Abstract
The treelike structures of many organs, including the mammary gland, are generated by branching morphogenesis, a reiterative process of branch initiation and invasion from a preexisting epithelium. Using a micropatterning approach to control the initial three-dimensional structure of mouse mammary epithelial tubules in culture, combined with an algorithm to quantify the extent of branching, we found that the geometry of tubules dictates the position of branches. We predicted numerically and confirm experimentally that branches initiate at sites with a local minimum in the concentration of autocrine inhibitory morphogens, such as transforming growth factor–β. These results reveal that tissue geometry can control organ morphogenesis by defining the local cellular microenvironment, a finding that has relevance to control of invasion and metastasis.
A burst of dichotomous and lateral branching at puberty transforms the mammary epithelial tubule rudiment present at birth into a fully elaborated ductal tree in the female adult. The overall process of branching morphogenesis is regulated globally by a number of cues, including growth factors, extracellular matrix (ECM) molecules, proteases, and morphogens (1-4). These global cues must be integrated locally within the context of the tissue to determine where branches are initiated; thus, a subgroup of epithelial cells is instructed to form a branch or to bifurcate, whereas neighboring cells are not (5). Current techniques to study this process, which is common to many organs including the lung, kidney, and salivary gland, do not allow for a precise quantitative understanding of how spatial positioning is determined. Given that the mammary ductal network branches out from preexisting epithelial tubules, we hypothesized that the position of cells within a tubule might provide contextual information to instruct branch site initiation.
To define the role of positional context, we developed a three-dimensional (3D) micropatterned assay for mammary epithelial branching morphogenesis that allowed us to mimic the mammary rudiment by controlling the initial geometry of tubules and to quantify the positions at which they branched. We engineered epithelial tubules of defined geometry by embedding functionally normal mouse mammary epithelial (EpH4) cells in cavities of collagen gel generated by molding unpolymerized collagen I around a patterned elastomeric stamp (Fig. 1A) (6). Embedded epithelial cells formed hollow tubules (7) conforming to the size and shape of the collagen cavities (Fig. 1, B and C, and fig. S1). To quantify branching and to represent its magnitude and position statistically, we stained nuclei and stacked multiple fluorescent images in registration such that the stacked image revealed the average spatial distribution of cells within, and branching from, the tubules (Fig. 1D). Stacked images were depicted as frequency maps (Fig. 1E) (8).
Fig. 1.

Characteristics of branching from engineered mammary epithelial tubules. (A) Schematic of 3D microfabrication method to engineer tubules. (B) Phase contrast image and (C) confocal image of tubules stained for actin (green) and nuclei (blue) before induction of branching. The position of cells was quantified by (D) stacking images of nuclei from 50 tubules to generate (E) a frequency map before induction of branching. (F) Phase contrast image and (G) frequency map of tubules 24 hours after adding EGF to induce branching. (H) Confocal image of tubule of primary mammary epithelial cells stained for luminal epithelial keratin-8 (green), myoepithelial keratin-14 (red), and nuclei (blue); (inset) shows z section through tubule. (I) Frequency map of primary mammary epithelial tubules 24 hours after adding EGF. (J) Fluorescent image of vimentin gene promoter-GFP (green) and nuclei (blue) and (K) frequency map of vimentin gene promoter-GFP expression 8 hours after adding EGF. Scale bars, 50 μm.
The tubules remained quiescent (Fig. 1E) until induced to undergo branching morphogenesis by addition of epidermal growth factor (EGF) (Fig. 1F) or hepatocyte growth factor (HGF) (fig. S1) (9). Within 24 hours after induction, multicellular branches extended into the surrounding collagen. These branches initiated only from the ends and not from the sides of the tubules (Fig. 1, F and G), a pattern reminiscent of the dichotomous branching of mammary end buds in vivo (10). Primary mammary epithelial cells or organoids formed correctly polarized bilayered tubules with myoepithelial cells and basement membrane surrounding an inner layer of luminal epithelial cells (Fig. 1H and fig. S1). Primary tubules also branched solely from the ends of the polarized bilayer (Fig. 1I).
To initiate a branch, cells must deform from their positions within the polarized epithelial tubule and must invade the surrounding tissue. Mammary and kidney epithelial cells in 3D cultures undergo a transient epithelial-to-mesenchymal transition (EMT) at the tips of branches as they invade the surrounding ECM (10-13). Because we could predict the position of branching with high certainty (~96 to 99%), we were able to determine whether acquisition of the invasive mesenchymal phenotype was restricted to these positions in the tubule before branch extension or merely correlated with the branches themselves. The appearance of mesenchymal attributes was assayed in situ in real time by monitoring expression of green fluorescent protein (GFP) under the control of the human vimentin gene promoter (11, 14, 15), and was found to be activated specifically in cells at positions that later branched (Fig. 1, J and K). Invasion during mammary branching morphogenesis requires matrix metalloproteinases (MMPs) and mesenchymally derived morphogens, such as epimorphin (1, 2, 9); blocking these activities prevented branching from the tubules, but did not prevent spatially localized activation of the vimentin gene promoter (fig. S2). These data reveal that the invasive phenotype is spatially restricted before extension of branches and that the cells are instructed to branch depending on their position within the tubule.
We confirmed that positional context controls branch sites by examining morphogenesis of tubules of varying geometry. Increasing the length of the tubules increased the magnitude of branching, although cells still branched exclusively from the ends (fig. S3). Curved tubules branched preferentially from the convex side of the curve (Fig. 2A). Asymmetric branching was also observed in bifurcated tubules and trees (Fig. 2, B and C), which preferentially branched from distal positions. The position of branching thus depended on the initial geometry of the tubule. Branching required cellular proliferation (fig. S4) (9), but the pattern of branching was not due to localized proliferation or locally enhanced signaling by growth factor receptors (fig. S4).
Fig. 2.

Branching position is determined by tubule geometry and is consistent with the concentration profile of secreted diffusible inhibitor(s). Frequency maps 24 hours after induction of branching for (A) curved tubules, (B) bifurcated tubules, and immunofluorescence staining of actin (red) and nuclei (green) of (C) fractal trees. Branch sites in (C) are denoted by arrows; image stitched from multiple fields. Calculated concentration profiles of diffusible inhibitors for (D) curved tubules, (E) bifurcated tubules, and (F) fractal trees predict lowest local concentration of inhibitors where branching was found to be induced experimentally. Scale bars, 50 μm.
That branch sites depended on the initial tubule geometry suggested that positional context was encoded by the preexisting structure. Positional information during branching morphogenesis might be encoded by stimulatory morphogens secreted by adjacent mesenchymal tissues, which act as chemoattractants for growing epithelial branches (4, 16). This mechanism would require an initial prepattern of signal within the mesenchyme. Alternatively, stimulatory cues released distally (such as the EGF or HGF, which we provided exogenously) might be balanced by inhibitory cues released locally by epithelial cells (17). In this scenario, branching would initiate only at tubule positions that were surrounded by a local minimum or subthreshold concentration of autocrine inhibitors. Because the epithelial cells within our engineered tubules exhibited patterned behavior in the absence of adjacent mesenchymal cells, we considered the possibility that branching position was determined by autocrine inhibitory morphogens secreted locally by the epithelial cells themselves.
To test this hypothesis, we generated a computational model of the concentration profile of hypothetical inhibitory molecules produced by epithelial cells in each of the engineered geometries by constructing a 3D model of diffusion. We assumed a constant flux (uniform rate of secretion) of inhibitors from the surface of the tubule and passive isotropic diffusion through the collagenous ECM. At steady state, a concentration gradient of inhibitors was found across each of the tubule geometries (Fig. 2, D to F, and figs. S1 and S3). Notably, the concentration of inhibitors was lowest in positions where branching was induced, in agreement with our hypothesis.
We thus investigated whether increasing the local concentration of inhibitors would be sufficient to block branching from the ends of tubules—regions that usually branched (Fig. 1G). One way to effect a local increase in inhibitor concentration is to decrease the distance between tubules (Fig. 3, A and B). Consistent with the predicted concentration profile, mammary epithelial cells branched from distant tubule ends, but not from ends near neighboring tubules (Fig. 3, C and D, and fig. S5). Varying the spacing between tubules revealed that branching was inhibited at distances up to ~75 μm (fig. S5); a similar ductal spacing is found in the mouse mammary gland (18). Therefore, inducing a change in concentration profile by altering the initial geometry of the tissue alters the position of branching.
Fig. 3.
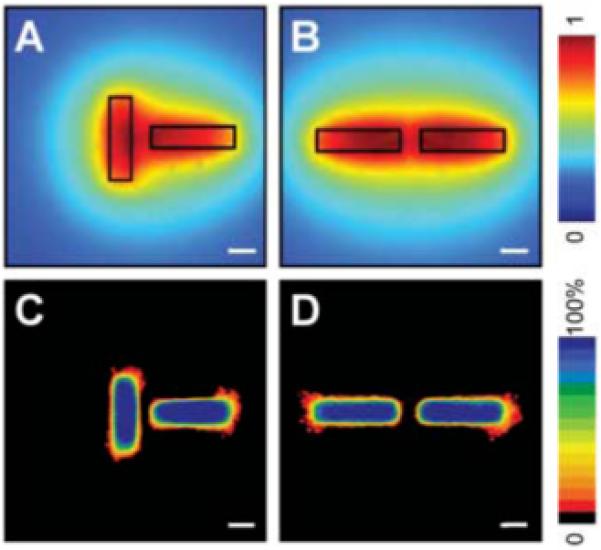
Position of branching can be predicted by calculated concentration profile. Calculated profiles of diffusible inhibitors in tubules oriented perpendicular (A) and parallel (B) to each other. Frequency maps 24 hours after induction of branching confirm that branching is inhibited in regions predicted to be surrounded by a high concentration of inhibitors in perpendicular (C) and parallel (D) tubules. Scale bars, 50 μm.
A number of proteins inhibit mammary branching morphogenesis in vivo, including transforming growth factor–β (TGFβ) (19, 20), which signals through a tetrameric complex of type I and type II receptors. Engineered tubules expressed TGFβ and its receptors (fig. S6), and immunofluorescence staining revealed a gradient in the concentration of TGFβ1 that correlated with our numerical predictions (Fig. 4A). Overexpression of active TGFβ1 completely inhibited branching (Fig. 4, B and C). To determine whether TGFβ acts as an endogenous inhibitor in this system, we used three approaches to disrupt its signaling: treatment with a function-blocking antibody against TGFβ1, treatment with a pharmacological inhibitor of TGFβ type I receptor kinase activity, and overexpression of a dominant negative TGFβ type II receptor. All treatments resulted in uniform branching from tubules of EpH4 or primary mammary epithelial cells (Fig. 4, D and E, and fig. S6), effectively abolishing the positional information.
Fig. 4.
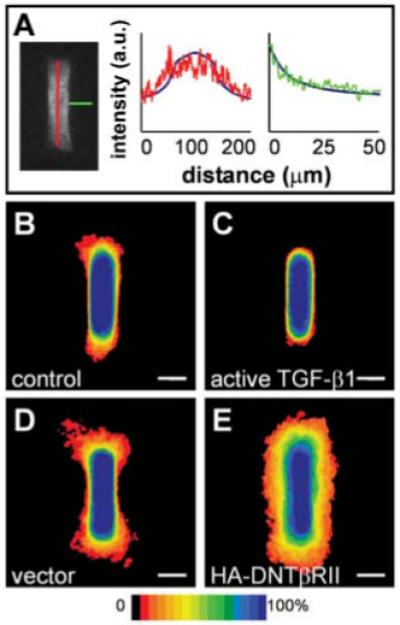
Inhibitory activity is mediated in part by autocrine TGFβ in cultured cells. (A) Confocal section of primary mammary epithelial tubule stained for TGFβ1, with graphs representing relative pixel intensity (arbitrary units, a.u.) as a function of distance along tubules (red) and away from tubules (green). Numerical predictions are superimposed as solid blue curves to fit the intensity range. Frequency maps 24 hours after induction of branching in tubules of (B) control cells and (C) cells overexpressing active TGFβ1 confirm that TGFβ1 inhibits branching. (D and E) Positional control of branching is disrupted by blocking signaling of endogenous TGFβ1. Shown are frequency maps 24 hours after induction of branching in tubules of (D) vector control cells and (E) cells overexpressing dominant negative TGFβ receptor type II (HA-DNTβRII). Scale bars, 50 μm.
These data demonstrate that tissue form and context—including the geometry of multicellular tubules, as well as their proximity to neighbors—can control the position of branching during morphogenesis of mammary epithelial cells and primary organoids. That this simple ex vivo system exhibits complex bifurcating and lateral branching behaviors suggests that the geometry of the duct may act as an instructive cue during its morphogenesis in vivo. This mechanism could explain how the mammary gland achieves its open architecture during development, a possibility to be explored further. We found that positional context is determined at least in part by the local concentration of autocrine TGFβ, an inhibitory morphogen in the mammary gland and other branched organs (21-23). Because TGFβ is secreted in an inactive latent form (24) and because overexpression of wild-type (latent) TGFβ1 had no effect on branching (fig. S6), we speculate that additional signals are required to sculpt the concentration profile of inhibitory activity that determines branching position. These signals may be chemical or mechanical in nature: MMPs and tissue inhibitors of MMPs (TIMPs) are known to affect branching (1, 2, 25), and intercellular tension can alter the response of cells to morphogens or the activity of morphogens themselves (26, 27).
The balance between stimulatory and inhibitory morphogen gradients was postulated long ago by a number of theoretical scientists as a mechanism to explain pattern formation during development of other tissues (28-30). The model system presented here allows direct quantitative testing of the positional integration of these cues in a relevant developmental context and can be extended to investigate the mechanisms that control morphogenesis of any branched organ system.
Supplementary Material
Acknowledgments
We thank J. Tien, D. Radisky, and M. LaBarge and other members of the Bissell laboratory for insightful discussions and C. Gilles, Y. Hirai, K. Luo, A. Roberts, and L. Sorokin for generously providing reagents. Supported by grants from the U.S. Department of Energy (DE-AC03-76SF00098 and a Distinguished Fellow Award to M.J.B.), the NIH (CA64786 and CA57621 to M.J.B., GM72736 to D.A.F.), the U.S. Department of Defense (an Innovator Award to M.J.B. and W81XWH-04-1-0582 to C.M.N.), a Lawrence Berkeley National Laboratory Laboratory Directed Research and Development grant (D.A.F.), an NSF Career Award (D.A.F.), and a QB3 fellowship (M.V.D.). We dedicate this paper to the memory of Anita Roberts, codiscoverer of TGFβ.
Footnotes
Supporting Online Material
References and Notes
- 1.Simian M, et al. Development. 2001;128:3117. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiseman BS, et al. J. Cell Biol. 2003;162:1123. doi: 10.1083/jcb.200302090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai T, Larsen M, Yamada KM. Nature. 2003;423:876. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 4.Weaver M, Dunn NR, Hogan BL. Development. 2000;127:2695. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- 5.Daniel CW, Smith GH. J. Mammary Gland Biol. Neoplasia. 1999;4:3. doi: 10.1023/a:1018796301609. [DOI] [PubMed] [Google Scholar]
- 6.Tang MD, Golden AP, Tien J. J. Am. Chem. Soc. 2003;125:12988. doi: 10.1021/ja037677h. [DOI] [PubMed] [Google Scholar]
- 7.Hall HG, Farson DA, Bissell MJ. Proc. Natl. Acad. Sci. U.S.A. 1982;79:4672. doi: 10.1073/pnas.79.15.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson CM, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11594. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirai Y, et al. J. Cell Biol. 1998;140:159. doi: 10.1083/jcb.140.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fata JE, Werb Z, Bissell MJ. Breast Cancer Res. 2004;6:1. doi: 10.1186/bcr634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen CS, Radisky D, Bissell MJ. in preparation. [Google Scholar]
- 12.Affolter M, et al. Dev. Cell. 2003;4:11. doi: 10.1016/s1534-5807(02)00410-0. [DOI] [PubMed] [Google Scholar]
- 13.O’Brien LE, et al. Dev. Cell. 2004;7:21. doi: 10.1016/j.devcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Gilles C, et al. J. Cell Sci. 1999;112:4615. doi: 10.1242/jcs.112.24.4615. [DOI] [PubMed] [Google Scholar]
- 15.Radisky DC, et al. Nature. 2005;436:123. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzger RJ, Krasnow MA. Science. 1999;284:1635. doi: 10.1126/science.284.5420.1635. [DOI] [PubMed] [Google Scholar]
- 17.Hogan BL. Cell. 1999;96:225. doi: 10.1016/s0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 18.Faulkin LJ, Jr., Deome KB. J. Natl. Cancer Inst. 1960;24:953. [PubMed] [Google Scholar]
- 19.Silberstein GB, Daniel CW. Science. 1987;237:291. doi: 10.1126/science.3474783. [DOI] [PubMed] [Google Scholar]
- 20.Ewan KB, et al. Am. J. Pathol. 2002;160:2081. doi: 10.1016/s0002-9440(10)61158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serra R, Pelton RW, Moses HL. Development. 1994;120:2153. doi: 10.1242/dev.120.8.2153. [DOI] [PubMed] [Google Scholar]
- 22.Ritvos O, et al. Mech. Dev. 1995;50:229. doi: 10.1016/0925-4773(94)00342-k. [DOI] [PubMed] [Google Scholar]
- 23.Bush KT, et al. Dev. Biol. 2004;266:285. doi: 10.1016/j.ydbio.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence DA, Pircher R, Kryceve-Martinerie C, Jullien P. J. Cell. Physiol. 1984;121:184. doi: 10.1002/jcp.1041210123. [DOI] [PubMed] [Google Scholar]
- 25.Fata JE, Leco KJ, Moorehead RA, Martin DC, Khokha R. Dev. Biol. 1999;211:238. doi: 10.1006/dbio.1999.9313. [DOI] [PubMed] [Google Scholar]
- 26.Ingber DE. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11571. doi: 10.1073/pnas.0505939102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munger JS, et al. Cell. 1999;96:319. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 28.Turing A. Philos. Trans. R. Soc. London Ser. B. 1952;237:37. [Google Scholar]
- 29.Crick F. Nature. 1970;225:420. doi: 10.1038/225420a0. [DOI] [PubMed] [Google Scholar]
- 30.Tabata T. Nat. Rev. Genet. 2001;2:620. doi: 10.1038/35084577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


