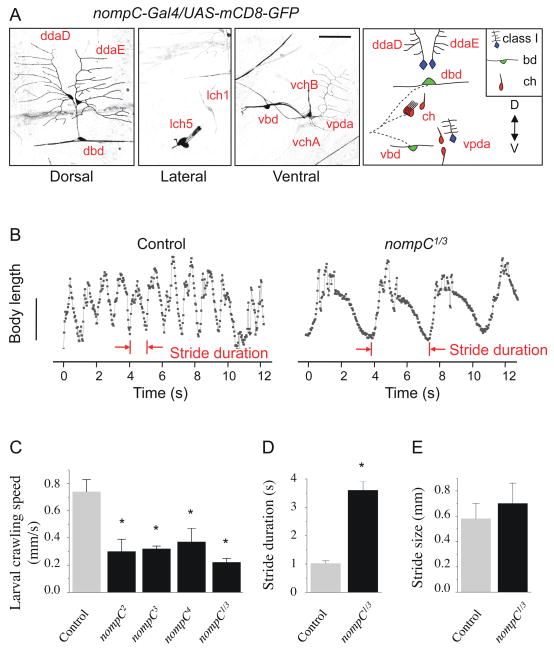
To search for ion channels that are expressed in bd and class I da neurons, the two types of sensory neurons implicated in larval crawling, we began with RNA in situ hybridization to examine PNS expression of candidate ion channels. We found that one TRP channel, TRPN1/NompC, was expressed in a subset of sensory neurons at the embryonic stage (Fig. S1A). To further define the expression pattern of nompC, we generated a nompC-Gal4 line by using the nompC promoter region. In nompC-Gal4/UAS-mCD8-GFP larvae, expression of mCD8-GFP is mainly detected in PNS sensory neurons (Fig. S1B). Specifically, chordotonal organs (type I neurons), bd neurons (type II neurons) and class I da neurons (type II neurons) are labeled (Fig. 1A). On the dorsal side, the two NompC-expressing class I da neurons (ddaD and ddaE) project their fan-shaped dendrites anteriorly and posteriorly, respectively, and the NompC-expressing bd neuron (dbd) extends two longitudinal dendrites that span the entire length of the segment (Fig. 1A, left panel). On the lateral and ventral side, expression is detected in chordotonal neurons (lch1, lch5, vchA, vchB), one ventral bd neuron (vbd), and one class I da neuron (vpda) (Fig. 1A, right panels). While it is possible that the NompC expression pattern is not fully reproduced by this nompC-Gal4 line, expression of a nompC cDNA driven by nompC-Gal4 is sufficient to rescue locomotion defects of nompC mutants (see below, Fig. 3B), indicating that this nompC-Gal4 driver is active in those neurons that require NompC for their function in locomotion.
Figure 1. NompC is expressed in larval sensory neurons and controls the pace of larval crawling.
(A) NompC expression examined in nompC-Gal4/UAS-mCD8-GFP larvae. Expression was detected in class I da neurons (ddaD, ddaE, vpda), bd neurons (dbd, vbd) and chordotonal (ch) organs (lch1, lch5, vchA, vchB) in the abdominal segments. Scale bar, 50 μm.
(B) Plot of body length (mm) over time (s) during peristaltic contractions for control and nompC1/3 larvae. Scale bar, 2 mm.
(C–E) Quantification of crawling defects in nompC mutant larvae. Crawling speed is the length of a path (mm) divided by the time (s) it takes for the larva to complete the path. Stride duration is the average time for a larva to complete a single peristaltic contraction cycle. Stride size is the length of a path (mm) divided by the number of strides. Error bars represent SEM, n≥20. * P<0.01. See also Figure S1 and Movie S1.
Figure 3. A full-length nompC cDNA rescues the locomotion defects in nompC1/3 mutant larvae and adult flies.
(A) The exon-intron structures of nompC-L and nompC-A aligned with the genomic sequence. Exons are indicated by shaded boxes/triangles and introns by dotted lines.
(B, C) Quantification of larval crawling speed and stride duration with the expression of NompC-L or NompC-L-GFP by different Gal4 drivers. SN, sensory neurons; MD, multi-dendritic neurons; bd/I, bd and class I da neurons.
(D) Quantification of adult fly walking speed with NompC-L expression.
(E) Quantification of stride duration with expression of the truncated NompC, Δ12ANK-GFP and Δ29ANK-GFP. Error bars represent SEM, n≥20. * P<0.01. See also Figure S2, Movie S3 and Table S1.
nompC mutations impair locomotion of larval and adult flies
Having found that NompC is expressed in bd and class I da neurons, we went on to test the role of NompC in larval locomotion behavior. Larvae homozygous for null mutations (significant truncations) in nompC (nompC1, nompC2, nompC3) (Walker et al., 2000) as well as larvae with heteroallelic combinations of nompC null mutations exhibited dramatically reduced crawling speed (Fig. 1C and Movie S1). We noticed that while most nompC2 and nompC3 homozygous larvae die at the third instar stage, nompC1 homozygous larvae die at the first instar stage, indicating that there might be additional background mutations in nompC1. Therefore, to avoid potential confounding effects of background mutations that could manifest in homozygotes, we used primarily nompC1/3 trans-heterozygotes to abolish NompC function in our study. To explore the involvement of NompC in locomotion control, we analyzed the length and contour of larval locomotion strides during peristaltic contraction cycles. Figure 1B shows the typical patterns of change in larval body length during locomotion for wild-type and nompC1/3 mutant larvae. Similar to silencing of bd/class I da neurons (Hughes and Thomas, 2007), loss of function of nompC leads to a prolonged stride duration with normal stride size (Fig. 1D, E), revealing that NompC is crucial for larval locomotion.
To test whether the role of NompC in locomotion depends on its function as a putative mechanotransduction channel, we examined the locomotion behavior of the nompC4 allele. nompC4 contains a missense mutation of C1400Y between the third and fourth transmembrane segments, which impairs the adaptation to mechanical stimuli in fly bristles (Walker et al., 2000). The nompC4 mutants display a similar locomotion defects as nompC null mutants (Fig. 1C), suggesting that proper function of NompC is important for its role in locomotion.
nompC null mutant larvae have markedly reduced viability and most nompC flies fail to eclose from their pupal cases (Kernan et al., 1994). A small percentage (~5%) of nompC mutant flies eclose and these adult escapers show severe locomotion defects (Walker et al., 2000). Adult nompC mutant flies exhibited uncoordinated leg/wing twisting movements and the walking speed was dramatically reduced (Fig. 3D, Movie S3). Consistent with the reported nompC transcript expression in proprioceptors in leg joints (Walker et al., 2000), we found that the nompC-Gal4 driver labeled chordotonal organs spanning the coxa-trochanter joint and external sensory neurons associated with bristles (Fig. S1C). These studies thus establish that NompC is required for both larval and adult fly locomotion behavior.
nompC mutations impair activation of MD neurons during peristaltic muscle contractions
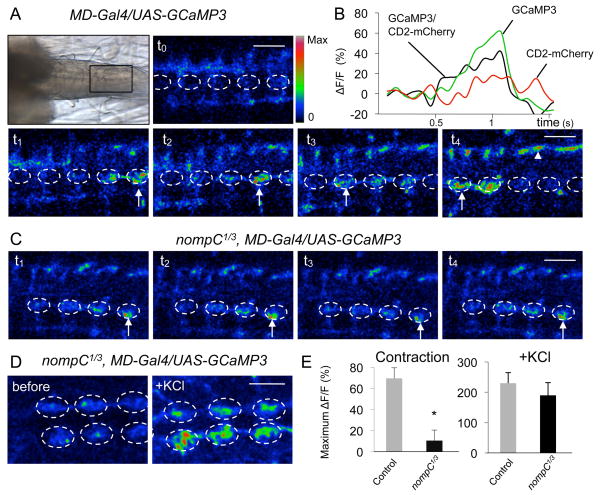
To provide physiological evidence that NompC is required for locomotion, we developed a larval preparation that maintains peristaltic muscle contractions and is amenable to calcium imaging. We expressed the genetically encoded calcium sensor GCaMP3 (Tian et al., 2009) in larval sensory neurons using the MD neuron driver (MD-Gal4). A CD2-mCherry fusion protein was co-expressed as an internal reference marker. In the larval central nervous system, the axons of the sensory neurons from each body segment project to the corresponding segment in the ventral nerve cord (VNC), where information is relayed to the second order neurons (Grueber et al., 2007). In wild-type larvae, spontaneous muscle contractions activated the sensory neurons and generated Ca2+ signals at the axon terminals in the VNC (Fig. 2A, B). Similar to the peristaltic muscle contractions, the Ca2+ signal propagated from the posterior to the anterior segments (Fig. 2A, arrows). Comparable Ca2+ signals were observed when GCaMP3 was expressed using the bd and class I da neuron-specific driver (bd/I-Gal4) (Movie S2). In both cases, the Ca2+ signal was only detected in the nerve bundles entering the VNC (arrowhead) and the dorsalmedial neuropil (dashed circle), consistent with the site of axonal projections from bd and class I da neurons (Grueber et al., 2007). The Ca2+ signal at each VNC segment presumably represents response to muscle contractions of each body segment. In nompC1/3 mutant larvae, spontaneous muscle contractions and GCaMP activation were greatly reduced and no segmental propagation of the Ca2+ signal could be observed (Fig. 2C, E). In contrast, membrane depolarization induced by KCl generated similar Ca2+ signals in control and nompC1/3 sensory neurons (Fig. 2D, E). These data suggest that NompC is required for the activation of bd and class I da neurons during peristaltic muscle contractions.
Figure 2. NompC is required for the activation of MD neurons during peristaltic muscle contractions.
(A) Ca2+ response of MD neurons to peristaltic muscle contractions in control larvae. Top left panel, a transmitted light image showing the ventral nerve cord in a larval preparation. Anterior is to the left and posterior to the right. (t0 to t4) Time-lapse images of Ca2+ signals (arrows) generated by spontaneous muscle contractions. The dorsalmedial neuropil is outlined with a dashed circle. The nerve bundles are marked with arrowheads. Color scale indicates activation level (red is the highest). Scale bar, 30 μm.
(B) Representative fluorescence change (ΔF/F) of GCaMP3 and CD2-mCherry (internal reference control) in control larvae.
(C) Ca2+ response to muscle contractions in nompC1/3 mutant larvae. Scale bar, 30 μm.
(D) Representative images before and after adding KCl to nompC1/3 mutant larvae. Scale bar, 50 μm.
(E) Quantification of GCaMP responses. Left panel, responses to muscle contractions for control (n=8) and nompC1/3 larvae (n=10). Right panel, responses to KCl for control (n=3) and nompC1/3 larvae (n=3). The GCaMP signal is normalized to CD2-mCherry reference signal. * p<0.01. See also Movie S2.
Rescue of nompC mutant locomotion defects in larvae and adult flies
To determine if the locomotion defect is caused by loss of nompC function in sensory neurons, we isolated a full-length nompC cDNA from larval body wall tissue and tested its ability to rescue the defects of nompC mutants. The nompC cDNA we isolated diverges substantially from the predicted nompC cDNA reported previously (Walker et al., 2000), which is expected to code for a protein of 1619 amino acids (NompC-A). Instead, the cDNA of 5199 nt isolated in our study codes for a protein of 1732 amino acids, including amino acids 1 to 1565 of NompC-A and a distinct C-terminus of 167 amino acids that contains a conserved TRP domain (Fig. 3A, S2). We named this nompC cDNA nompC-L to denote its longer coding sequence. It should be noted that NompC-A is a predicted isoform that may or may not exist; conservation analysis suggests that the 3′ sequence of the predicted nompC-A mRNA is intronic in nature (data not shown). We did not detect transcription of nompC-A by RT-PCR from RNA samples prepared from either larvae or adult flies. To test whether NompC-L is a functional isoform of NompC, we expressed NompC-L using the nompC-Gal4 driver and found that the lethality of nompC1/3 mutant larvae was fully rescued. This in vivo test confirms that NompC-L is functional.
Next, we examined the ability of NompC-L to rescue the locomotion defects of nompC mutant larvae. Expression of NompC-L using different Gal4 drivers that are expressed in sensory neurons (SN-Gal4), MD neurons (MD-Gal4), bd and class I da neurons (bd/I-Gal4) or NompC-expressing neurons (nompC-Gal4) rescued locomotion speed and stride duration of nompC mutant larvae (Fig. 3B, C). The detailed expression patterns of the Gal4 drivers used in these experiments are summarized in Table S1. Combined with results showing that silencing bd and class I da neurons results in slow locomotion (Hughes and Thomas, 2007), these results indicate that bd and class I da neurons rely on NompC for their function in larval locomotion.
The walking defects in adult nompC flies were also rescued by expression of NompC-L in all sensory neurons via SN-Gal4, or NompC-positive neurons via nompC-Gal4 (Fig. 3D and Movie S3), confirming that NompC function in sensory neurons is also essential for adult locomotion. In addition to chordotonal organs in leg joints, SN-Gal4 and nompC-Gal4 are also expressed in mechanosensitive neurons located in wing hinges, bristles, halteres and Johnston’s organs, which could also contribute to adult fly walking behavior.
NompC protein is localized to the chordotonal ciliary tip and MD neuron dendrites
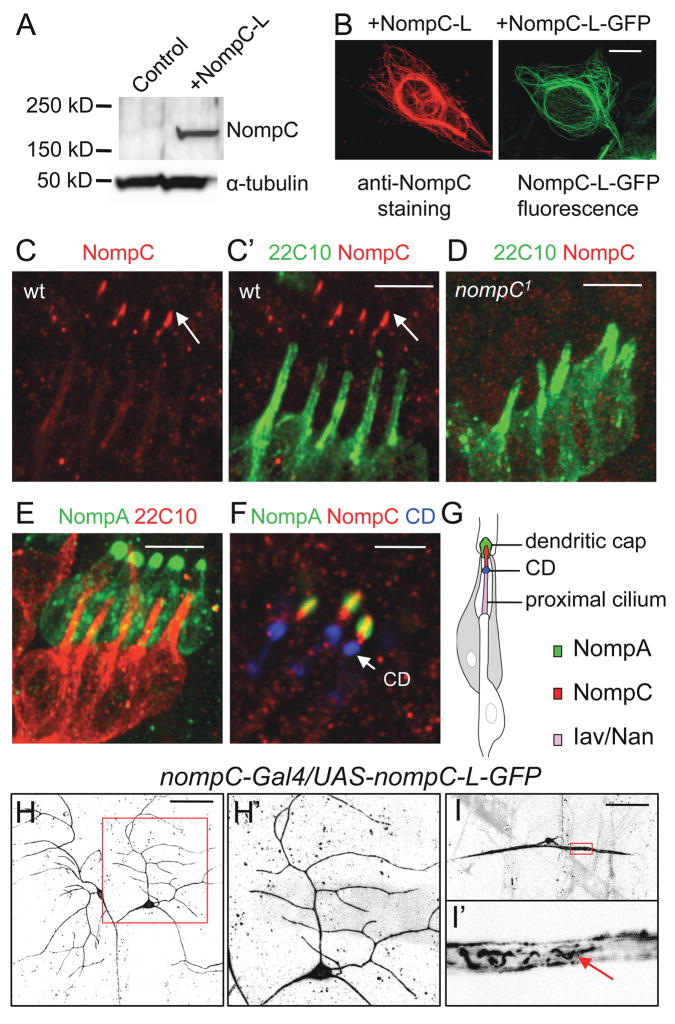
Although members of the TRP family have been implicated in mechanosensation, little is known about how mechanical forces are detected by these channels. As a first step to approach this question, we set out to characterize the localization of the channel in sensory neurons. To visualize the NompC protein, we generated a polyclonal antibody against the cytoplasmic C-terminal region of NompC-L that is absent in NompC-A (Fig. 3A). The antibody detected a protein of ~180 kDa, close to the expected size (189 kDa) of the NompC-L protein, on Western blots of NompC-L-expressing human embryonic kidney (HEK) 293 cells, but not of control cells (Fig. 4A). In addition, staining of NompC-L-transfected cells with anti-NompC yielded a pattern similar to the distribution of GFP fluorescence in cells transfected with NompC-L-GFP (Fig. 4B), indicating that the NompC antibody recognizes the NompC-L protein.
Figure 4. NompC distribution in embryonic and larval sensory neurons.
(A) Western blots with the NompC antibody in NompC-L-transfected HEK 293 cells.
(B) Immunostaining with the NompC antibody in NompC-L-transfected cells and GFP fluorescence in NompC-L-GFP-transfected cells. Scale bar, 10 μm.
(C, D) Co-staining of chordotonal organs (lch5) with the NompC antibody (red) and neural-specific cytoskeletal maker mAb 22C10 (green) in wild-type and nompC1 mutant embryos. Scale bar, 5 μm.
(E) In embryos expressing GFP-NompA, co-staining of GFP (green) and mAb 22C10 (red) marks the NompA-enriched dendritic caps. Scale bar, 5 μm.
(F) Co-staining of embryos expressing GFP-NompA (green) with mAb 21A6/anti-EYS (blue), a marker for the ciliary dilation (CD), and the NompC antibody (red). Scale bar, 3 μm.
(G) A schematic of a single chordotonal organ showing the localization of NompA in dendritic caps, NompC in distal ciliary tips and Iav/Nan in the proximal cilium.
(H, I) Localization of NompC-L-GFP in larval class I da neurons (H) and bd neurons (I). The arrow indicates NompC-L-GFP fluorescence in filamentous structures within the proximal dendrites. The boxed area in H and I is enlarged and shown in H′ and I′, respectively. Scale bar, 50 μm. See also Figure S3.
In fly embryos, staining for endogenous NompC-L yielded the strongest signals in chordotonal neurons. In lch5, co-staining with the neural-specific cytoskeletal marker mAb 22C10/anti-Futsch (Roos et al., 2000), showed that NompC-L is specifically localized at distal ciliary tips (arrow, Fig. 4C′), which are subject to stretch or shear under mechanical stimulation. The staining was absent in nompC1 mutant embryos (Fig. 4D), demonstrating the specificity of the antibody to NompC. The chordotonal cilium can be subdivided into distinct functional domains, the distal ciliary tip and proximal cilium, that are separated by the ciliary dilation (Lee et al., 2008). A previous study has suggested that NompA, an extracelluar protein localized to the dendritic cap wrapping the distal ciliary tip (Fig. 4E), forms part of a mechanical linkage that transmits mechanical stimuli to the transduction apparatus (Chung et al., 2001). In larvae expressing GFP-NompA, we found that NompC-L was localized distal to the ciliary dilation, at the ciliary tips that are wrapped by NompA-enriched dendritic caps (Fig. 4F). Two other TRP channels, Nan and Iav, have been shown to localize to the proximal cilium (Gong et al., 2004). Therefore, NompC and Nan/Iav are localized to distinct domains in chordotonal cilia (Fig. 4G). The close proximity of NompC and NompA is compatible with the model that they form a transcelluar complex to mediate mechanotransduction (Gillespie and Walker, 2001).
In order to visualize NompC localization in living larval neurons, we generated a UAS-nompC-L-GFP transgene. Expression of NompC-L-GFP was sufficient to rescue the locomotion defects of nompC mutants, demonstrating that NompC-L-GFP is a functional protein (Fig. 3B). Consistent with the staining pattern for endogenous NompC, NompC-L-GFP fluorescence was observed in the ciliary tips of chordotonal organs in fly embryos (data not shown) and larvae (Fig. 5B, upper left panel). In class I da neurons, NompC-L-GFP fluorescence was observed along the sensory dendrites (Fig. 4H, H′). In bd neurons, NompC-L-GFP fluorescence was observed along the cell membrane and in filamentous structures within the proximal dendrites (arrow, Fig. 4I, I′). The localization of NompC-L at the ciliary tips of chordotonal organs and along sensory neuron dendrites supports the notion that NompC is a putative mechanosensitive channel.
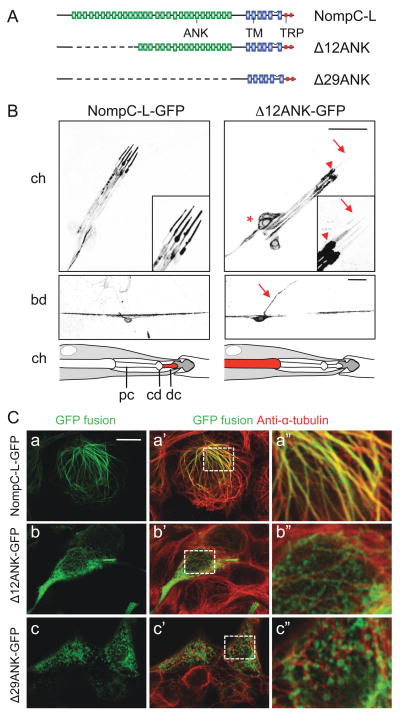
Figure 5. Ankyrin repeats of NompC are required for ciliary localization and microtubule association.
(A) A schematic of full-length and truncated NompC-L protein structures. ANK, ankyrin domain; TM, transmembrane domain; TRP, TRP box domain.
(B) Localization of NompC-L-GFP and Δ12ANK-GFP in chordotonal organs (upper panels) and bd neurons (middle panels). The lch5 sensory neurons are indicated by *, the base of the cilium by an arrowhead and the ciliary tip by an arrow. The bd axon is indicated by an arrow. A schematic interpretation of the localization of NompC-L-GFP and Δ12ANK-GFP in chordotonal organs are shown in the bottom panels. pc, proximal cilium; cd, ciliary dilation; dc, distal ciliary tip. Scale bars, 20 μm.
(C) Localization of NompC-L-GFP, Δ12ANK-GFP and Δ29ANK-GFP in HEK 293 cells. Full-length and truncated NompC-L protein was visualized by the GFP fluorescence (green). The cells were co-stained with anti-α-tubulin (red). Boxed area in a′, b′, c′ is enlarged and shown in a″, b″, c″ respectively. Scale bar, 10 μm. See also Figure S4.
Ciliary localization and function of NompC depends on ankyrin repeats
Several TRP subfamilies (TRPA, TRPC, TRPN and TRPV) have ankyrin (ANK) repeats in their cytoplasmic N-terminus, but the role of these ANK repeats remains unclear. NompC contains 29 consecutive ANK repeats at its N-terminus and it has been proposed that 29 ANK repeats could form one helical turn, and thus function as a gating spring in mechanotransduction (Howard and Bechstedt, 2004). To test the role of the ANK repeats in NompC function, we generated two forms of NompC that delete all or a subset of the ANK repeats. Δ29ANK-GFP has all 29 ANK repeats removed and Δ12ANK-GFP has ANK repeats 1 to 12 removed (Fig. 5A). The amino acids preceding the ANK repeats at the N- terminus are retained and GFP is fused to the C-terminus of the truncated protein. In contrast to the full-length NompC-L-GFP, both Δ29ANK-GFP and Δ12ANK-GFP failed to rescue the locomotion defects of nompC mutant larvae (Fig. 3E) and mutant flies (Movie S3). Next, we examined the localization of the truncated proteins in vivo. In contrast to the localization of the full-length NompC-L-GFP to the distal ciliary tips, Δ12ANK-GFP showed abnormal accumulation in the cell body (marked by *, Fig. 5B, upper right panel) and near the base of the cilium (arrowhead), but failed to be transported to distal ciliary tips (arrows). In bd neurons, NompC-L-GFP is preferentially distributed in dendrites (Fig. 5B, middle left panel). In contrast, Δ12ANK-GFP is present at low levels in dendrites, as well as the cell body and the axon (Fig. 5B, middle right panel, the axon is indicated by an arrow). Δ29ANK-GFP showed a similar distribution as Δ12ANK-GFP, but with lower expression levels (data not shown). These results suggest that ANK repeats are required for proper targeting of NompC protein in chordotonal organs. In addition, ANK repeats may also contribute to NompC protein stability in bd and class I da neurons.
Ankyrin repeats mediate association of NompC with microtubules
To further explore the role of the ANK repeats in NompC localization and function, we expressed the full-length NompC-L protein and truncated NompC with ANK deletions in HEK 293 cells. The full-length NompC-L protein displayed a cytoskeleton-like distribution and double labeling with anti-α-tubulin revealed that NompC-L-GFP co-localizes with microtubules (Fig. 5C–a). Similar results were obtained in Chinese Hamster Ovary (CHO) cells, Drosophila Schneider (S2) cells, and ciliary inner medullary collecting duct (IMCD) cells (data not shown). In fly embryos and larvae, co-localization of NompC-L with α-tubulin was also detected (Fig. S3).
To test whether NompC associates with microtubules, we monitored the localization of NompC-L-GFP following microtubule depolymerization after nocodazole treatment. Treatment of cells for 20 minutes with nocodazole (10 μM) caused NompC-L-GFP to redistribute to the ER (Fig. S4), suggesting that the localization of NompC depends on intact microtubule structures. Truncated NompC-L with ANK deletions failed to co-localize with microtubules. Δ12ANK-GFP displayed a partial filamentous pattern and a large fraction of Δ12ANK-GFP failed to co-localize with microtubules (Fig. 5C–b) whereas Δ29ANK-GFP displayed a punctate expression pattern and showed very little, if any, co-localization with microtubules (Fig. 5C–c). These experiments demonstrate that the ANK repeats are required for the association of NompC with microtubules. The disruption of this association may contribute to the defects in localization and function of the truncated proteins in vivo.
DISCUSSION
Here we report the identification and characterization of NompC as a critical regulator of Drosophila locomotion. Proper NompC function in bd and class I da neurons is crucial for larval peristaltic crawling behavior; the locomotion speed is greatly reduced in both null-mutants and mutants carrying a missense mutation. Importantly, NompC is required for activating bd and class I da neuron central projections during muscle contractions. Moreover, NompC protein is localized to chordotonal ciliary tips and along sensory neuron dendrites, the potential sites of mechanosensation. Finally, the ankyrin repeats in NompC are required for its association with microtubules and for the proper localization and function of NompC in vivo.
Previous studies (Hughes and Thomas, 2007) and our own work showed that two types of sensory neurons, namely the bd and class I da neurons, are required for Drosophila larval locomotion. Indeed, our calcium imaging revealed that bd and class I da neurons were activated during peristaltic muscle contractions, suggesting that they may function as proprioceptor neurons during locomotion. Structurally, bd and class I da neurons are suitable to function as proprioceptor neurons. The bd neuron is found in many different insects (Finlayson and Lowenstein, 1958). It is situated between muscles in the dorsal body wall. The two dendrites of a bd neuron span the length of the segment and reach the segmental folds on either end, where the dendritic tips are attached to the epidermal cells (Schrader and Merritt, 2007). Class I da neurons have a relatively simple dendrite branching pattern. Two dorsal class I da neurons project their fan-shaped dendrites anteriorly and posteriorly, respectively (Grueber et al., 2002). Similar to other da neurons, class I da neurons form a subepidermal plexus sandwiched between the epidermis and muscles. The dendrite morphology of bd and class I da neurons, their location in the segment, as well as their positions relative to muscles and the epidermis suggest that bd and class I da neurons could sense the stretch in muscles and/or epidermis during larval locomotion movement, thus function as proprioceptors. Consistent with the requirement of the function of bd and class I da neurons in locomotion, their axons project to the same dorsal regions of the neuropil (Grueber et al., 2007), indicating that signals from bd and class I da neurons might be processed in the same circuitry for the feedback regulation of motor neuron activity (Hughes and Thomas, 2007; Song et al., 2007).
The TRP family of cation channels plays an important role in the detection of various sensory stimuli (Venkatachalam and Montell, 2007). We found that NompC is required for activating bd and class I da neurons during muscle contractions and controls the pace of larval crawling. NompC is also involved in mechanosensation in bristles (Walker et al., 2000) and hearing in fly (Gopfert et al., 2006; Kim et al., 2003). How does a single channel contribute to multiple mechanosensation modalities in different neurons? While NompC is presumably gated by mechanical forces, the specificity in the sensory modality could arise from different dendrite morphology of the neurons in which NompC is expressed. For example, the ciliary structure in hair cells makes NompC responsive to sound stimuli, whereas the two longitudinal dendrites of bd neurons may allow NompC to be tuned to body stretch or bending. Another possibility is that NompC interacts with different partners to achieve the specificity in different neurons.
A critical question in the study of mechanosensation is to understand how the ion channels are gated mechanically. Ankyrin repeats are a common feature in many TRP channels, yet the function of ankyrin repeats remains unclear (Gaudet, 2008). Previous studies implicate NompC as a mechanosensitive channel because nompC null mutants lack the mechanoreceptor potential, whereas a point mutation of nompC alters adaptation of mechanoreceptor potential (Walker et al., 2000). One question raised by Walker et al. concerns the ability of the ankyrin repeats of NompC to mediate interactions with the cytoskeleton. Here, we show that the ankyrin repeats of NompC are essential for NompC-microtubule interaction in transfected cells and for proper targeting of NompC in vivo to the distal ciliary tips, which are wrapped by NompA-enriched dendritic caps. It is noteworthy that two other TRP channels that mediate hearing in Drosophila, Nan and Iav, are localized to the proximal cilium and are absent from the ciliary tips that are in contact with the dendritic caps (Gong et al., 2004). Therefore, if Nan and Iav are mechanically gated, the forces that gate the channels would have to be transmitted down the axoneme through the ciliary membrane or via other extracellular material. Localization of NompC at distal ciliary tips and its interaction with microtubules raise the possibility that NompC may be anchored to both the cytoskeleton and to the extracelluar matrix of the dendritic caps at the ciliary tip. With mechanical stimulation, the relative displacement between extracelluar matrix and the cytoskeleton might “pull” the channel open as suggested by the prevailing model for direct mechanogating (Albert et al., 2007; Ernstrom and Chalfie, 2002; Sukharev and Corey, 2004). Although it remains to be tested whether NompC can be directly activated by mechanical forces, studies using atomic force microscopy have demonstrated the elastic properties of ankyrin repeats in response to pulling on both ends of the domain (Lee et al., 2006). In larval bd neurons, NompC-L-GFP is distributed throughout the two longitudinal dendrites, which are wrapped by glial processes (Schrader and Merritt, 2007). We speculate that the shearing forces between the dendrites and the glial wrapping could provide the relevant stimulation to activate the NompC channel in bd neurons, thus leading to Ca2+ signals in their central projections in the central nervous system.
EXPERIMENTAL PROCEDURES
Flies and stocks
MD (5–40)-Gal4, SN (21–7)-Gal4, Class I (2–21)-Gal4 drivers were provided by U. Heberlein. The bd-Gal4 driver was provided by K. Emoto. nompC-Gal4 was generated using a 1.5 kb promoter located immediately upstream of the first exon of nompC. Detailed larval expression patterns of each driver are summarized in Table S1. nompC mutant lines were provided by C. Zuker. The GFP-NompA line was provided by M. Kernan. The GCaMP3 line was provided by L. Looger. All flies were raised on standard media.
Locomotion assay
Locomotion was analyzed using Digital Imaging Analysis Software (DIAS, Solltech, Oakdale, IA) as previously described (Song et al., 2007; Wang et al., 1997). Briefly, a wandering-stage larva was placed on a fresh 2% agar plate and videotaped using a digital camcorder attached to a microscope. Only continuous forward locomotion (≥5 strides without turns) was used to calculate crawling speed, stride duration, and stride length. For adult locomotion, a fly was placed on an evenly illuminated fluorescent light box and covered with a 10cm petri dish. Each individual fly was recorded for 20 seconds or until it moved out of the recording field.
Calcium Imaging
Third instar foraging larvae were dissected in haemolymph-like (HL3) solution that contains 2mm Ca2+. The larva was dissected through the dorsal midline, and the brain lobes, ventral nerve cord and the segmental nerves were left intact. The ventral nerve cord was imaged using a Leica SP5 confocal microscope equipped with a 40× water lens. Time-lapse series were collected at 70 ms/frame. Data analysis was performed using the LAS AF software. The GCaMP3 signal was normalized over CD2-mCherry signal and the ratio (FGCaMP/FCD2-mCherry) was used to calculate ΔF/F. A Gaussian filter with radius of 2 was applied to all images presented. The images were then pseudo-colored using ImageJ (version 1.43, available from NIH).
Isolation of nompC cDNA
nompC cDNA was cloned from larval RNA by RT-PCR using two sets of primers that amplify the N-terminal (primer set 1: 5′-ATGTCGCAGCCGCGCGGAGGGCGT-3′ and 5′-CTGCTGCAGGAAGAGTTTGGCCACTTC-3′) and C-terminal (primer set 2: 5′-TGAGAAAGCAAACGCCGCTC-3′ and 5′-CTATTTTGTAGTGTCTGCCAGGGT-3′) region of nompC respectively. The primers were designed based on the sequence of a predicated isoform of nompC (nompC-D, NM_001103619.2). The N-terminal and C terminal fragments were then joined using an internal PstI site to generate the full-length nompC-L cDNA. The sequence of nompC-L is available through Genbank (accession number: HM582118). nompC-L and nompC-L-GFP was inserted into the pUAST vector to generate transgenic flies and the pcDNA3 vector (Invitrogen) for expression in cells.
Production of anti-NompC antibody, Western blotting and immunohistochemisty
Polyclonal anti-NompC antibodies were raised against the C-terminal NompC-L peptide CGRRAIKATLADTTK (Open Biosystems). Lipofectamine 2000 (Invitrogen) was used to transfect cells as instructed by the manufacturer. For immunohistochemisty, the following antibodies were used: anti-NompC (1:500 for fly embryos and 1: 2000 for cultured cells), mAB 22C10 (1:300), mAB 21A6 (1:300), rabbit anti-GFP (Invitrogen, 1:1000 for embryos). Secondary antibodies were Cy3-, Cy5, or TRX-conjugated goat anti-rabbit or anti-mouse IgG (Jackson ImmunoResearch). For Western blotting, anti-NompC (1:2000) and anti-α-tubulin (DM1A; AbCam, 1:2000) were used. Blots were developed with ECL reagents (Amersham) and documented with a VersaDoc imaging system (BioRad).
Supplementary Material
Acknowledgments
We thank Ulrike Heberlein, Kazuo Emoto, Charles Zuker and Maurice Kernan for fly stocks; members of the Jan lab for experimental advice; Jay Parrish and Peter Soba for comments on the manuscript. This work was supported by a Javits Neuroscience Investigator Award R37NS040929 from NINDS (Y.N. J). L.Y.J and Y.N.J are investigators of the Howard Hughes Medical Institute.
Footnotes
Note added in proof
As this manuscript was under review, a publication by Lee at al. (Lee et al., 2010) reported NompC localization to the distal end of mechanosensory cilia using a different antibody generated against the N-terminus of NompC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ainsley JA, Pettus JM, Bosenko D, Gerstein CE, Zinkevich N, Anderson MG, Adams CM, Welsh MJ, Johnson WA. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr Biol. 2003;13:1557–1563. doi: 10.1016/s0960-9822(03)00596-7. [DOI] [PubMed] [Google Scholar]
- Albert JT, Nadrowski B, Gopfert MC. Mechanical signatures of transducer gating in the Drosophila ear. Curr Biol. 2007;17:1000–1006. doi: 10.1016/j.cub.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Jan YL. Morphological differentiation of the embryonic peripheral neurons in Drosophila. Roux’s Archives of Developmental Biology. 1987;196:69–77. doi: 10.1007/BF00402027. [DOI] [PubMed] [Google Scholar]
- Chung YD, Zhu J, Han Y, Kernan MJ. nompA encodes a PNS-specific, ZP domain protein required to connect mechanosensory dendrites to sensory structures. Neuron. 2001;29:415–428. doi: 10.1016/s0896-6273(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Ernstrom GG, Chalfie M. Genetics of sensory mechanotransduction. Annu Rev Genet. 2002;36:411–453. doi: 10.1146/annurev.genet.36.061802.101708. [DOI] [PubMed] [Google Scholar]
- Finlayson LH, Lowenstein O. The structure and function of abdominal stretch receptors in insects. Proc R Soc Lond B Biol Sci. 1958;148:433–449. doi: 10.1098/rspb.1958.0037. [DOI] [PubMed] [Google Scholar]
- Gaudet R. A primer on ankyrin repeat function in TRP channels and beyond. Mol Biosyst. 2008;4:372–379. doi: 10.1039/b801481g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PG, Walker RG. Molecular basis of mechanosensory transduction. Nature. 2001;413:194–202. doi: 10.1038/35093011. [DOI] [PubMed] [Google Scholar]
- Gong Z, Son W, Chung YD, Kim J, Shin DW, McClung CA, Lee Y, Lee HW, Chang DJ, Kaang BK, et al. Two interdependent TRPV channel subunits, inactive and Nanchung, mediate hearing in Drosophila. J Neurosci. 2004;24:9059–9066. doi: 10.1523/JNEUROSCI.1645-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopfert MC, Albert JT, Nadrowski B, Kamikouchi A. Specification of auditory sensitivity by Drosophila TRP channels. Nat Neurosci. 2006;9:999–1000. doi: 10.1038/nn1735. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Yang CH, Younger S, Borden K, Jan LY, Jan YN. Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development. 2007;134:55–64. doi: 10.1242/dev.02666. [DOI] [PubMed] [Google Scholar]
- Howard J, Bechstedt S. Hypothesis: a helix of ankyrin repeats of the NOMPC-TRP ion channel is the gating spring of mechanoreceptors. Curr Biol. 2004;14:R224–226. doi: 10.1016/j.cub.2004.02.050. [DOI] [PubMed] [Google Scholar]
- Hughes CL, Thomas JB. A sensory feedback circuit coordinates muscle activity in Drosophila. Mol Cell Neurosci. 2007;35:383–396. doi: 10.1016/j.mcn.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, Tracey WD. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikouchi A, Inagaki HK, Effertz T, Hendrich O, Fiala A, Gopfert MC, Ito K. The neural basis of Drosophila gravity-sensing and hearing. Nature. 2009;458:165–171. doi: 10.1038/nature07810. [DOI] [PubMed] [Google Scholar]
- Kernan M, Cowan D, Zuker C. Genetic dissection of mechanosensory transduction: mechanoreception-defective mutations of Drosophila. Neuron. 1994;12:1195–1206. doi: 10.1016/0896-6273(94)90437-5. [DOI] [PubMed] [Google Scholar]
- Kernan MJ. Mechanotransduction and auditory transduction in Drosophila. Pflugers Arch. 2007;454:703–720. doi: 10.1007/s00424-007-0263-x. [DOI] [PubMed] [Google Scholar]
- Kim J, Chung YD, Park DY, Choi S, Shin DW, Soh H, Lee HW, Son W, Yim J, Park CS, et al. A TRPV family ion channel required for hearing in Drosophila. Nature. 2003;424:81–84. doi: 10.1038/nature01733. [DOI] [PubMed] [Google Scholar]
- Lee E, Sivan-Loukianova E, Eberl DF, Kernan MJ. An IFT-A protein is required to delimit functionally distinct zones in mechanosensory cilia. Curr Biol. 2008;18:1899–1906. doi: 10.1016/j.cub.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Abdi K, Jiang Y, Michaely P, Bennett V, Marszalek PE. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- Lee J, Moon S, Cha Y, Chung YD. Drosophila TRPN(=NOMPC) channel localizes to the distal end of mechanosensory cilia. PLoS One. 2010;5:e11012. doi: 10.1371/journal.pone.0011012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Feng Z, Sternberg PW, Xu XZ. A C. elegans stretch receptor neuron revealed by a mechanosensitive TRP channel homologue. Nature. 2006;440:684–687. doi: 10.1038/nature04538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin EA, Caterina MJ. Mechanisms of sensory transduction in the skin. Nature. 2007;445:858–865. doi: 10.1038/nature05662. [DOI] [PubMed] [Google Scholar]
- Martinac B. Mechanosensitive channels in prokaryotes. Cell Physiol Biochem. 2001;11:61–76. doi: 10.1159/000047793. [DOI] [PubMed] [Google Scholar]
- Roos J, Hummel T, Ng N, Klambt C, Davis GW. Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron. 2000;26:371–382. doi: 10.1016/s0896-6273(00)81170-8. [DOI] [PubMed] [Google Scholar]
- Schrader S, Merritt DJ. Dorsal longitudinal stretch receptor of Drosophila melanogaster larva - fine structure and maturation. Arthropod Struct Dev. 2007;36:157–169. doi: 10.1016/j.asd.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Smetacek V, Mechsner F. Making sense. Nature. 2004;432:21. doi: 10.1038/432021a. [DOI] [PubMed] [Google Scholar]
- Song W, Onishi M, Jan LY, Jan YN. Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc Natl Acad Sci U S A. 2007;104:5199–5204. doi: 10.1073/pnas.0700895104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev S, Corey DP. Mechanosensitive channels: multiplicity of families and gating paradigms. Sci STKE. 2004;2004:re4. doi: 10.1126/stke.2192004re4. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu L, Ben-Shahar Y, Jacobs JS, Eberl DF, Welsh MJ. TRPA channels distinguish gravity sensing from hearing in Johnston’s organ. Proc Natl Acad Sci U S A. 2009;106:13606–13611. doi: 10.1073/pnas.0906377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N, Shreffler W, Wang S, Driscoll M. unc-8, a DEG/ENaC family member, encodes a subunit of a candidate mechanically gated channel that modulates C. elegans locomotion. Neuron. 1997;18:107–119. doi: 10.1016/s0896-6273(01)80050-7. [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RG, Willingham AT, Zuker CS. A Drosophila mechanosensory transduction channel. Science. 2000;287:2229–2234. doi: 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- Windhorst U. Muscle proprioceptive feedback and spinal networks. Brain Res Bull. 2007;73:155–202. doi: 10.1016/j.brainresbull.2007.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.