Abstract
The molecular pathways that participate in regulation of gene expression are being progressively unraveled. Extracellular signals, including the binding of extracellular matrix and soluble molecules to cell membrane receptors, activate specific signal transducers that process information inside the cell leading to alteration in gene expression. Some of these transducers when translocated to the cell nucleus may bind to transcription complexes and thereby modify the transcriptional activity of specific genes. However, the basic molecules involved in the regulation of gene expression are found in many different cell and tissue types; thus, the mechanisms underlying tissue-specific gene expression are still obscure.
In this review we focus on the study of signals that are conveyed to the nucleus. We propose that the way in which extracellular signals are integrated may account for tissue-specific gene expression. We argue that the integration of signals depends on the nature of the structural organization of cells (i.e., extracellular matrix, membrane proteins, cytoskeleton, nucleus) that defines a particular cell type within a tissue. Thus, gene expression can be envisioned as being regulated by the mutual influence of extracellular and intracellular organizations, i.e., in context.
Keywords: extracellular signal, signal transducer, gene expression, cellular structure, tissue specificity
I. Introduction
Regulation of tissue-specific gene expression, defined here broadly as the transcriptional and posttranscriptional control of gene products, depends on the integration of extracellular and intracellular signals. Extracellular signals include cell-extracellular matrix (ECM) and cell-cell contacts, and the binding of soluble molecules (e.g., growth factors and hormones) to their cellular receptors. Intracellular signals are composed of translocating signal transducers and cytoskeletal rearrangements that convey information elicited by extracellular signals, and the variety of enzymes that biochemically alter both signal transducers and cytoskeletal proteins.
Recent studies have begun to show how gene expression could be regulated at the nuclear level. Nuclear functions are operated by the assembly of proteins into supramolecular complexes characterized by their composition and activities. Alteration of the composition and/or distribution of these complexes has been linked directly to changes in gene expression. The sequential assembly of activator/transcription factors and his-tone acetyltransferase complexes, co-activator and mediator-holoenzyme complexes, and repressor complexes (e.g., histone deacetylase complexes) (reviewed by Berk, 1999; Razin, 1998) has been shown to regulate gene transcription. Furthermore, modifications of PML protein and NuMA protein distribution in the nucleus have been shown to be associated with alterations in the cellular pheno-type (Weis et al., 1994; Lelièvre et al., 1998).
The great majority of extracellular and intracellular signals and molecules that form nuclear supramolecular complexes can be found in different cell and tissue types; therefore, an additional level of regulation is necessary for tissue-specific gene expression. In this brief review we analyze how the “context” resulting from the nature of the extracellular signals, the nature of the cell type, and finally the nature of the organization of cell assemblies that gives rise to tissues contributes to tissue-specific gene expression.
II. Specificity of the nuclear response to extracellular signals
Numerous studies have reported that cells respond specifically to different extracellular signals by altering the expression of defined genes. For instance, mammary epithelial cells cultured in the presence of prolactin and the ECM component laminin turn on the expression of the milk protein beta-casein, while hepatocytes cultured in the presence of laminin turn on the expression of the liver-specific protein albumin. In those cases, tissue-specific gene expression in response to ECM signaling is illustrated by the fact that laminin-cell membrane interaction activates a response element in the promoter of the beta-casein gene in mammary epithelial cells (Schmidhauser et al., 1990) and a response element in the promoter of the albumin gene in hepatocytes (DiPersio et al., 1991). The presence of response elements in promoters of genes (which exist for many other types of extracellular signals, including hormones and growth factors) suggests a simple mechanism by which the pattern of tissue-specific gene transcription is determined by the extracellular milieu. However, the mechanisms that control the activation of these promoters involve ubiquitous regulatory molecules. To follow up on the example of the mammary gland, it has been shown that transcription factor STAT5 binds to the promoter of the beta-casein gene in mammary epithelial cells (Myers et al., 1998). STAT5 also binds the milk protein beta-lactoglobulin promoter after interactions of mammary epithelial cells with both ECM and prolactin (Streuli et al., 1995). In addition, STAT5 is found in many different cell types and tissues (Leonard and O'Shea, 1998). Tissues are also enriched with less ubiquitous types of transcription factors, but so far no transcription factor has been found to define tissue specificity by itself.
Studies focusing on chromatin structure have unraveled yet another level for the mechanism of regulation of extracellular context-dependent gene expression. Transcription factors STAT5 and C/ EBPβ were shown to be bound to the ECM response element of the beta-casein gene in a functional mammary epithelial cell line even in the absence of ECM and prolactin. Nevertheless, there was no beta-casein gene expression in the absence of ECM. Beta-casein expression could be triggered either by addition of ECM or after treatment with histone deacetylase inhibitors, suggesting that ECM may allow transcription of the beta-casein gene also via alteration of histone acetylation (Myers et al., 1998; Pujuguet and Bissell, unpublished results). This effect on chromatin structure involves apparently ubiquitous regulatory molecules, such as histone acetylases, that interact with the entire genome and have been shown to participate in many signaling pathways. For instance, activated RhoA triggers the expression of integrated serum-response element-controlled reporter genes by cooperating with other pathways, including SAPK/JNK-dependent pathways, to induce histone H4 acetylation (Alberts et al., 1998). Other studies have shown that neural growth-factor (NGF)-induced differentiation of PC 12 cells is accompanied by the accumulation of transcriptional co-activator and histone acetylase p300 in complexes associated with the NGF-responsive DNA sequence (Billon et al., 1999). These data suggest that a number of extracellular signals are capable of interfering with the organization of histones bound to specific DNA sequences.
The specificity of the response to extracellular signals could result from the type of signal transducers involved. Signal transducers can be defined as the molecules that convey initial signals to different locations inside the cells. Many signals originating at the cell membrane will lead to the nuclear translocation of defined signal transducers, including transcription factors and kinases. Nuclear translocation can be regulated by signaling-induced tyrosine phosphorylation, as is the case for STAT transcription factors where phosphorylation is necessary for dimerization and subsequent unmasking of a latent nuclear localization sequence (Milocco et al., 1999). Nuclear translocation of mitogen-activated protein (MAP) kinases, which has been proposed to require piggy-backing with transcription factors, is also activated by phosphorylation (Brunet et al., 1999). Other ways to initiate translocation of signal transducers include ligand-binding in the cytoplasm, as demonstrated for some steroid receptors (Htun et al., 1996) or liberation from cytoplasmic complexes as is the case for beta-catenin and NF-κB (Novak et al., 1998; Venkataraman et al., 1996). Once in the nucleus, signal transducers can bind directly to response elements of genes, as shown for STATs and ligand-steroid complexes, or form complexes with transcription factors already in place. For instance, translocated beta-catenin heterodimerizes with members of the LEF/TCF family of transcription factors to induce gene expression (Tetsu and McCormick, 1999). NF-κB interacts with histone acetylases p300 and CPB to drive gene activation (Shen et al., 1998). The same situation has been described for signal transducer Smad3, where once phosphorylated by TGF beta it can bind to p300 (Gerritsen et al., 1997). Therefore, a number of translocating signal transducers can be envisioned as triggering signals for the modification or recruitment of the molecular machines during the initiation of transcription. Again, none of these signal transducers, by themselves, can define tissue specificity, but some of them can be associated with certain cell behaviors. Translocation of MAP kinases has been associated with proliferation and differentiation and NF-κB plays a role in protection against apoptosis.
III. Cellular organization regulates gene expression and cell behavior
It is easily apparent, by reading the tremendous amount of literature available, that the response induced by specific extracellular signals depends on the cell type, and that different extracellular signal-triggered pathways share common components at all levels of the signaling cascade. Therefore, part of the answer to tissue-specific gene expression may lie elsewhere, for example, in the organization of the cell type. What defines a particular cell type? In addition to differences in protein composition at the cell surface and throughout the cell, other characteristics include cellular shape, itself determined by the composition and organization of the cytoskeleton, the nature of neighboring cells, and the composition and organization of the extracellular milieu.
Several studies have suggested that the way the cytoskeleton and the nucleus are organized can influence gene expression and cell behavior. Disruption of microtubules organization was shown to disturb insulin-induced cell cycle regulation (Teng et al., 1977). More recently, disassembly of actin filaments was shown to abolish insulin-induced phosphorylation of signal transducers, including MAP kinase, and c-fos expression in muscle cells (Tsakiridis et al., 1998). PMA-mediated NF-κB activation and induction of NF-κB-dependent reporter gene activity was demonstrated to be mediated via microtubules depolymerization in NIH3T3 cells (Spencer et al., 1999). Alteration in cytoskeleton organization has been shown to accompany changes in cell shape and thus is likely to be responsible for the modification of cell behavior observed following modification of cellular form. About two decades ago manipulations of cell shape via alteration of the surface contact had suggested that shape participated in the regulation of RNA translation and DNA synthesis (Folkman and Moscona, 1978; Farmer et al., 1983). Similar manipulations of simply culturing cells on an inert substratum that forces them to round up allowed rabbit synovial fibroblasts to express collagenase (Aggeler et al., 1984) and mammary epithelial cells to express lactoferrin, a milk protein (Roskelley et al., 1994).
What triggers modifications in gene expression after alteration of cytoskeletal organization? Many factors, including transcription factors and other signal transducers are known to be bound to the cytoskeleton. Thus, the reorganization of the cytoskeleton following the receipt of extracellular signals could release these transducers and alter gene expression. The relation between changes in the organization of cytoskeletal components and signal transduction is beginning to be addressed at the molecular level (Kheradmand et al., 1998).
The same scenario is starting to emerge from studies undertaken at the level of the nuclear structure. For example, the disruption of the nuclear distribution of the structural protein NuMA in mammary epithelial cells organized into glandular structures (acini) led to the induction of metalloproteinase activity and the reorganization of the distribution pattern of histone H4 acetylated (Lelièvre et al., 1998). DNA binding activities identical to several transcription factors were found in the nuclear matrix fraction of different cell types, suggesting a role for the nuclear skeleton or nuclear matrix in the regulation of gene expression (van Wijnen et al., 1993).
The formation of higher-order cell assemblies into specific tissues defines a specific context for the regulation of gene expression that is illustrated by the rearrangement of internal cellular organization and the expression of tissue-specific genes. This was shown using three-dimensional (3-D) cultures of mammary epithelial cells that recapitulate the formation of mammary tissue structures (acini). For example, expression of the whey acidic protein, a marker of lactation, was only achieved following the complete formation of mouse mammary acini (Chen and Bissell, 1989). The nuclear protein NuMA was shown to reorganize its distribution from a diffuse pattern in non-acinar human mammary epithelial cells (i.e., cells cultured as monolayers on plastic or before completion of acinar morphogenesis in early 3-D cultures) to a multifocal pattern at the nuclear periphery only after the formation of an organized acinus (Lelièvre et al., 1998). Finally, formation of tissue structure appeared to modify the way signaling pathways interact with each other, as shown by the coupling between the beta-1 integrin and the epidermal growth factor receptor (EGFR) pathways following acini formation, measured by the downregulation of one pathway after the inhibition of the other (Wang et al., 1998). Such coupling was not observed in cells cultured as a monolayer.
IV. What determines tissue-specific gene expression?
Given that unique tissue-specific nuclear components have not yet been found, it has been suggested that the restricted amounts of common nuclear transcriptional regulators may be determinant for the regulation of tissue-specific gene expression at the nuclear level. Notably, it has been proposed that competition for the limited amount of histone acetyl transferase CREB-binding protein (CBP) plays a key role in the modulation of cortical activity in the brain because CBP immunoreactivity was found to be localized to cholecystokinin mRNA-expressing neurons in the hippocampus and the thalamus (Stromberg et al., 1999). Similarly, northern blot analysis of mRNA from various tissues demonstrated that the heavy ferritin gene is expressed at high levels in cells containing a high amount of P/CAF transcriptional regulator, which is known to bind p300/ CBP at gene promoters. Hence, it was argued that the relative amount of P/CAF in different cell types could be responsible for the cell-specific control of the ferritin gene (Bevilacqua et al., 1998). Interestingly, the concept of limiting nuclear regulatory factors can also be applied for the determination of cell behavior in a specific cell type. For instance, the choice between induction of apoptosis or resistance to apoptosis, stimulated by p53 or NF-κB, respectively, has been shown to be determined by the competition of p53 and NF-κB for the binding of limiting amounts of CBP (Webster and Perkins, 1999).
The specificity of action of these general regulators of gene expression may be further determined by the gene's local environment. For example, the CBP protein, referred to above, can interact with many transcription factors and components of the basal transcriptional machinery. Therefore, the effect of CBP may depend on the type of multiprotein complexes that are present around gene promoters. The dynamics of the organization of gene promoters is well illustrated by studies done on the osteocalcin gene promoter, the regulatory elements of which have been shown to vary depending on the cellular microenvironment and phenotype (reviewed by Lian et al., 1998). The emerging concept that the composition of promoter-bound protein complexes varies helps better understand the specificity of the response to extracellular signals. Recently, it was shown that the presence of ski oncogenic protein at smad-dependent gene promoters was responsible for the recruitment of the transcriptional co-repressor N-CoR and the inhibition of TGF-induced, smad-dependent transcriptional activation (Luo et al., 1999). Therefore, the balance in the type and amount of nuclear regulatory components that also participate in nuclear organization may account for the specific response to extracellular signals and ultimately for tissue-specific gene expression.
Another current explanation that may be adjunct to the above is that cell type-specific nuclear co-activators, the molecules that bridge activator complexes and the core of the transcription machinery (holoenzyme), may play a critical role in tissue-specific gene expression. This was proposed almost a decade ago when the study of stimulation of transcription by E2 enhancers revealed that the number of promoters stimulated was cell type specific (Forsberg and Westin, 1991). This concept was further supported by the identification of the co-activator TAFII 105, which appears to be highly expressed at the protein level in B cells compared with other cell types (Dikstein et al., 1996). However, a co-activator that would be found exclusively in one particular cell type remains to be discovered.
Finally, as discussed above, cells within a tissue are characterized by their arrangement, the composition of the membrane receptors, the nature of the cytoskeleton, and the nuclear compartments. Each of these parameters is likely to contribute to the specificity of the response to extracellular signals (Figure 1). Therefore, we propose that the way a cell is organized may create the context that determines how universal and more restricted regulators of gene expression participate in tissue-specific gene expression. Accordingly, it is expected that studies that bridge extracellular signaling and intracellular structure would help elucidate the mechanisms of regulation of gene expression in tissues.
Figure 1.
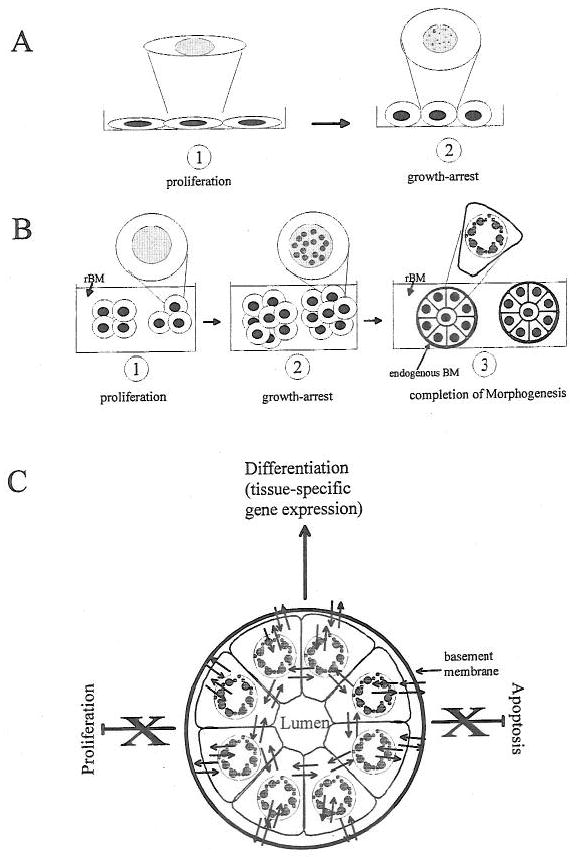
Nuclear organization in context. Taking the example of mammary epithelial cells we describe how nuclear organization and function are influenced by the extracellular milieu and the formation of organized cell assemblies. (A) Proliferation vs. growth-arrest in monolayer cultures. Mammary epithelial cells cultured on plastic surfaces grow as a monolayer (1). The removal of the epidermal growth factor from the culture medium induces growth arrest (2). This manipulation alters both gene expression (by controlling gene products responsible for cell proliferation) and nuclear structure. The latter is exemplified by the redistribution of the nuclear structural protein NuMA from a diffuse pattern to a more punctate pattern (1 and 2; see inserts) and the retinoblastoma protein Rb from a diffuse pattern to a multifocal pattern (not drawn, Lelièvre et al., 1998). The relationship between these alterations in nuclear organization and the modulation of gene expression remains to be demonstrated. (B) The hierarchy of nuclear structural alterations during tissue morphogenesis. The formation of mammary gland epithelial structures (acini) can be recapitulated by culturing mammary epithelial cells in the presence of reconstituted basement membrane (rBM). Isolated cells embedded in rBM proliferate to form small clusters of cells (1). They stop proliferating after a few days (2) and finally become organized into tight and rounded cell assemblies (3). In stage (1) the nuclear structural protein NuMA is diffusely distributed (see insert); in stage (2) it is more punctate (see insert) and it becomes organized into enlarged peripheral foci in stage (3) (see insert). Both the cytoplasmic and nuclear architectures are modified during morphogenesis and signaling pathways interact differently in acini compared with monolayer cultures. Indeed, it appears that the nuclear architecture itself may have signaling information (see text and Lelièvre et al., 1998). (C) The schematic presentation of the interactions within a breast structural and functional unit. We take the example of the formation of an organized mammary acinus (only the epithelial cells are represented here) to illustrate how gene expression can be regulated in tissue-like structure. The continuous bi-directional communication between cytoplasmic and nuclear compartments is influenced by the cellular microenvironment defined by extracellular signals (basement membrane, soluble signals, and cell-cell interactions). The integrity of this multicellular structural and functional unit coordinates incoming signals and maintains the acinar phenotype. If the integrity is destroyed the acinar phenotype is altered and cells may be induced to proliferate or undergo apoptosis. (Reviewed in Lelièvre, S. A., Weaver, V. M., Larabell, C. A., and Bissell, M. J., 1997: extracellular matrix and nuclear matrix interactions may regulate apoptosis and tissue-specific gene expression: a concept whose time has come”. In: Getzenberg, R. H., Ed., “Advances in Molecular and Cell Biology.” Greenwich, CT: JAI Press Inc., pp. 1–55.)
V. Conclusion and perspectives
We can envision gene expression as being regulated in context by the mutual influence of extracellular and intracellular organizations that are typical of a particular cell type. As many cell types share the same signal transducers and transcriptional regulators, it appears that what may play a determining role in tissue-specific gene expression is the way the extracellular signals are integrated by the cellular organization. Thus we propose that rather than single molecules being the “master regulators”, the organization of the cellular structure and machinery together with the balance of regulatory molecules may be playing the primary role in tissue-specific gene expression. What determines such a balance in a specific cell or tissue type remains to be elucidated.
Acknowledgments
We thank Amy Ukena for secretarial assistance. This work was supported by the U.S. Department of Energy, Office of Biological and Environmental Research (contract DE-AC03-76SF00098) and the National Institutes of Health (grants CA-57621 and CA64786) to MJB and by the U.S. Department of Defense/Breast Cancer Research Program to SAL and the University of California/Breast Cancer Research Program to PP.
References
- Aggeler J, Frisch SM, Werb Z. Changes in cell shape correlate with collagenase gene expression in rabbit synovial fibroblasts. J Cell Biol. 1984;98:1662–1671. doi: 10.1083/jcb.98.5.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts AS, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- Berk JA. Activation of RNA polymerase II transcription. Curr Opin Cell Biol. 1999;11:333–335. doi: 10.1016/S0955-0674(99)80045-3. [DOI] [PubMed] [Google Scholar]
- Bevilacqua MA, Faniello MC, Russo T, Cimino F, Costanzo F. P/CAF/p300 complex binds the promoter for the heavy subunit of ferritin and contributes to its tissue-specific expression. Biochem J. 1998;335:521–525. doi: 10.1042/bj3350521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon N, Carlisi D, Datto MB, van Grunsven LA, Watt A, Wang XF, Rudkin BB. Cooperation of Spl and p300 in the induction of the CDK inhibitor p21WAFl/CIP1 during NGF-mediated neuronal differentiation. Oncogene. 1999;18:2872–2882. doi: 10.1038/sj.onc.1202712. [DOI] [PubMed] [Google Scholar]
- Brunet A, Roux D, Lenormand P, Dowd S, Keyse S, Pouyssegur J. Nuclear translocation of p42/p44 milogen-activated protein kinase is required for growth factor-induced gene expression and cell cycle entry. EMBO J. 1999;18:664–674. doi: 10.1093/emboj/18.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LH, Bissell MJ. A novel regulatory mechanism for whey acidic protein gene expression. Cell Regul. 1989;1:45–54. doi: 10.1091/mbc.1.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikstein R, Zhou S, Tjian R. Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- DiPersio CM, Jackson DA, Zaret KS. The extracellular matrix coordinately modulates liver transcription factors and hepatocyte morphology. Mol Cell Biol. 1991;11:4405–4414. doi: 10.1128/mcb.11.9.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR, Wan KM, Ben-Ze'ev A, Penman S. Regulation of actin mRNA levels and translation responds to changes in cell configuration. Mol Cell Biol. 1983;3:182–189. doi: 10.1128/mcb.3.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkman J, Moscona A. Role of cell shape in growth control. Nature. 1978;273:345–349. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- Forsberg M, Westin G. Enhancer activation by a single type of transcription factor shows cell type dependence. EMBO J. 1991;10:2543–2551. doi: 10.1002/j.1460-2075.1991.tb07794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H, Barsony J, Renyi I, Gould DL, Hager GL. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci USA. 1996;93:4845–4850. doi: 10.1073/pnas.93.10.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradmand F, Werner E, Tremble P, Symons M, Werb Z. Role of Racl and oxygen radicals in collagenase-1 expression induced by cell shape change. Science. 1998;280:898–902. doi: 10.1126/science.280.5365.898. [DOI] [PubMed] [Google Scholar]
- Lelièvre SA, Weaver VM, Nickerson JA, Larabell CA, Bhaumik A, Petersen OW, Bissell MJ. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proc Natl Acad Sci USA. 1998;95:14711–14716. doi: 10.1073/pnas.95.25.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard WJ, O'Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- Lian JB, Stein GS, Stein JL, van Wijnen AJ. Osteocalcin gene promoter: unlocking the secrets for regulation of osteoblast growth and differentiation. J Cell Biochem. 1998;31(Suppl):62–72. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<62::AID-JCB10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, Zhou Q. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milocco LH, Haslam JA, Rosen J, Seidel HM. Design of conditionally active STATs: insights into STAT activation and gene regulatory function. Mol Cell Biol. 1999;19:2913–2920. doi: 10.1128/mcb.19.4.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CA, Schmidhauser C, Mellentin-Michelotti J, Fragoso G, Roskelley CD, Casperson G, Mossi R, Pujuguet P, Hager G, Bissell MJ. Characterization of BCE-1, a transcriptional enhancer regulated by prolactin and extracellular matrix and modulated by the state of historic acetylation. Mol Cell Biol. 1998;18:2184–2195. doi: 10.1128/mcb.18.4.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A, Hsu SC, Leung-Hagesteijn C, Radeva G, Papkoff J, Montesano R, Roskelley C, Grosschedl R, Dedhar S. Cell adhesion and the integrin-linked kinase regulate the LEF-1 and beta-catenin signaling pathways. Proc Natl Acad Sci USA. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A. CpG methylation, chromatin structure and gene silencing a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskelley CD, Desprez PY, Bissell MJ. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci USA. 1994;91:12378–12382. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser C, Bissell MJ, Myers CA, Casperson GF. Extracellular matrix and hormones transcriptionally regulate bovine beta-casein 5′ sequences in stably transfected mouse mammary cells. Proc Natl Acad Sci USA. 1990;87:9118–9122. doi: 10.1073/pnas.87.23.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Hu PP, Liberati NT, Datto MB, Frederick JP, Wang XF. TGF-beta-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol Biol Cell. 1998;9:3309–3319. doi: 10.1091/mbc.9.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer W, Kwon H, Crepieux P, Leclerc N, Lin R, Hiscott J. Taxol selectively blocks microtubule dependent NF-κB activation by phorbol ester via inhibition of IκBα phosphorylation and degradation. Oncogene. 1999;18:495–505. doi: 10.1038/sj.onc.1202335. [DOI] [PubMed] [Google Scholar]
- Streuli CH, Edwards GM, Delcommenne M, Whitelaw CB, Burdon TG, Schindler C, Watson CJ. Stat5 as a target for regulation by extracellular matrix. J Biol Chem. 1995;270:21639–21644. doi: 10.1074/jbc.270.37.21639. [DOI] [PubMed] [Google Scholar]
- Stromberg H, Svensson SP, Hermanson O. Distribution of CREB-binding protein immunoreactivity in the adult rat brain. Brain Res. 1999;818:510–514. doi: 10.1016/s0006-8993(98)01219-0. [DOI] [PubMed] [Google Scholar]
- Teng MH, Bartholomew JC, Bissell MJ. Synergism between anti-microtubule agents and growth stimulants in enhancement of cell cycle traverse. Nature. 1977;268:739–741. doi: 10.1038/268739a0. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Tsakiridis T, Bergman A, Somwar R, Taha C, Aktories K, Cruz TF, Klip A, Downey GP. Actin filaments facilitate insulin activation of the src and collagen homologous/mitogen-activated protein kinase pathway leading to DNA synthesis and c-fos expression. J Biol Chem. 1998;273:28322–28331. doi: 10.1074/jbc.273.43.28322. [DOI] [PubMed] [Google Scholar]
- van Wijnen AJ, Bidwell JP, Fey EG, Penman S, Lian JB, Stein JL, Stein GS. Nuclear matrix association of multiple sequence-specific DNA binding activities related to SP-1, ATF, CCAAT, C/EBP, OCT-1, and AP-1. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- Venkataraman L, Wang W, Sen R. Differential regulation of c-Rel translocation in activated B and T cells. J Immunol. 1996;157:1149–1155. [PubMed] [Google Scholar]
- Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between beta 1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster GA, Perkins ND. Transcriptional cross talk between NF-κB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]


