Abstract
The structure and function of each individual mammary epithelial cell (MEC) is largely controlled by a bidirectional interchange of chemical and mechanical signals with the microenvironment. Most of these signals are tissue-specific, since they arise from the three-dimensional (3D) tissue organization and are modulated during mammary gland development, maturation, pregnancy, lactation, and involution. Although the important role played by structural and mechanical signals in mammary cell and tissue function is being increasingly recognized, quantitative biomechanical approaches are still scarce. Here we review currently available biomechanical tools that allow quantitative examination of individual cells, groups of cells or full monolayers in two-dimensional cultures, and cells in 3D cultures. Current technological limitations and challenges are discussed, with special emphasis on their potential applications in MEC biology. We argue that the combination of biomechanical tools with current efforts in mathematical modeling and in cell and molecular biology applied to 3D cultures provides a powerful approach to unravel the complexity of tissue-specific structure-function relationships.
Keywords: microenvironment, mammary epithelial cells, structure-function, cell shape, cell biomechanics, 3D cultures
INTRODUCTION
The mammary gland is a highly organized organ comprised of a branched ductal network of bi-layered epithelium embedded in a complex mesenchymal stroma. Luminal epithelial cells lining the ducts are surrounded by an outer layer of myoepithelial cells that attach to the basement membrane (BM) (1). Full functional differentiation occurs during pregnancy and leads to the formation of acinar structures at the end of the ducts, which produce the milk (2). To achieve and maintain this remarkable level of tissue organization, mammary epithelial cells (MECs) and their surrounding extracellular matrix (ECM) must integrate their structure in a highly coordinated fashion. We hypothesized more than 20 years ago (3) that MECs accomplish this task by dynamically coordinating physical and biochemical signals from their microenvironment (defined by neighboring cells, surrounding ECM, and local soluble factors) (4). Some of the major elements involved in this bidirectional communication have been identified (1,2,5,6). However, the mechanisms by which MECs integrate biochemical and physical signals to perform tissue-specific functions remain largely unknown.
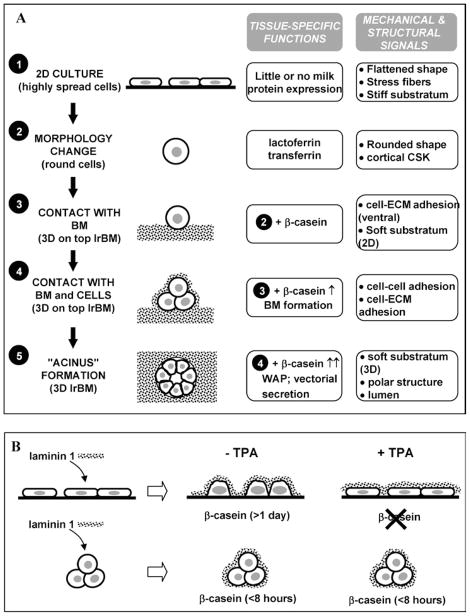
Our laboratory has developed experimental models based on the culture of MECs in designer microenvironments to dissect the chemical and physical signals that regulate cell fate during mammary gland development (4,7). This approach has allowed the identification of a hierarchical set of signals in which each additional level of functional differentiation (e.g., expression of milk protein genes) could be directly related to an additional level of change in cellular and tissue structure (Fig. 1(A)) (8). MECs cultured on standard two-dimensional (2D) tissue culture plastic exhibited a flattened morphology and failed to express milk proteins in the presence of lactogenic hormones (when laminin or BM is not present). Prerounding the cells by culturing them on nonadhesive substrata was sufficient to induce the expression of the milk protein lactoferrin (9). Unlike lactoferrin, additional signaling by laminin 1 was required to trigger the expression of β-casein in addition to the shape change. β-casein expression was inhibited when ECM-mediated cell rounding was prevented by the phorbol ester TPA (Fig. 1(B)) (9). Interestingly, β-casein expression could be induced in single MECs even in the absence of cell–cell interaction and morphological polarity as long as laminin 1 was present. However, expression of whey acidic protein (WAP) was only achieved when MECs were cultured in a laminin-rich BM (lrBM), in which they form three-dimensional (3D) acinus-like structures that resemble those seen in vivo. Beyond milk protein genes (8), we have identified other sets of genes that are differentially expressed at each tier of organization (7,10).
Fig. 1.
Studies performed with MECs cultured in designer microenvironments have allowed the identification of different mechanical and structural signals involved in functional differentiation. (A) Hierarchy of structure-function relationships in MECs in culture. Each additional level of tissue-specific function (e.g., expression of milk proteins) correlates with the acquisition of additional mechanical and structural features (7,8,10). Different aspects of these structural features can be probed and manipulated in a quantitative fashion with currently available biomechanical tools. (B) Cell shape plays a central role in modulating functional differentiation in MECs. Cells cultured on a plastic dish (2D) undergo a slow morphological change (become rounded) and start expressing β-casein >24 h after treatment with laminin 1 and lactogenic hormones. In contrast, prerounded cells express abundant β-casein much more rapidly (in less than 8 h) (B, central column). Inhibition of laminin-induced cell rounding with TPA also inhibits β-casein expression. However, TPA alters neither shape nor β-casein expression in prerounded cells (B, right column adapted from Roskelley et al. (9)).
Studies in vivo and in cell culture have provided further evidence of the role of mechanical and structural signals in regulating MEC function. In studies performed in the 1970s and 1980s, different groups modified the mechanical stresses generated in the ECM by culturing primary MECs in floating or attached collagen gels. These studies reported that expression of milk proteins was dramatically enhanced in floating gels (11). Lee and coworkers (12) proved that this increase was due to de novo synthesis. Using a similar approach, Wozniak and coworkers recently found that tubulogenesis of MECs is induced in floating but not in attached gels (13). In other mechanical studies, Pitelka and Taggart showed that mechanical forces generated in MEC sheets regulate the orientation of tight junctional components (14). Furuya and coworkers reported that distension of MECs markedly affects intracellular Ca2+ concentration and ATP release (15). A set of structurally-oriented studies revealed that different biochemical signaling pathways are coordinated in MECs in 3D but not in 2D cultures (10). Wang and coworkers showed that β1 integrin and epidermal growth factor receptor (EGFR) cross-talk in 3D but not in 2D (16). In a recent work, Weaver and coworkers found that both normal and malignant MECs acquire an apoptosis-resistant phenotype when they recapitulate a 3D-polarized architecture, in a mechanism involving β4 integrin–laminin interaction and NFκB activation (17). In vivo, mechanical signals associated with suckling are thought to be important for the control of lactogenesis (onset of milk secretion), galactopoiesis (maintenance of lactation), and mammary gland growth (5). The rate of milk secretion in particular has been related to changes in intramammary pressure that directly depend on milk accumulation in the gland (5). Additionally, physical deformation of milk-burdened acini could trigger the apoptotic response that initiates mammary gland involution (e.g., regression of the milk producing epithelial compartments) after weaning (18). While the above studies exemplify the important role that structural and, by inference, mechanical signals play in mammary cell and tissue function, physical signals have received little attention in general and quantitative biomechanical approaches are still scarce.
Cellular phenotype is commonly characterized by examining cellular morphology, the distribution of certain markers, and the level of expression of tissue-specific genes or genes involved in cellular organization. These approaches only provide qualitative or semiquantitative information, and do not measure the physical signals involved in the regulation of tissue-specific gene expression. This gap in the study of the relationships between form and function can be reduced by applying quantitative biomechanical and bioengineering techniques. The purpose of this review is to introduce currently available tools to probe and manipulate mechanical properties of adherent cells in culture, with special emphasis on their potential for mammary gland biology. Techniques are introduced according to their suitability to probe single cells, groups of cells or full monolayers in 2D cultures, and cells in 3D gels. For each technique, we provide a description of the basic operating principle, its capabilities, and limitations for cellular studies in general and reported applications in mammary gland biology in particular.
BIOMECHANICAL TOOLS FOR PROBING AND MANIPULATING SINGLE CELLS
How do MECs stabilize their shape within the tissue? To what extent does basal contact with BM and basolateral interactions with other cells reorganize the nucleus and the cytoplasm of MECs? What are the adhesion forces that MECs establish with neighboring cells and the surrounding matrix during mammary gland development? How do MECs modulate membrane elasticity during secretion of milk proteins? How do myoepithelial cells generate and transmit contractile forces to facilitate milk expulsion from acinar structures? These are just a few examples of fundamental questions that can now be addressed by an array of tools that have emerged in the last decade capable of measuring and applying forces (F) and deformations (d) to cells and biomolecules (19). The more widely used tools are listed in Table I along with their different cellular applications. These tools have dramatically expanded our knowledge of different cellular biophysical properties including mechanical properties of cell membranes, cortical cytoskeletons, the cytoplasm and the nucleus, as well as adhesion forces to other cells and to ECM molecules. However, very few studies on MECs have been reported. Most, if not all, of these mechanical properties are likely to depend on the cell type and its differentiated state, and therefore are useful to characterize cellular phenotype in a quantitative fashion. These techniques employ different physical principles to measure and/or apply F and d in living cells; in some cases, F and d can be transformed into stress (force per unit area) and strain (deformation per unit length), respectively. Stress-strain relationships allow calculation of absolute mechanical parameters (20). Each methodology is also unique in its resolution, the type of stress applied (tension/compression, shear), the distribution of stress (local or global), and the part of the cell being examined. For simplicity, each technique is introduced according to its physical operating principle (see schematic illustration in Fig. 2).
Table I.
List of Biomechanical Techniques for Different Cell Applications
| Application | Cell region | Techniquea | |
|---|---|---|---|
| Single cells | Cell deformability | Cell membrane (dorsal); cortical cytoskeleton | MN, AFM, MBMc, OT |
| cytoplasm | PTC, MBM | ||
| nucleus | TPC | ||
| Whole cell body | MN,b OTb | ||
| Cell-ECM adhesion | Dorsal | MN, AFM, OT, MBM | |
| Ventral | AFM,b MN,b SMC,c TPSc | ||
| Cell-cell adhesion | AFM, MNb | ||
| Clusters and Monolayers | Cell deformability | Dorsal | MTC, shear stress, compression |
| Ventral | Elastomeric membranes (tension) | ||
| Cell-ECM adhesion | Dorsal | MTC | |
| Ventral | electrical impedance | ||
| Cell shape | Whole cell body | polyHEMA, Micropatterning | |
| 3D cultures | Cell deformability | Whole cell body | Floating gels, stretched gels |
Technique abbreviations: AFM, atomic force microscopy; MBM, magnetic bead manipulation; MN, microneedles; MTC, magnetic twisting cytometry; OT, optical tweezers; SMC, substrates with micropatterned cantilevers; TPC, tracking of particles inside cells; TPS, tracking of particles inside the substratum.
Modified from standard setup.
Allow simultaneous detection of several cells.
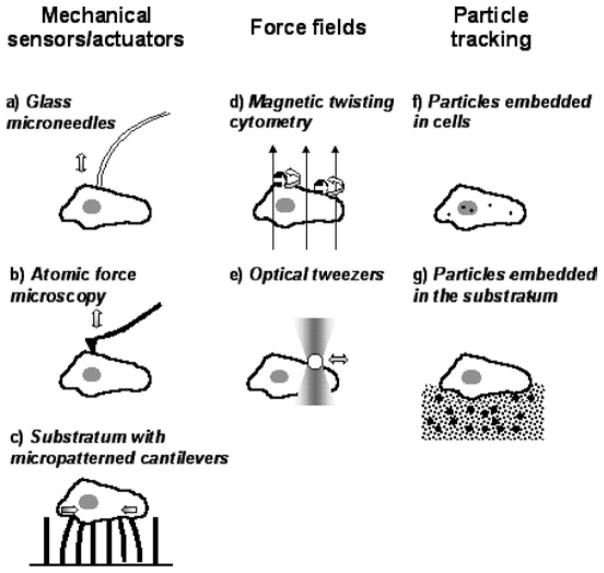
Fig. 2.
lllustration of the operating principle of currently available tools to probe and manipulate single cells based on mechanical sensors/actuators (a–c), on force fields (d,e) and on tracking the motion of embedded particles (f,g) (detailed description is provided in the text).
Techniques Based on Mechanical Sensors/Actuators
These techniques investigate cells by directly probing the cellular surface with different types of cantilevers or bendable beams that can be thought of as “fingers” or “hands” acting at molecular and cellular scales. These tools take advantage of the fact that, for small deformations, a cantilever acts like a spring and, as such, the bending force is easily calculated by measuring its deformation x and spring constant k, which are related by Hooke’s law: F = kx.
Glass Microneedles (MN) and Related Techniques
A bendable glass microneedle attached to a micromanipulator is brought into contact with the dorsal cellular surface and moved vertically or horizontally to apply either compression/tension (pushing/pulling) or shear (dragging) forces (Fig. 2(a)). k of the needle is usually assessed by bending a second calibrated needle (21). Bending of the needle is monitored by optical imaging and used to calculate F and d applied to the cell. At least two different instruments based on flexible glass beams have been reported to apply local (22) and global (23) compression to cells. Common applications of standard MN include measurement of the apparent cellular stiffness and shear modulus. Current limitations are the lack of standardized equipment and the semiquantitative mechanical information obtained. Furuya and coworkers used MN to show that intracellular Ca2+ increases in MECs after local mechanical deformation and that this local effect spreads rapidly to neighboring cells, a process that may contribute to the coordinated control of casein phosphorylation and secretion in the mammary gland (15).
Atomic Force Microscopy (AFM)
A micrometer-sized pyramidal or spherical tip located at the free end of a flexible silicon-based microcantilever is brought into contact with the dorsal cell surface using a piezoactuator to apply compression, tension, or shear forces (Fig. 2(b)). The bending of the cantilever is measured by focusing a laser beam on the back of the cantilever and collecting the light reflected off the surface onto a 4-segment photodetector. F is obtained by recording the imbalance between the light detected within the four segments. Cellular deformation is assessed by subtracting the cantilever bending from the vertical displacement of the piezoactuator. Currently available contact viscoelastic models allow the assessment of the cell-tip contact geometry and accurate calculation of absolute values of elastic and shear moduli (24) (Fig. 3(A)). A map of viscoelastic parameters over the whole cell can be obtained by recording force-deformation data at different locations. AFM can also be used to image cellular topography and major variations in cellular stiffness (25). Forces involved in cell–cell and cell–matrix adhesion have been measured by using tips coated with cells or specific ligands (26). The main advantages of AFM are its versatility and commercial availability. However, there is a need for a standard method for calibrating k of the cantilever and for a more realistic viscoelastic contact models. Our laboratory is currently using this technique to determine how MECs modulate their mechanical properties during ECM-induced functional differentiation.
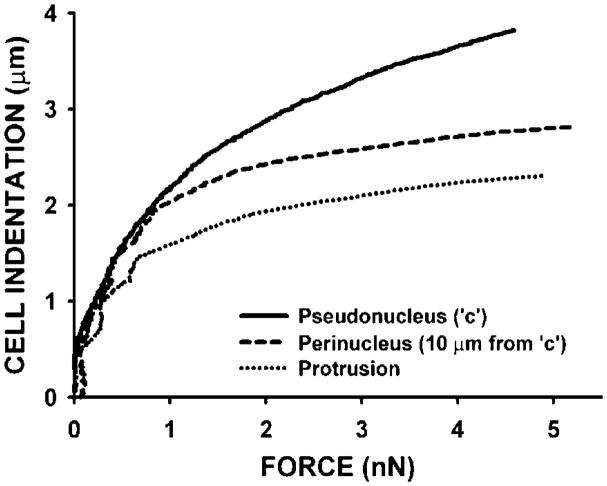
Fig. 3.
Example of mechanical manipulations of single MEC in 2D cultures (unpublished data). Illustration of the different mechanical response (indentation or cell deformation) of the center (pseudonucleus), the perinuclear region, and a protrusion of the same single MEC (Scp2 cell line) subjected to similar range of compressive forces with an AFM tip. The estimated tip-cell surface of contact is in the range 1–10 μm2.
Substrates With Micropatterned Cantilevers (SMC)
Cells are cultured on a microfabricated array of flexible elastomeric cantilever posts of known geometry and k. As cells adhere and spread on top of the posts, they exert traction forces that deflect the cantilevers in different directions (Fig. 2(c)). Bending of individual cantilevers is assessed by optical microscopy and used to calculate a 2D map of traction forces on the substrate (27). Unlike other traction techniques (see below), SMC provides a straightforward measurement of the traction forces. A different design using horizontal cantilever beams has been used to monitor traction forces during cellular locomotion (28). A major limitation of these approaches is that they require access to microfabrication facilities, since molds are not commercially available.
Techniques Based on Force Fields
These techniques use external force fields (electric, magnetic, or photonic) to manipulate cells by acting either on the cells themselves or on micrometer-sized beads that are attached to the cellular surface or internalized in the cytoplasm. In most setups these tools act like “tweezers” capable of manipulating specific parts of the cell.
Magnetic Bead Manipulation (MBM)
There are different approaches that probe the mechanical properties of individual cells by manipulating magnetic beads with magnetic fields (29,30). In a setup known as Magnetic Twisting Cytometry (MTC), micrometer-sized ferromagnetic beads are coated with ligands to allow their binding to the cell surface (30). Each bead behaves like a small magnet and therefore their magnetic dipoles can be aligned in the same direction by applying a brief and strong magnetic field parallel to the substrate. When a second, weaker magnetic field is applied perpendicular to the substrate, the beads will rotate to orient their magnetic dipoles parallel to the external field (Fig. 2(d)), just as a compass rotates to become parallel to the earth’s magnetic field. As the bead rotates, a torque is applied at the site of cell-bead attachment. The torque is calculated from the theoretical applied magnetic field and the calibrated average magnetic moment of the beads (31). In the standard MTC method, bead rotation is indirectly measured by monitoring the decrease in the magnetic field created by the beads in the direction of initial bead magnetization using an in-line magnetometer (30). Under these conditions, information about the rotation of individual beads is lost and therefore is not useful for single-cell studies. In a recent modification for single-cell applications, the lateral displacement of the beads is optically detected and quantified using an algorithm that tracks the center of mass (centroid) of the bead (31). This optical-MTC has been used to probe the changes in force transmission of cells subjected to pharmacological treatments and cells probed with different ligand-coated beads (31,32). Current challenges include theoretical analysis of the torque-induced forces and of the contact geometry.
Optical Tweezers (OT)
A ligand-coated micrometer-sized polystyrene or silica bead with high refractive index is trapped at the focal point of a laser beam focused using a high numerical aperture objective. Under these conditions, the density of photons in the focused beam develops a radiation pressure strong enough to trap the bead with a spring-like force (33). The trapped bead is brought into contact with the cell surface and held to allow its attachment. Moving the laser beam parallel to the substrate applies a shear force at the site of cell-bead attachment (Fig. 2(e)). Bead movement is usually measured by tracking the bead’s centroid or by collecting the refraction of the beam on a photodetector. The effective k of the laser trap is calibrated by flowing solution past a trapped bead at a known velocity and calculating the force from Stoke’s law. OT has been mainly used to study ligand-receptor adhesion forces (34,35) and the mechanical properties of cell membranes (36). Assessment of absolute mechanical values is limited by the difficulty of determining bead-cell contact geometry.
Techniques Based on Tracking the Motion of Embedded Particles
This group of techniques has been developed most recently and is based on tracking the motion of nanometer- or micrometer-sized particles embedded either inside the cell or in the substratum.
Tracking of Particles Inside Cells (TPC)
These particles can either be endogenous (lipid granules or mitochondria) (37,38) or synthetic sub-micrometer beads internalized through phagocytosis (29) or microinjected into the cytoplasm or the nucleus (39). The Brownian motion of several beads is tracked by analyzing videomicroscopy images or by detecting the refraction of a low-intensity laser focused on the particle (37); these data are used to calculate the mean-squared displacement (MSD) of the bead’s centroid, which is directly related to the viscoelastic properties of the media surrounding the bead (38). By tracking multiple particles inside a cell, it is possible to probe the mechanical heterogeneity of the cytoplasm and the nucleus (Fig. 2(f)) (39). Since this technique does not require force calibration, investigators can directly compare data obtained in different cell types as well as in cellular extracts. However, care must be taken that the injected particles do not perturb normal cellular functions and that particles that adhere to cellular structures are discarded (40).
Tracking of Particles Embedded Inside the Substratum (TPS)
Traction forces exerted on the substratum were first visualized as wrinkles when individual cells were cultured on distortable sheets of silicone rubber (41). More quantitative approaches have recently been developed, in which cells are plated on flexible polymeric substrates (silicone elastomers or polyacrylamide gels) containing several embedded beads (42). The surface of the polymer is usually coated with a thin layer of ECM to facilitate cellular attachment. As cells attach and spread, they pull the substrate at different points and thereby change the positions of the beads (Fig. 2(g)). A deformation map can be obtained by measuring the displacement of several beads using optical imaging. Computational analysis of the displacements in terms of elastic models provides a map of the traction forces developed in the substrate and at the cell-substrate interface (43). These so-called traction microscopy techniques have been used to study ventral cellular adhesion and migration (42). A major challenge in TPS is to develop more realistic theoretical models that overcome the assumption that the substrate has pure elastic behavior and that the layer of ECM does not affect the transmission of traction forces. Rabinovitz and coworkers (44) used this approach to study the forces that breast carcinoma cells exert on BM gels specifically through α6β4 integrins. They found that these integrins can transmit forces to laminin without engaging other integrins and suggested that these traction forces are important in BM remodeling.
BIOMECHANICAL TOOLS FOR PROBING AND MANIPULATING GROUPS OF CELLS ON FLAT SUBSTRATA (2D)
Several tools have been developed to probe and manipulate groups of cells or full monolayers in 2D cultures. Unlike those for single cells, many of the techniques are commercially available and usually do not require very sophisticated instrumentation, as larger forces are needed to deform cells in bulk. By studying large numbers of cells simultaneously, these techniques can easily be combined with standard bulk biological assays to measure changes at protein and mRNA levels. We present each technique based on its application (schematized in Fig. 4), highlighting existing and potential applications in the mammary gland.
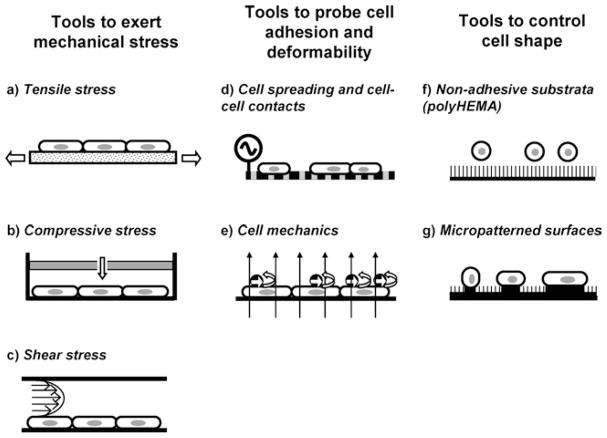
Fig. 4.
Schematic description of the operating principle of tools for probing and manipulating groups of cells in 2D cultures. Techniques that directly exert forces on cell populations are specifically designed to apply tension (a), compression (b) or shear (c) stresses. Cell–cell and cell-substrate adhesion can be assessed by monitoring the changes in electrical impedance as cells spread across a substrate coated with microelectrodes and subjected to an alternate current (d). Cell deformability can be probed in bulk by applying local torques through magnetic beads bound to the cell surface subjected to external magnetic fields (e). Cell shape can be controlled qualitatively by culturing cells in substrates coated with a non-adhesive substance (f) and quantitatively by using micropatterned surfaces (g).
Tools Used to Exert Forces on Cells
Cells in the mammary gland are subjected to a dynamic mechanical environment. During pregnancy and lactation, MECs experience changes in tension/stretch from suckling, compression due to engorgement from the increased fluid in the ductal network, and shear from the flow of milk through the gland. Although each of these stresses (tension/compression and shear) can be simulated in culture, the effects of these physical signals on MECs are largely unknown. Moreover, none of these stresses have been properly quantified in vivo. The following tools apply stresses to populations of cells using direct mechanical approaches. Cellular response is monitored by traditional biochemical and microscopy methods either during or after the application of the stress. Thus, these tools are very useful to study systematically the effect of different stresses on tissue-specific function. These 2D techniques can be classified according to the type of stress (tension/compression or shear) applied.
Several designs have been developed to apply stretch or strain to groups of cells, some of which are commercially available. Most require culturing cells on a flexible elastomeric membrane and subsequently distorting that membrane using vacuum or mechanical actuators to pull the substratum in one (uniaxial strain) or two (biaxial strain) directions (Fig. 4(a)) (45). Controlling the duration and rate of the membrane’s distortion allows the investigator to apply both steady and oscillating stretch to the cells. Although the stretch applied to the cells is a function of the strain applied to the elastomeric membrane, they may differ dramatically (32). In addition, the membrane deformations achieved, and therefore the strains applied to the cells, may be nonuniform across the substrate (45). Our laboratory is currently using a vacuum-operated stretching device to study the effect of exogenous forces in regulating tissue-specific gene transcription, expression, and secretion.
To apply a compressive stress, cells are usually grown on membrane-based cell culture inserts, which separate apical and basolateral compartments, and subsequently subjected to a hydrostatic or trans-membrane pressure change in the upper (apical) portion of the chamber (46) (Fig. 4(b)). The method produces a uniform stress across the monolayer, and requires no special equipment. To apply a shear stress, monolayers of cells are typically grown on slides that are subsequently used as the bottom plane of a parallel-plate flow chamber (Fig. 4(c)). This simple geometry allows control of the shear stress on the surface of the cells by defining the laminar flow rate through the chamber and the distance between the top and bottom planes. The flow of fluid through the chamber can be controlled either with pumps (47) or gravity (48). To date, the study of the effects of shear stress has been limited primarily to endothelial cells, since this cell type is subjected to shear due to blood flow. These studies have reported that shear stresses affect signaling through Erk, Rho GTPases, and reactive oxygen species (49).
Tools to Measure Cellular Adhesion and Mechanics
A popular method to monitor changes in cellular adhesion is based on plating cells on substrates coated with microelectrodes, applying a small alternating current, and measuring changes in transcellular electrical resistance or impedance. Since plasma membranes constrain the current, increased cell-ECM adhesion will reduce the current measured, thereby increasing the electrical impedance (Fig. 4(d)). Importantly, increasing cell–cell adhesion will also reduce the current that passes through the cellular monolayer; to circumvent this complication, algorithms have been developed to calculate contributions from cell-ECM and cell–cell adhesion (50). The microelectrode substrates are commercially available and have been used to monitor temporally the adhesion of different cell types and cells subjected to different perturbations. This approach does not provide absolute values, since its measurements are relative to a baseline level. Mechanical properties of a population of cells can be probed by magnetic twisting cytometry (MTC; Fig. 4(e)) (30). Coating the magnetic beads with ligands specific for different cell-surface receptors has allowed the probing of the efficiency of different receptors in transmitting forces to the cytoplasm. MTC is subject to the same limitations as MBM, in addition to those derived from the magnetic detection approach. The combination of biochemical bulk measurements with the ability of MTC and similar magnetic bead-based devices to apply mechanical stresses to specific cell surface receptors represents a valuable approach to identify proteins and signaling molecules that are directly regulated by mechanical signals. The p38 mitogen-activated protein kinase and the transcription factor Sp1 were found to be modulated by external forces in fibroblasts using this experimental approach (51). The measurement of the average spreading, adhesion, and mechanical properties of a cell population provides quantitative approaches to characterize a given cellular phenotype. These quantitative approaches may allow distinguishing normal and malignant MECs in terms of their biophysical properties, as well as testing the effect of different treatments such as agents that revert the malignant phenotype.
Tools to Control Cell Shape
Cell shape plays a central role in the regulation of functional differentiation of MECs. As mentioned above, flattened MECs fail to express any milk proteins in the presence of lactogenic hormones, while prerounded cells express lactoferrin and transferrin. Moreover, transcription of β-casein is triggered much more rapidly in prerounded than in flattened cells upon exposure to laminin (Fig. 1(B)) (8). Cell shape has similarly been shown to regulate cellular uptake, differentiation, proliferation, and cell death of several cell types (17,52–55). All of the methods that have been developed to date to control cell shape operate by controlling the extent of adhesion between cells and the underlying substratum. Folkman and Moscona pioneered the use of poly (2-hydroxyethyl methacrylate) (polyHEMA), a chemically inert material, over 25 years ago (53). Tissue culture plastic coated with a layer of polyHEMA is nonadhesive to proteins and cells (Fig. 4(f)); decreasing the density of polyHEMA adsorbed on the surface increases the amount of ECM proteins that can subsequently adsorb from solution, and thereby increases the extent of spreading of cells. However, polyHEMA neither controls the shape of individual cells per se nor the amount of cell–cell contacts, thereby introducing undesired variability. Greater control over the shape of individual cells can be obtained using more sophisticated methods based on one of the many micropatterning techniques developed in the past 10 years (56). Micropatterning uses technologies from the semiconductor industry to control the location, geometry, and size of adhesive and nonadhesive moieties on the underlying cell-culture surface with micrometer precision. Cells seeded on these surfaces can only attach and spread on the adhesive islands and not on the surrounding nonadhesive regions; by controlling the geometry of the islands, investigators can indirectly control the shape of cells on the surface (Fig. 4(g)). A primary advantage of micropatterning is its ability to specify a priori the shape of the cells. However, it is limited by the fact that substrates are not commercially available, and therefore access to microfabrication facilities is required. We anticipate that the application of micropatterning techniques to control cell shape may help in dissecting how cell shape participates in the regulation of tissue-specific gene expression in MECs.
BIOMECHANICAL TOOLS FOR PROBING AND MANIPULATING CELLS IN 3D CULTURES
Although it is now well recognized that cellular behavior in 3D culture is a closer reflection of natural physiology than that in 2D culture (57), there are surprisingly few techniques to investigate and manipulate cells under 3D conditions. Likewise, little is known about the mechanical properties of ECM gels themselves. A better knowledge of the forces developed within ECM gels may help elucidate the role that physical properties of different ECM components play in MEC function in vivo. In particular, it has been suggested that differences in the polymerization properties of laminin 1, laminin 5, and laminin 10/11 may account for the failure of the latter two to promote the acquisition of functional polarity of MECs in 3D cultures (6). The relative tension cells experience in 3D cultures can be manipulated by altering the attachment of the matrix to the walls of the culture vessel. Attached and floating gels are expected to have the same composition, but different local density and internal stresses. Although it is clear that the internal forces in attached (restrained) gels will be dramatically different from those in floating (unrestrained) gels, the differences in mechanical stresses developed within the matrix have yet to be quantitatively and theoretically analyzed. Nevertheless, it is presumed that floating gels overall are more compliant than attached gels (58) and therefore cells cultured in floating gels experience less resistance. As already mentioned, MEC function and morphogenesis in 3D collagen are dramatically altered, depending on whether or not the gel is restrained (11–13). In a different and more recent approach, tension has been directly exerted on cells in 3D gels by applying a strain to the gel in a modification of a 2D stretching technique (59). The forces exerted within collagen gels have been investigated by measuring the ability of the cells to contract a slab of gel over time (22). Overall, none of these methods provides a direct quantitative measurement or manipulation of the mechanical properties of cells in 3D cultures. Moreover, it is important to note that it is very difficult to modify the mechanical properties of a material without altering its chemical properties, and cells can undoubtedly sense differences in the chemistry of the materials in which they are embedded.
CONCLUSIONS AND FUTURE DIRECTIONS
The use of biomechanical tools has dramatically expanded our knowledge of the mechanical properties of different cell structures as well as the effect of mechanical signals on gene expression in different cell types (26,35,39,40,60). In particular, the expression, activity, and localization of several molecules, including transcription factors, cytoskeletal proteins, growth factors, ECM molecules, matrix metalloproteinases, and reactive-oxygen species, have been identified as being dramatically modulated by physical signals in MECs (8,12,61) (Radisky et al., submitted for publication) and other cell types (46,62,63). Our knowledge of structure-function relationships in mammary epithelia in particular can further benefit from biomechanical approaches in at least three different ways. First, these tools allow a systematic study of the effect of mechanical forces on the regulation of transcription, translation, and secretion of tissue-specific genes or any gene of interest. Second, it is possible to determine quantitatively different biophysical properties that may define cell phenotype, such as membrane elasticity (involved in secretory activity) (64), cell viscoelasticity (related to cell shape and plasticity) or cell-ECM adhesion (60). Finally, physical signals involved in the bidirectional interaction between epithelial and stromal cells or luminal and myoepithelial cells can be probed and manipulated.
There are several intrinsic features of living cells that distinguish them from inert matter and that need to be considered to facilitate the biological interpretation of biomechanical data. Unlike most engineered materials, cells are dynamic. Thus, they continuously undergo internal rearrangements to perform their different tasks and to respond to mechanochemical signals from the microenvironment (3,10,55). Furthermore cells may sense and respond differently to different physical stimuli, and these responses will probably depend on cell type as well as its differentiated state. In addition, whereas structural changes triggered by the probe itself are usually ignored, they can be quite important, particularly in techniques that use ligand-coated beads (65). Therefore, a meaningful interpretation of biomechanical data requires their combination with biochemical “read-outs” to identify the functional differentiated state of the cell being investigated. Otherwise, it becomes very difficult to dissect what is a general cellular property and what is cell type- or function-specific. Likewise, biochemical data become much more meaningful when the inter- or extra-cellular physical signals involved are probed as well. Since cell function is controlled by context through integration of both biochemical and biophysical signals, we believe it is crucial to take a more integrative approach in mammary gland biology in which biomechanical and bioengineering tools are combined with biochemical techniques.
Current biomechanical approaches are still subject to several technological and theoretical limitations that offer challenges for the future. Most of these tools are not commercially available and require some degree of expertise in the fields of physics, engineering, and mathematics. Moreover, although cells develop stresses and strains in three dimensions, currently available techniques only measure forces and deformations in one or two dimensions. In addition, most techniques rely on simplified theoretical models, usually based on unrealistic idealizations of cellular behavior to extract quantitative data. Perhaps the most urgent technological need is to develop tools to interrogate cells embedded in 3D cultures. Currently existing methodologies based on external force fields have the potential to fill this technological gap (66). Above and beyond technological improvements, there is also an urgent need to develop conceptual frameworks and models to aid the integrative analysis of genomic, proteomic, and mechanochemical data. Although the discussion of possible conceptual frameworks is beyond the scope of this review, we briefly highlight two groups of data that could provide some clues. The first set of data providing a clue is based on the fact that connections occur among ECM, cell-surface receptors, CSK, and nuclear skeleton (3,67,68). These connected structures form an architectural scaffold (commonly referred to as tissue matrix) capable of transmitting physical signals outside-in (67) and inside-out (34) in a dynamic and reciprocal fashion (3). The existence of this scaffold provides a mechanism of integrating physical signals with cellular function by modulating the different signal transduction pathways overlain within it, although other mechanisms for cellular mechanotransduction are also possible (69). Another set of data provides a clue based on the findings that different signaling pathways, including those downstream of β1 integrin and EGFR (10,70), laminin and prolactin receptors (71), or β4 integrin–laminin interaction, and NFκB activation (17) are properly coordinated in 3D but not 2D cultures. This structure-dependent functional coordination of signaling pathways suggests that the 3D environment provides the correct spatial organization and compartmentation to enhance and/or inhibit the formation of signaling modules or nodes necessary to achieve a given cellular function. From this perspective, it was recently suggested that aspects of this integration appear to mimic features of the nodal-based organization used in different complex networks (10).
We expect that our current understanding of the intimate structure-function relationship that occurs in the mammary gland will benefit enormously from the combination of biomechanical technologies with biochemical, bioengineering, and computational methodologies. In particular, it is anticipated that such multidisciplinary research will allow more and more aspects of mammary gland biology to be expressed in mathematical terms. Undoubtedly, current efforts to develop techniques to study cells cultured in conditions that recapitulate behavior in vivo will facilitate this very challenging yet fascinating endeavor, as is already happening in other tissues and for single molecules (60).
Acknowledgments
We apologize to all those investigators whose papers could not be quoted due to space limitations. We thank Carlos Bustamante for helpful guidance and support (to J.A.). We also thank Jamie Bascom and other members of the Bissell Laboratory for helpful suggestions. This work was supported by grants from the U.S. Department of Energy, Office of Environmental and Biological Research (DE-AC0376 SF00098), from the National Cancer Institute of the National Institutes of Health (CA64786-02 and CA57621), and from an Innovator Award from the U.S. Department of Defense Breast Cancer Research Program (DAMD17-02-1-0438) (to M.J.B.); by postdoctoral fellowships from the Ministry of Universities, Research and Information Society – Generalitat of Catalonia, and a grant from the Lawrence Berkeley National Laboratory LDRD Program (to J.A.); and by a postdoctoral fellowship from the Department of Defense Breast Cancer Research Program (W81XWH-04-1-0582) (to C.M.N.).
Abbreviations
- AFM
atomic force microscopy
- BM
basement membrane
- CSK
cytoskeleton
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- lrBM
laminin-rich basement membrane
- MBM
magnetic bead manipulation
- MEC
mammary epithelial cell
- MN
microneedles
- MTC
magnetic twisting cytometry
- OT
optical tweezers
- SMC
substrates with micropatterned cantilevers
- TPA
12-O-tetradecanoyl phorbol 13-acetate
- TPC
tracking of particles inside cells
- TPS
tracking of particles inside the substratum
- WAP
whey acidic protein
- 2D
two-dimensional
- 3D
three-dimensional
- d
cell deformation
- F
force
- k
spring constant
References
- 1.Hagios C, Lochter A, Bissell MJ. Tissue architecture: The ultimate regulator of epithelial function? Philos Trans R Soc Lond B Biol Sci. 1998;353:857–70. doi: 10.1098/rstb.1998.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neville MC, McFadden TB, Forsyth I. Hormonal regulation of mammary differentiation and milk secretion. J Mammary Gland Biol Neoplasia. 2002;7:49–66. doi: 10.1023/a:1015770423167. [DOI] [PubMed] [Google Scholar]
- 3.Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 4.Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci. 2003;116:2377–88. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight CH, Peaker M, Wilde CJ. Local control of mammary development and function. Rev Reprod. 1998;3:104–12. doi: 10.1530/ror.0.0030104. [DOI] [PubMed] [Google Scholar]
- 6.Bissell MJ, Bilder D. Polarity determination in breast tissue: Desmosomal adhesion, myoepithelial cells, and laminin 1. Breast Cancer Res. 2003;5:117–9. doi: 10.1186/bcr579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bissell MJ, Weaver VM, Lelievre SA, Wang F, Petersen OW, Schmeichel KL. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59:1757s–63s. discussion 63s–64s. [PubMed] [Google Scholar]
- 8.Roskelley CD, Srebrow A, Bissell MJ. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr Opin Cell Biol. 1995;7:736–47. doi: 10.1016/0955-0674(95)80117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roskelley CD, Desprez PY, Bissell MJ. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci USA. 1994;91:12378–82. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bissell MJ, Rizki A, Mian IS. Tissue architecture: The ultimate regulator of breast epithelial function. Curr Opin Cell Biol. 2003;15:753–62. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerman JT, Pitelka DR. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro. 1977;13:316–28. doi: 10.1007/BF02616178. [DOI] [PubMed] [Google Scholar]
- 12.Lee EY, Lee WH, Kaetzel CS, Parry G, Bissell MJ. Interaction of mouse mammary epithelial cells with collagen substrata: Regulation of casein gene expression and secretion. Proc Natl Acad Sci USA. 1985;82:1419–23. doi: 10.1073/pnas.82.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583–95. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitelka DR, Taggart BN. Mechanical tension induces lateral movement of intramembrane components of the tight junction: Studies on mouse mammary cells in culture. J Cell Biol. 1983;96:606–12. doi: 10.1083/jcb.96.3.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuya K, Enomoto K, Yamagishi S. Spontaneous calcium oscillations and mechanically and chemically induced calcium responses in mammary epithelial cells. Pflugers Arch. 1993;422:295–304. doi: 10.1007/BF00374284. [DOI] [PubMed] [Google Scholar]
- 16.Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, et al. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: A different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95:14821–6. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, et al. beta4 Integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–16. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marti A, Feng Z, Altermatt HJ, Jaggi R. Milk accumulation triggers apoptosis of mammary epithelial cells. Eur J Cell Biol. 1997;73:158–65. [PubMed] [Google Scholar]
- 19.Van Vliet K, Bao G, Suresh S. The biomechanics toolbox: Experimental approaches for living cells and biomolecules. Acta Materialia. 2003;51:5881–905. [Google Scholar]
- 20.Fung YC. Mechanical properties of living tissues. 2. New York: Springer-Verlag; 1993. [Google Scholar]
- 21.Heidemann SR, Kaech S, Buxbaum RE, Matus A. Direct observations of the mechanical behaviors of the cytoskeleton in living fibroblasts. J Cell Biol. 1999;145:109–22. doi: 10.1083/jcb.145.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakatsuki T, Wysolmerski RB, Elson EL. Mechanics of cell spreading: Role of myosin II. J Cell Sci. 2003;116:1617–25. doi: 10.1242/jcs.00340. [DOI] [PubMed] [Google Scholar]
- 23.Thoumine O, Ott A, Cardoso O, Meister JJ. Microplates: A new tool for manipulation and mechanical perturbation of individual cells. J Biochem Biophys Methods. 1999;39:47–62. doi: 10.1016/s0165-022x(98)00052-9. [DOI] [PubMed] [Google Scholar]
- 24.Alcaraz J, Buscemi L, Grabulosa M, Trepat X, Fabry B, Farre R, et al. Microrheology of human lung epithelial cells measured by atomic force microscopy. Biophys J. 2003;84:2071–9. doi: 10.1016/S0006-3495(03)75014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You HX, Yu L. Atomic force microscopy imaging of living cells: Progress, problems and prospects. Methods Cell Sci. 1999;21:1–17. doi: 10.1023/a:1009876320336. [DOI] [PubMed] [Google Scholar]
- 26.Baumgartner W, Hinterdorfer P, Ness W, Raab A, Vestweber D, Schindler H, et al. Cadherin interaction probed by atomic force microscopy. Proc Natl Acad Sci USA. 2000;97:4005–10. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100:1484–9. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galbraith CG, Sheetz MP. A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci USA. 1997;94:9114–8. doi: 10.1073/pnas.94.17.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bausch AR, Moller W, Sackmann E. Measurement of local viscoelasticity and forces in living cells by magnetic tweezers. Biophys J. 1999;76:573–9. doi: 10.1016/S0006-3495(99)77225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–7. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 31.Fabry B, Maksym GN, Butler JP, Glogauer M, Navajas D, Fredberg JJ. Scaling the microrheology of living cells. Phys Rev Lett. 2001;87:148102. doi: 10.1103/PhysRevLett.87.148102. [DOI] [PubMed] [Google Scholar]
- 32.Trepat X, Grabulosa M, Puig F, Maksym GN, Navajas D, Farre R. Viscoelasticity of human alveolar epithelial cells subjected to stretch. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1025–34. doi: 10.1152/ajplung.00077.2004. [DOI] [PubMed] [Google Scholar]
- 33.Grier DG. A revolution in optical manipulation. Nature. 2003;424:810–6. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- 34.Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88:39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 35.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raucher D, Sheetz MP. Characteristics of a membrane reservoir buffering membrane tension. Biophys J. 1999;77:1992–2002. doi: 10.1016/S0006-3495(99)77040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada S, Wirtz D, Kuo SC. Mechanics of living cells measured by laser tracking microrheology. Biophys J. 2000;78:1736–47. doi: 10.1016/S0006-3495(00)76725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau AW, Hoffman BD, Davies A, Crocker JC, Lubensky TC. Microrheology, stress fluctuations, and active behavior of living cells. Phys Rev Lett. 2003;91:198101. doi: 10.1103/PhysRevLett.91.198101. [DOI] [PubMed] [Google Scholar]
- 39.Tseng Y, Lee JS, Kole TP, Jiang I, Wirtz D. Micro-organization and viscoelasticity of the interphase nucleus revealed by particle nanotracking. J Cell Sci. 2004;117:2159–67. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 40.Heidemann SR, Wirtz D. Towards a regional approach to cell mechanics. Trends Cell Biol. 2004;14:160–6. doi: 10.1016/j.tcb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Harris AK, Wild P, Stopak D. Silicone rubber substrata: A new wrinkle in the study of cell locomotion. Science. 1980;208:177–9. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 42.Beningo KA, Wang YL. Flexible substrata for the detection of cellular traction forces. Trends Cell Biol. 2002;12:79–84. doi: 10.1016/s0962-8924(01)02205-x. [DOI] [PubMed] [Google Scholar]
- 43.Dembo M, Oliver T, Ishihara A, Jacobson K. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys J. 1996;70:2008–22. doi: 10.1016/S0006-3495(96)79767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabinovitz I, Gipson IK, Mercurio AM. Traction forces mediated by alpha6beta4 integrin: Implications for basement membrane organization and tumor invasion. Mol Biol Cell. 2001;12:4030–43. doi: 10.1091/mbc.12.12.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown TD. Techniques for mechanical stimulation of cells in vitro: A review. J Biomech. 2000;33:3–14. doi: 10.1016/s0021-9290(99)00177-3. [DOI] [PubMed] [Google Scholar]
- 46.Tschumperlin DJ, Dai G, Maly IV, Kikuchi T, Laiho LH, McVittie AK, et al. Mechanotransduction through growth-factor shedding into the extracellular space. Nature. 2004;429:83–6. doi: 10.1038/nature02543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacobs CR, Yellowley CE, Davis BR, Zhou Z, Cimbala JM, Donahue HJ. Differential effect of steady versus oscillating flow on bone cells. J Biomech. 1998;31:969–76. doi: 10.1016/s0021-9290(98)00114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frangos JA, McIntire LV, Eskin SG. Shear stress induced stimulation of mammalian cell metabolism. Biotech Bioeng. 1988;32:1053–60. doi: 10.1002/bit.260320812. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi M, Ishida T, Traub O, Corson MA, Berk BC. Mechanotransduction in endothelial cells: Temporal signaling events in response to shear stress. J Vasc Res. 1997;34:212–9. doi: 10.1159/000159225. [DOI] [PubMed] [Google Scholar]
- 50.Moy AB, Winter M, Kamath A, Blackwell K, Reyes G, Giaever I, et al. Histamine alters endothelial barrier function at cell–cell and cell-matrix sites. Am J Physiol Lung Cell Mol Physiol. 2000;278:L888–98. doi: 10.1152/ajplung.2000.278.5.L888. [DOI] [PubMed] [Google Scholar]
- 51.D’Addario M, Arora PD, Ellen RP, McCulloch CA. Interaction of p38 and Sp1 in a mechanical force-induced, beta 1 integrin-mediated transcriptional circuit that regulates the actin-binding protein filamin-A. J Biol Chem. 2002;277:47541–50. doi: 10.1074/jbc.M207681200. [DOI] [PubMed] [Google Scholar]
- 52.Bissell MJ, Farson D, Tung AS. Cell shape and hexose transport in normal and virus-transformed cells in culture. J Supramol Struct. 1977;6:1–12. doi: 10.1002/jss.400060102. [DOI] [PubMed] [Google Scholar]
- 53.Folkman J, Moscona A. Role of cell shape in growth control. Nature. 1978;273:345–9. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 54.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA. 2002;99:1972–7. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann Med. 2003;35:564–77. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 56.Folch A, Toner M. Microengineering of cellular interactions. Annu Rev Biomed Eng. 2000;2:227–56. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 57.Abbot A. Cell culture: Biology’s new dimension. Nature. 2003;424:870–2. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 58.Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 2003;13:264–9. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- 59.Garvin J, Qi J, Maloney M, Banes AJ. Novel system for engineering bioartificial tendons and application of mechanical load. Tissue Eng. 2003;9:967–79. doi: 10.1089/107632703322495619. [DOI] [PubMed] [Google Scholar]
- 60.Zhu C, Bao G, Wang N. Cell mechanics: Mechanical response, cell adhesion, and molecular deformation. Annu Rev Biomed Eng. 2000;2:189–226. doi: 10.1146/annurev.bioeng.2.1.189. [DOI] [PubMed] [Google Scholar]
- 61.Talhouk RS, Bissell MJ, Werb Z. Coordinated expression of extracellular matrix-degrading proteinases and their inhibitors regulates mammary epithelial function during involution. J Cell Biol. 1992;118:1271–82. doi: 10.1083/jcb.118.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kheradmand F, Werner E, Tremble P, Symons M, Werb Z. Role of Rac1 and oxygen radicals in collagenase-1 expression induced by cell shape change. Science. 1998;280:898–902. doi: 10.1126/science.280.5365.898. [DOI] [PubMed] [Google Scholar]
- 63.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633–9. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 64.Apodaca G. Modulation of membrane traffic by mechanical stimuli. Am J Physiol Renal Physiol. 2002;282:F179–90. doi: 10.1152/ajprenal.2002.282.2.F179. [DOI] [PubMed] [Google Scholar]
- 65.Chicurel ME, Singer RH, Meyer CJ, Ingber DE. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature. 1998;392:730–3. doi: 10.1038/33719. [DOI] [PubMed] [Google Scholar]
- 66.Voldman J. BioMEMS: Building with cells. Nat Mater. 2003;2:433–4. doi: 10.1038/nmat936. [DOI] [PubMed] [Google Scholar]
- 67.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94:849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maxwell CA, Hendzel MJ. The integration of tissue structure and nuclear function. Biochem Cell Biol. 2001;79:267–74. [PubMed] [Google Scholar]
- 69.Janmey PA, Weitz DA. Dealing with mechanics: Mechanisms of force transduction in cells. Trends Biochem Sci. 2004;29:364–70. doi: 10.1016/j.tibs.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 70.Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol. 2004;164:603–12. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zoubiane GS, Valentijn A, Lowe ET, Akhtar N, Bagley S, Gilmore AP, et al. A role for the cytoskeleton in prolactin-dependent mammary epithelial cell differentiation. J Cell Sci. 2004;117:271–80. doi: 10.1242/jcs.00855. [DOI] [PubMed] [Google Scholar]